Abstract
Chikungunya virus nonstructural protein nsP3 has an essential but unknown role in alphavirus replication and interacts with Ras-GAP SH3 domain-binding protein (G3BP). Here we describe the first known function of nsP3, to inhibit stress granule assembly by recruiting G3BP into cytoplasmic foci. A conserved SH3 domain-binding motif in nsP3 is essential for both nsP3-G3BP interactions and viral RNA replication. This study reveals a novel role for nsP3 as a regulator of the cellular stress response.
TEXT
Chikungunya virus (CHIKV; family Togaviridae, genus Alphavirus) can severely affect human health by causing debilitating disease with symptoms such as high fever, rash, and arthralgia and sometimes death (21). CHIKV is endemic in parts of Africa, the Indian Ocean area, and southern Asia (19, 22) and has recently been transmitted by the Asian tiger mosquito, Aedes albopictus, in Italy (2007) (20) and France (2010) (8). CHIKV has a positive-sense, single-stranded RNA genome (gRNA) that encodes two polyproteins. The first polyprotein is directly translated from the gRNA and produces nonstructural proteins nsP1 to nsP4, which are essential for viral replication. The structural proteins are translated from a subgenomic RNA, which is transcribed later in infection from the viral 26S promoter. nsP3 is the most enigmatic of all nsPs, with unclear yet essential roles in alphavirus RNA replication (11). nsP3 is highly phosphorylated (12, 23), is part of the viral replication complex (RC) (5), and has been shown to interact with Ras-GAP SH3 domain-binding proteins (G3BP1 and G3BP2) in two independent coimmunoprecipitation studies (3, 7). Infection with another Togaviridae family member; rubella virus, has been shown to alter the distribution of host cell G3BP, suggesting that G3BP plays an important role in the outcome of viral infection (16). G3BP is an essential factor in the assembly of stress granules (SGs) (1), which are nonmembranous cytoplasmic focal structures (foci) containing cytoplasmic mRNPs. SGs rapidly aggregate in response to different types of environmental stress and lead to the impaired translation of most mRNAs (2). SGs can have diverse anti- or proviral activities (10, 14, 24).
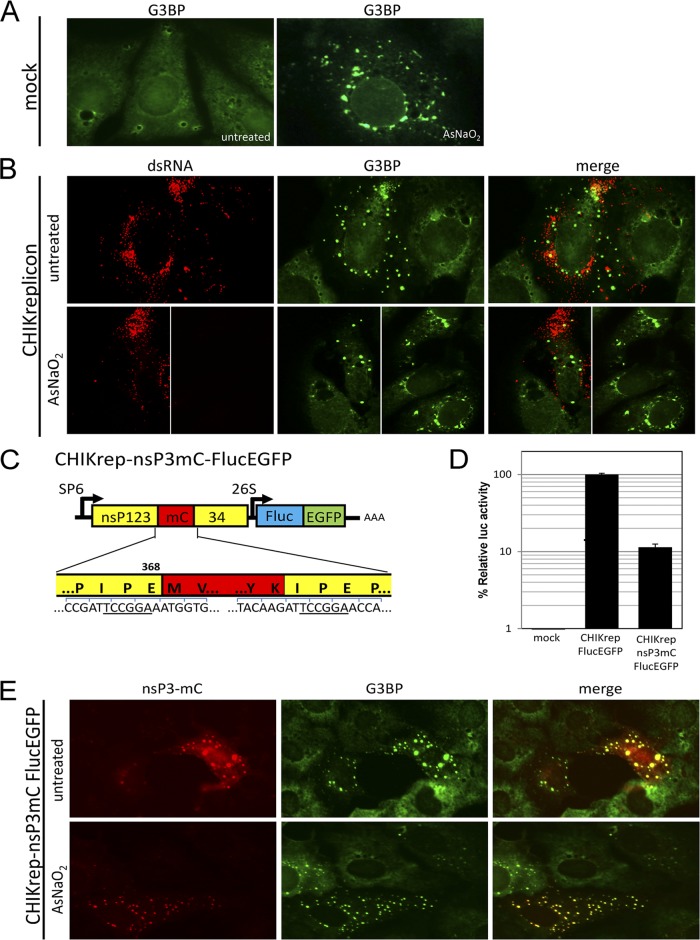
To investigate the relationship between CHIKV replication and SG formation, Vero cells were transfected with CHIKV replicon RNA in two independent experiments as described previously (6). Next, at 16 h posttransfection (hpt), cells were either exposed to oxidative stress by using arsenite to induce SGs or left untreated. Cells were immunostained for double-stranded RNA (dsRNA; a replication intermediate of viral RNA replication) and/or G3BP (Fig. 1). Cells that were treated with only transfection reagent (mock transfected) (Fig. 1A) responded to arsenite-induced stress with the formation of SGs. G3BP readily localized to these typical SGs, displayed as irregularly shaped granules that converge around the nucleus and extend from there into the cytoplasm and are often smaller when located farther away from the nucleus (Fig. 1A, right). In contrast to arsenite-induced SGs, the presence of replicating CHIKV replicon RNA (visualized by the dsRNA signal) (Fig. 1B) caused G3BP to localize into foci that displayed a more punctate morphology and were distributed throughout the cytoplasm in a seemingly random manner (Fig. 1B, top). Exposing cells that harbor replicating CHIKV replicon RNA to oxidative stress did not affect the morphology of these G3BP foci (Fig. 1B, bottom left). Cells from the same sample that did not harbor replicating CHIKV replicon RNA after transfection were still able to respond to arsenite with the formation of SGs (Fig. 1B, bottom right).
Fig 1.
During CHIKV RNA replication, nsP3 localizes to punctate cytoplasmic foci concurrently with G3BP. (A) Mock-transfected Vero cells were left untreated or were treated with sodium arsenite (AsNaO2). Cells were fixed in 4% paraformaldehyde in phosphate-buffered saline for 10 min at room temperature and permeabilized with ice-cold acetone-methanol (1:1) for 10 min at −20°C. SGs were stained with G3BP antibody (G6046; Sigma). (B, top) Vero cells transfected with in vitro-transcribed CHIK replicon RNA (CHIKrep) and stained with J2 anti-dsRNA and G3BP antibodies. (B, bottom) CHIKrep-transfected cells (left) and cells from the same sample that remained untransfected (right). (C) Schematic representation of the CHIKrep-nsP3mC-FlucEGFP replicon expressing nsP3 with an internal mCherry (mC) fusion and an FlucEGFP fusion protein from the subgenomic promoter. (D) Vero cells were transfected with a pRL-TK plasmid constitutively expressing Renilla luciferase (Rluc) together with either CHIKrep-FlucEGFP or CHIKrep-nsP3mC-FlucEGFP in vitro-transcribed RNA. Cells were lysed at 16 hpt, and Fluc/Rluc activities were measured. Luciferase activity in mock-transfected cells was also measured. The Fluc measurements were normalized to Rluc to compensate for differences in transfection efficiency. The values depicted are averages of triplicate samples and are expressed as the percentage of normalized Fluc activity relative to that of CHIKrep-FlucEGFP. Error bars represent standard deviations. (E) CHIKrep-nsP3mC-FlucEGFP in vitro-transcribed RNA was transfected into Vero cells, and at 16 hpt, cells were fixed and stained with G3BP antibodies. Where indicated, samples were treated with sodium arsenite (0.5 mM) for 30 min at 16 hpt.
To investigate the interaction between G3BP and the viral protein nsP3, an mCherry reporter protein was incorporated into nsP3 within the CHIKV replicon in a way similar to that described for Sindbis virus (3, 13), creating CHIKrep-nsP3mC-FlucEGFP (where FlucEGFP is firefly luciferase-enhanced green fluorescent protein) (Fig. 1C) (cloning details are available upon request). Fluc measurements of two independent experiments indicated that CHIKrep-nsP3mC-FlucEGFP RNA was still able to replicate, albeit to lower levels than wild-type CHIKrep-FlucEGFP (Fig. 1D). Immunofluorescence analysis revealed nearly complete colocalization between nsP3-mCherry and G3BP (Fig. 1E), indicative of a close interaction between nsP3 and G3BP during CHIKV RNA replication. These nsP3-G3BP foci were indistinguishable from G3BP foci observed in Fig. 1B in both their morphology and unresponsiveness to arsenite-induced stress. Note that only untransfected cells were able to form typical arsenite-induced SGs (Fig. 1E).
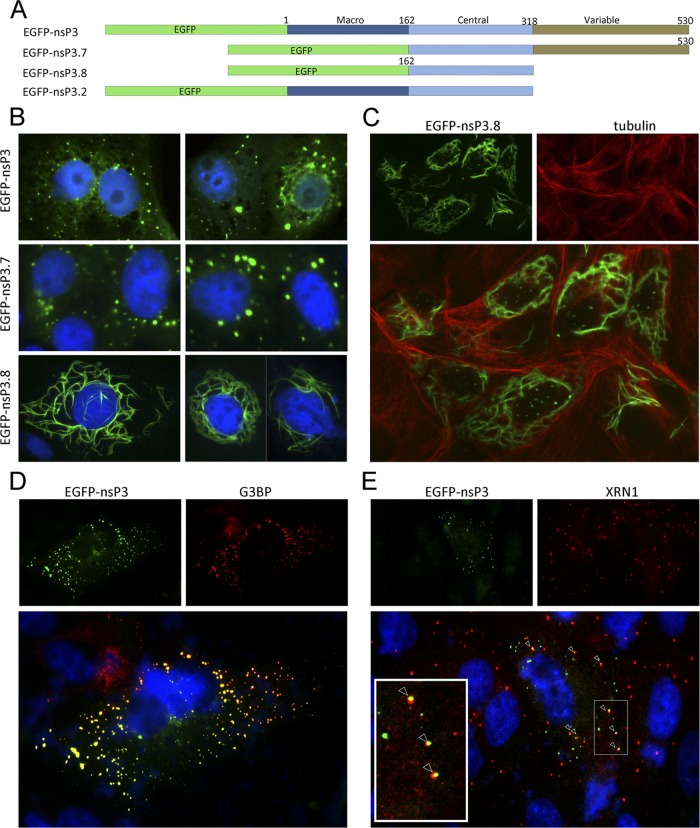
To determine whether nsP3 expressed alone also localizes into foci and which domain(s) in nsP3 is required for its specific subcellular localization, several expression plasmids with an N-terminal EGFP tag fused to nsP3 (Fig. 2A) were constructed (cloning details are available upon request) and transfected into Vero cells. Expression of EGFP-nsP3 resulted in foci that were indistinguishable from those generated during CHIKV replicon RNA replication, indicating that subcellular localization in cytoplasmic foci is an intrinsic property of nsP3 (Fig. 2B, top). Deletion of the N-terminal macrodomain (nsP3.7, Fig. 2A), which is conserved among the Togaviridae, Coronaviridae, and hepatitis E virus (15), did not result in a change in localization (Fig. 2B, middle). In contrast, deletion of the highly variable, C-terminal domain (nsP3.8 and nsP3.2, Fig. 2A) resulted in the formation of filaments instead of foci (Fig. 2B, bottom; data not shown). Since these filaments resembled the cytoskeleton, cells expressing nsP3.8 were stained for tubulin (Fig. 2C). No colocalization of nsP3.8 and the cytoskeleton was observed, indicating that multimerization is an intrinsic property of the conserved central domain of alphavirus nsP3.
Fig 2.
Individually expressed CHIKV nsP3 localizes to small punctate cytoplasmic foci that are juxtaposed with PBs. (A) Schematic representation of N-terminal EGFP-nsP3 fusion proteins expressing either full-length nsP3 (EGFP-nsP3) or truncated forms of nsP3 missing only the macrodomain (EGFP-nsP3.7), both the macrodomain and the variable domain (EGFP-nsP3.8), or only the variable domain (EGFP-nsP3.2). (B) Plasmids expressing these nsP3 variants from a cytomegalovirus promoter were transfected into Vero cells. Cells were fixed at 20 hpt, and nuclei were visualized by Hoechst staining. (C) Vero cells transfected with EGFP-nsP3.8 were fixed at 20 hpt and stained for tubulin (14-4502; eBioscience). Vero cells expressing EGFP-nsP3 were fixed and stained with either SG marker G3BP (D) or PB marker XRN1 (A300-443A; Bethyl Laboratories) (E) antibodies. Arrowheads indicate juxtaposed SGs and PBs.
Next, EGFP-nsP3-transfected cells were stained for the SG marker G3BP (Fig. 2D) or a marker for processing bodies (PBs), XRN1 (Fig. 2E). PBs are small cytoplasmic foci and have been shown to be modulated by various RNA viruses (4, 10, 14, 17). Here we show for the first time that individually expressed nsP3 displays complete colocalization with G3BP in Vero (Fig. 2D) and HEK293T (not shown) cells in the absence of viral RNA replication and other CHIKV proteins. XRN1 showed a punctate localization in untransfected cells similar to that in nsP3-transfected cells (Fig. 2E). Interestingly, the PBs observed did not colocalize but were sometimes juxtaposed to nsP3 foci (Fig. 2E, arrowheads). Juxtaposition of PBs and SGs is a well-known phenomenon (9). The observations that nsP3 foci include G3BP and are often juxtaposed to PBs suggest that nsP3/G3BP foci might be SGs.
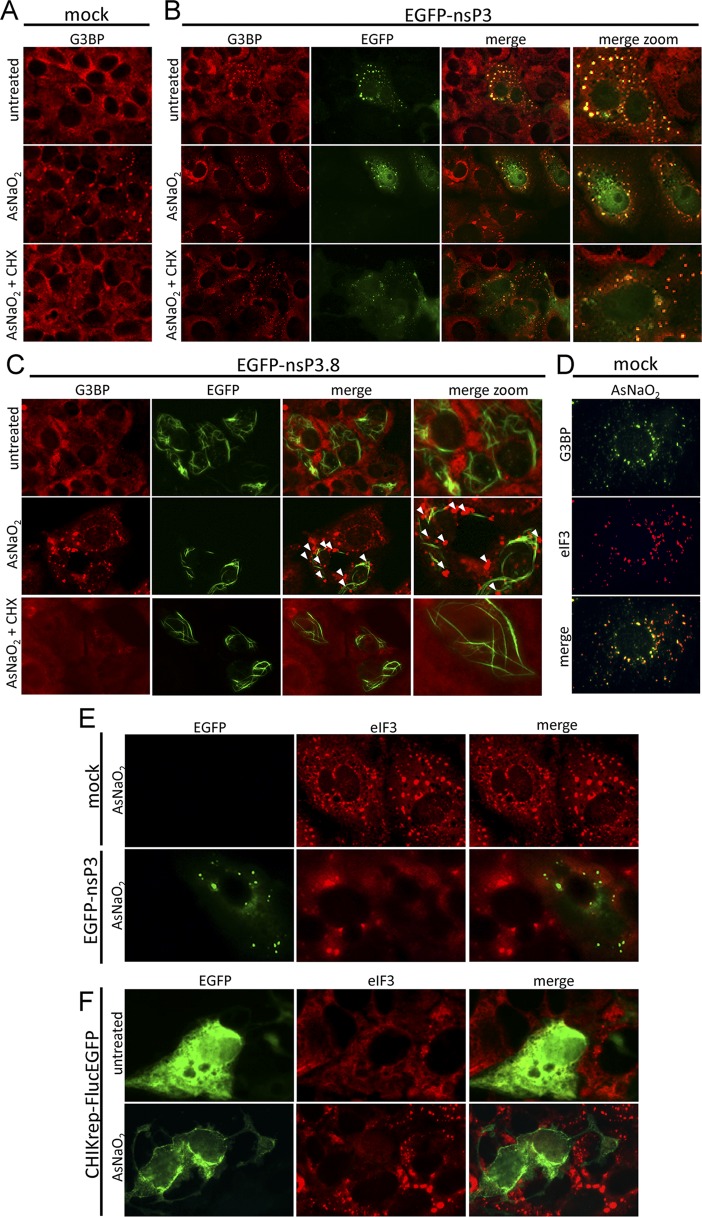
Since nsP3/G3BP foci have a morphology different from that of bona fide, arsenite-induced SGs, we set out to elucidate whether or not CHIKV nsP3/G3BP foci are indistinguishable from true SGs. In three independent experiments, cells expressing EGFP-nsP3 or EGFPns-P3.8 were treated with arsenite and subsequently treated with cycloheximide (CHX) before being immunostained for G3BP (Fig. 3A to C). CHX has been shown to disassemble arsenite-induced, bona fide SGs (Fig. 3A) (1). Cells that remained untransfected in the field (Fig. 3B) behaved similarly to mock-transfected cells (Fig. 3A), no longer displaying any bona fide SGs after CHX treatment. Cells transfected with EGFP-nsP3 displayed typical nsP3/G3BP foci, which were unaffected by either arsenite or CHX treatment and are therefore different from SGs (Fig. 3B). In contrast, when the same experiment was performed with EGFP-nsP3.8, arsenite induction resulted in G3BP-containing granules with typical SG morphology (described above) (Fig. 3C, arrowheads) and CHX treatment led to SG disassembly. This experiment indicated an essential role for the nsP3 C-terminal variable region in inhibiting bona fide SG assembly.
Fig 3.
nsP3/G3BP foci do not disassemble upon CHX treatment and do not colocalize with eIF3. (A to C) Vero cells were mock transfected (A) or transfected with either EGFP-nsP3 (B) or EGFP-nsP3.8 (C). After 16 h, samples were left untreated (top), treated with arsenite for 30 min (middle), or treated with arsenite for 30 min prior to CHX (10 μg/ml) treatment for 30 min (bottom). After treatment, samples were fixed and stained for G3BP. Arrowheads show bona fide SGs in cells transfected with EGFP-nsP3 variants. (D) Untransfected Vero cells were mock treated or treated with sodium arsenite, fixed, and stained for G3BP and eIF3 (sc-16377; Santa Cruz). (E) Vero cells were either mock transfected (top) or transfected with EGFP-nsP3 (bottom), and at 16 hpt, all samples were treated with arsenite for 30 min before being fixed and stained for eIF3. (F) Vero cells were transfected with CHIKrep-FlucEGFP in vitro-transcribed RNA, and at 16 hpt, cells were either left untreated (top) or treated with arsenite for 30 min (bottom) before being fixed and stained with anti-eIF3 antibodies.
In addition to CHX treatment, another well-known SG marker, eIF3 (1), was used to assess the composition of the nsP3/G3BP foci. Similar to G3BP, eIF3 readily localized to bona fide SGs and colocalized with G3BP in arsenite-treated cells (Fig. 3D). Cells were transfected with either EGFP-nsP3 alone (Fig. 3E) or CHIKV replicon RNA expressing FlucEGFP from its subgenomic promoter (Fig. 3F). Mock-transfected cells (Fig. 3E, top) and cells that remained untransfected (Fig. 3E, bottom) readily displayed arsenite-induced, eIF3-containing SGs. In contrast, cells transfected with either EGFP-nsP3 (Fig. 3E, bottom) or CHIKrep-FlucEGFP (Fig. 3F, bottom) did not show any eIF3-positive granules, strongly suggesting that nsP3/G3BP foci are different from SGs and that the formation of nsP3/G3BP foci inhibits the assembly of bona fide, eIF3-positive SGs.
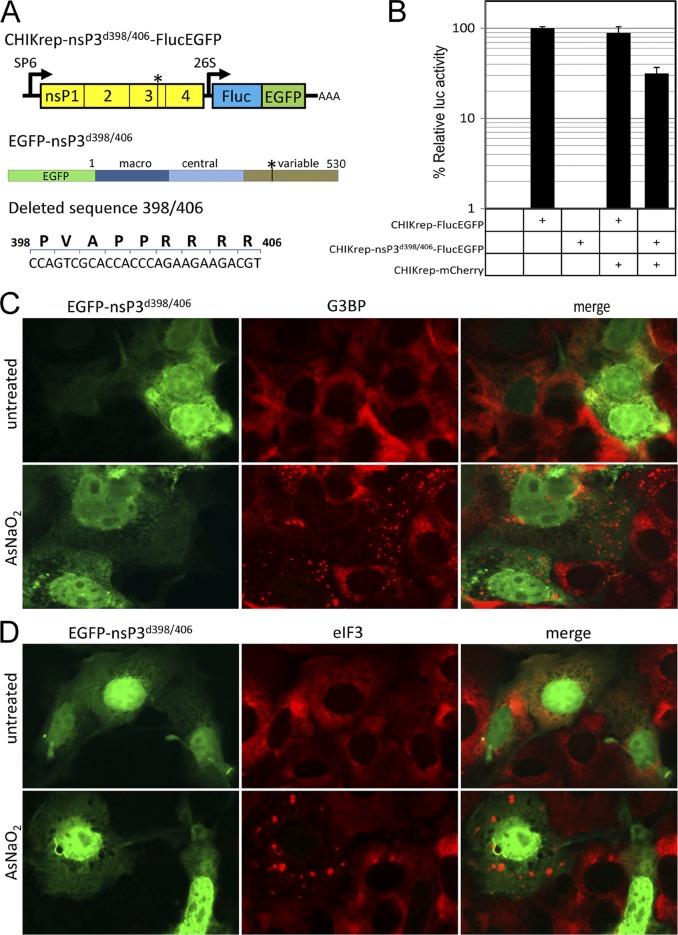
Since deletion of the nsP3 C-terminal variable domain inhibited the formation of foci and restored the ability of the host cell to form bona fide SGs, we set out to determine the element responsible within this domain. Interestingly, a Src homology 3 (SH3) domain-binding motif (PxxPxR) is conserved within the variable domains of many, if not all, alphaviruses. This element was demonstrated to be important for efficient Semliki Forest virus replication (18). Deletion of this element from a CHIKV replicon (CHIKrep-nsP3d398/406-FlucEGFP, Fig. 4A) rendered it unable to replicate (Fig. 4B). Cotransfection with a second (wild-type) CHIKV replicon in a trans complementation experiment rescued CHIKrep-nsP3d398/406-FlucEGFP replication (Fig. 4B). This confirmed that the RNA of CHIKrep-nsP3d398/406-FlucEGFP was unable to replicate in the absence of wild-type nsP3. Interestingly, nsP3 expressed with the SH3 domain-binding motif deleted (EGFP-nsP3d398/406) (Fig. 4A) no longer localized in foci but was diffusely distributed throughout the cytoplasm and the nucleus in two independent experiments. Staining with either anti-G3BP (Fig. 4C) or anti-eIF3 (Fig. 4D) antibodies did not show apparent colocalization with EGFP-nsP3d398/406. Treatment with arsenite readily induced SGs in both untransfected and EGFP-nsP3d398/406-transfected cells. These results indicate that deletion of the SH3 domain-binding motif from nsP3 resulted in the complete loss of association with G3BP.
Fig 4.
The SH3 domain-binding motif is necessary for CHIKV replicon RNA replication and the formation of nsP3/G3BP foci. (A) Schematic representation of the SH3 domain-binding motif deletion constructs CHIKrep-nsP3d398/406-FlucEGFP (top) and EGFP-nsP3d398/406 (middle). The schematic representation of the deleted sequence (bottom) depicts both the deleted amino acid sequence and the corresponding nucleotides. The numbers indicate the amino acid positions within CHIKV nsP3. (B) Vero cells were transfected with plasmid pRL-TK constitutively expressing Renilla luciferase (Rluc) together with in vitro-transcribed RNA from CHIKrep-FlucEGFP, CHIKrep-nsP3d398/406-FlucEGFP, CHIKrep-FlucEGFP together with CHIKrep-mCherry, or CHIKrep-nsP3d398/406-FlucEGFP together with CHIKrep-mCherry. Cells were lysed at 16 hpt, and Fluc/Rluc activities were measured. Luciferase activity in mock-transfected cells was also measured. The Fluc measurements were normalized to Rluc to compensate for differences in transfection efficiency. The values depicted are averages of triplicate samples and are expressed as percentages of normalized Fluc activity relative to that of CHIKrep-FlucEGFP. Error bars represent standard deviations. (C, D) Vero cells were transfected with EGFP-nsP3d398/406, and at 16 hpt, cells were either left untreated (top) or treated with arsenite for 30 min (bottom) before being fixed and stained with anti-G3BP (C) or anti-eIF3 (D) antibodies.
In conclusion, we show that G3BP localizes to CHIKV nsP3-containing foci both in the presence and in the absence of CHIKV RNA replication. Although reminiscent of SGs, these nsP3/G3BP foci differ from bona fide SGs in four ways; i.e., (i) they are morphologically different, (ii) they do not contain the hallmark SG marker eIF3, (iii) they do not disassemble upon CHX treatment, and (iv) they render cells that express these nsP3/G3BP foci unable to form bona fide SGs in response to oxidative stress. The observation that CHIKV replication inhibits SG assembly may suggest that SGs have antiviral activity. However, it cannot be completely ruled out that G3BP is part of the alphavirus RC, although the limited overlap between viral dsRNA and G3BP does not support this possibility. Furthermore, deletion of the SH3 domain-binding motif from nsP3 restores bona fide SG assembly but abolishes CHIKV RNA replication. We propose that nsP3 blocks SG assembly by the sequestration of G3BP into foci via an interaction with the SH3 domain-binding motif in nsP3. The nature of the binding (direct versus indirect) between G3BP and nsP3 and the relative contributions of other nsP3-binding proteins are important avenues for follow-up studies. This study provides new insights into the function of CHIKV nsP3 at the molecular level and suggests that its C-terminal variable domain plays an important role in modulating SG assembly during CHIKV RNA replication.
ACKNOWLEDGMENTS
We thank Martijn Langereis and Frank van Kuppeveld for sharing the eIF3 antibody.
This work is supported by the European Community's Seventh Framework Programme (FP7 VECTORIE project number 261466).
Footnotes
Published ahead of print 25 July 2012
REFERENCES
- 1. Anderson P, Kedersha N. 2009. Stress granules. Curr. Biol. 19:R397–R398 [DOI] [PubMed] [Google Scholar]
- 2. Buchan JR, Parker R. 2009. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell 36:932–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cristea IM, et al. 2006. Tracking and elucidating alphavirus-host protein interactions. J. Biol. Chem. 281:30269–30278 [DOI] [PubMed] [Google Scholar]
- 4. Emara MM, Brinton MA. 2007. Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc. Natl. Acad. Sci. U. S. A. 104:9041–9046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Frolova EI, Gorchakov R, Pereboeva L, Atasheva S, Frolov I. 2010. Functional Sindbis virus replicative complexes are formed at the plasma membrane. J. Virol. 84:11679–11695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fros JJ, et al. 2010. Chikungunya virus nonstructural protein 2 inhibits type I/II interferon-stimulated JAK-STAT signaling. J. Virol. 84:10877–10887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gorchakov R, Garmashova N, Frolova E, Frolov I. 2008. Different types of nsP3-containing protein complexes in Sindbis virus-infected cells. J. Virol. 82:10088–10101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Grandadam M, et al. 2011. Chikungunya virus, southeastern France. Emerg. Infect. Dis. 17:910–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kedersha N, et al. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169:871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Khong A, Jan E. 2011. Modulation of stress granules and P bodies during dicistrovirus infection. J. Virol. 85:1439–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. LaStarza MW, Lemm JA, Rice CM. 1994. Genetic analysis of the nsP3 region of Sindbis virus: evidence for roles in minus-strand and subgenomic RNA synthesis. J. Virol. 68:5781–5791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li GP, La Starza MW, Hardy WR, Strauss JH, Rice CM. 1990. Phosphorylation of Sindbis virus nsP3 in vivo and in vitro. Virology 179:416–427 [DOI] [PubMed] [Google Scholar]
- 13. Liang Z, Li G. 2005. Recombinant Sindbis virus expressing functional GFP in the nonstructural protein nsP3. Gene Ther. Mol. Biol. 9:317–324 [Google Scholar]
- 14. Lindquist ME, Lifland AW, Utley TJ, Santangelo PJ, Crowe JE., Jr 2010. Respiratory syncytial virus induces host RNA stress granules to facilitate viral replication. J. Virol. 84:12274–12284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Malet H, et al. 2009. The crystal structures of chikungunya and Venezuelan equine encephalitis virus nsP3 macro domains define a conserved adenosine binding pocket. J. Virol. 83:6534–6545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Matthews JD, Frey TK. 2012. Analysis of subcellular G3BP redistribution during rubella virus infection. J. Gen. Virol. 93:267–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McInerney GM, Kedersha NL, Kaufman RJ, Anderson P, Liljestrom P. 2005. Importance of eIF2alpha phosphorylation and stress granule assembly in alphavirus translation regulation. Mol. Biol. Cell 16:3753–3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Neuvonen M, et al. 2011. SH3 domain-mediated recruitment of host cell amphiphysins by alphavirus nsP3 promotes viral RNA replication. PLoS Pathog. 7:e1002383 doi:10.1371/journal.ppat.1002383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Powers AM, Logue CH. 2007. Changing patterns of chikungunya virus: re-emergence of a zoonotic arbovirus. J. Gen. Virol. 88:2363–2377 [DOI] [PubMed] [Google Scholar]
- 20. Rezza G, et al. 2007. Infection with chikungunya virus in Italy: an outbreak in a temperate region. Lancet 370:1840–1846 [DOI] [PubMed] [Google Scholar]
- 21. Schwartz O, Albert ML. 2010. Biology and pathogenesis of chikungunya virus. Nat. Rev. Microbiol. 8:491–500 [DOI] [PubMed] [Google Scholar]
- 22. Thiboutot MM, et al. 2010. Chikungunya: a potentially emerging epidemic? PLoS Negl. Trop. Dis. 4:e623 doi:10.1371/journal.pntd.0000623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vihinen H, Saarinen J. 2000. Phosphorylation site analysis of Semliki Forest virus nonstructural protein 3. J. Biol. Chem. 275:27775–27783 [DOI] [PubMed] [Google Scholar]
- 24. White JP, Lloyd RE. 2012. Regulation of stress granules in virus systems. Trends Microbiol. 20:175–183 [DOI] [PMC free article] [PubMed] [Google Scholar]