Abstract
Mutations that reduce the efficiency of deoxynucleoside (dN) triphosphate (dNTP) substrate utilization by the HIV-1 DNA polymerase prevent viral replication in resting cells, which contain low dNTP concentrations, but not in rapidly dividing cells such as cancer cells, which contain high levels of dNTPs. We therefore tested whether mutations in regions of the adenovirus type 5 (Ad5) DNA polymerase that interact with the dNTP substrate or DNA template could alter virus replication. The majority of the mutations created, including conservative substitutions, were incompatible with virus replication. Five replication-competent mutants were recovered from 293 cells, but four of these mutants failed to replicate in A549 lung carcinoma cells and Wi38 normal lung cells. Purified polymerase proteins from these viruses exhibited only a 2- to 4-fold reduction in their dNTP utilization efficiency but nonetheless could not be rescued, even when intracellular dNTP concentrations were artificially raised by the addition of exogenous dNs to virus-infected A549 cells. The fifth mutation (I664V) reduced biochemical dNTP utilization by the viral polymerase by 2.5-fold. The corresponding virus replicated to wild-type levels in three different cancer cell lines but was significantly impaired in all normal cell lines in which it was tested. Efficient replication and virus-mediated cell killing were rescued by the addition of exogenous dNs to normal lung fibroblasts (MRC5 cells), confirming the dNTP-dependent nature of the polymerase defect. Collectively, these data provide proof-of-concept support for the notion that conditionally replicating, tumor-selective adenovirus vectors can be created by modifying the efficiency with which the viral DNA polymerase utilizes dNTP substrates.
INTRODUCTION
Mutations that reduce the efficiency of deoxynucleoside (dN) triphosphate (dNTP) substrate utilization by viral DNA polymerases (Pols) can result in the selective loss of viral replicative activity in resting cells, which contain low dNTP concentrations, but not in rapidly dividing cells, which contain high levels of dNTPs (10, 13, 16, 17). Actively dividing tumor cells can contain between 10- and 30-times-higher dNTP concentrations than primary cells (16), providing a biochemical basis on which it might be possible to develop viral vectors that can selectively replicate in tumor cells but not normal cells. Since adenovirus vectors have been well studied as candidate oncolytic agents (8, 9, 11, 14, 19), we decided to focus our analysis on this virus and to determine the feasibility of generating modified adenoviruses containing mutated DNA polymerases with reduced dNTP binding affinities.
The adenovirus DNA polymerase (AdPol) has been identified as a 140-kDa DNA polymerase of the alpha-like, Pol B family of polymerases (6, 12). Such polymerases contain several conserved motifs that are essential for polymerase function and contain amino acid residues that are necessary for dNTP binding (2, 29, 33), template DNA binding (4, 7, 15), and polymerase activity (5, 23, 30). It was previously shown that amino acid substitutions in the conserved (I/Y)xGG motif of AdPol result in the mispositioning of the template DNA in the polymerase active site, leading to a reduced affinity of the polymerase for the dNTP substrate (7). Additionally, residues from the highly conserved motif B have been shown to play a direct role in the initial binding and stabilization of the incoming dNTP (4, 29). We therefore evaluated these and other mutations in key AdPol residues, including conserved motif A residues, shown to alter the dNTP binding affinity in the related Pol B family Pfu DNA polymerase (20). The goal of creating this panel of mutations was to disrupt dNTP utilization by these mutant polymerases and examine how this might affect virus replication.
MATERIALS AND METHODS
Cells.
The human cancer cell lines A549 (lung epithelial carcinoma), HeLa (cervical carcinoma), MCF-7 (breast cancer), and H1299 (non-small-cell lung carcinoma) were purchased from the American Type Culture Collection (ATCC) and grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) or RPMI 1640 medium supplemented with 10% FBS. The normal human cell lines Wi38 and MRC5 (lung fibroblasts) and BJ and HCA-2 cells (normal foreskin fibroblasts) were also purchased from the ATCC. These cells were grown in DMEM plus 10% FBS. Hek 293A cells were purchased from Stratagene and grown in DMEM plus 10% FBS. The human SF-539 cell line (glioma) was obtained from the DCTD Tumor/Cell Line Repository of the National Cancer Institute (NCI).
Plasmids.
The replication-competent E3-deleted adenoviral vectors were generated by using the AdEasy system (Stratagene). First, the region of AdEasy-1 represented by the ClaI-PmeI restriction fragment (nucleotides [nt] 3699 to 13437) was inserted into the cloning plasmid pLitmus (NEB), to generate pLitmus V.1. This plasmid was used to introduce mutations into the DNA polymerase. Second, the adenovirus type 5 (Ad5) domain represented by the XhoI-NotI fragment (nt 505 to 4070), and containing the E1 region, was PCR amplified from wild-type (WT) adenovirus VR5 DNA (ATCC) and inserted into the multiple-cloning site of pAdtrack-CMV/eGFP. The simian virus 40 (SV40) splice acceptor and late poly(A) signal (nt 1772 to 1993) from the pGL3-basic vector (Promega) were incorporated downstream of the E1 region via overlapping PCR. This generated plasmid pAdtrack-E1SV40/eGFP.
DNA manipulations and cloning procedures.
The appropriate nucleotide substitutions for each adenovirus molecular clone were introduced into the Ad5 DNA polymerase (AdPol) by employing PCR-based site-directed mutagenesis. This created a PCR product flanked by PshAI-SapI. The mutated AdPol PCR product was then inserted into the pLitmus V.1 cloning vector. The pLitmus V.1 cloning vector containing the mutated AdPol was then digested with ClaI-PmeI. This generated a ∼10-kb fragment that was then inserted into the AdEasy-1 ΔE1/E3 backbone. The AdEasy mutant AdPol plasmids were then homologously recombined with pAdTrack-E1SV40/eGFP in BJ5183 cells (Stratagene). This final step yielded a full-length adenovirus genome containing the E1 region, a green fluorescent protein (GFP) reporter cassette, and the mutant Ad5 DNA polymerase. All viral molecular clones were sequence verified.
Transfections and virus purification.
The viral molecular clones were then digested with PacI (to linearize them) and transfected into Hek 293A cells. To ensure that the availability of dNTPs would not become a rate-limiting factor for viral replication, all transfections were carried out in the presence or absence of 0.5 mM exogenous dNs (deoxynucleosides) (Sigma). Transfected cultures were observed daily by utilizing bright-field and fluorescence microscopy to track the growth and spread of the virus via the GFP reporter, viral plaque formation, and cytopathic effects (CPEs) (i.e., signs of viable replication-competent viral molecular clones). Large-scale virus stocks were then prepared for all viable mutants by growing them in Hek 293A cells and then banding the viruses in CsCl equilibrium density gradients for purification. Once virus stocks were prepared, we employed a 50% tissue culture infective dose (TCID50) assay to determine infectious virus titers in Hek 293A cells.
Replication kinetics.
To study the kinetics of viral replication, normal or cancer cells were seeded into 6-well plates and grown in either DMEM or RPMI 1640 medium plus 5% FBS. BJ and HCA-2 cells were infected at a multiplicity of infection (MOI) of 20. Wi38 and MRC5 cells were infected at an MOI of 5. A549, HeLa, and H1299 cells were infected at MOIs of 0.5, 5, and 5, respectively. For infections conducted in the presence of exogenous dNs, 0.25 mM or 0.50 mM each dN was added to cultures every other day for the duration of the experiment. Infections were allowed to proceed for 7 days, and the cells were monitored daily via bright-field and fluorescence microscopy (Olympus Ix71). Afterwards, cells were harvested and stained with eFluor 660 cell viability dye (eBiosciences), according to the manufacturer's protocol. Samples were fixed in a 4% formaldehyde solution and analyzed by using the Accuri C6 flow cytometer system and software (AccuriCytometers).
Construction of recombinant baculoviruses.
The Ad5 fragment (nt 5197 to 8784) containing the viral DNA polymerase was PCR amplified from the previously constructed adenovirus vectors and inserted into the multiple-cloning site of the baculovirus transfer vector pAcGP67A (BD Bioscience). The adenovirus polymerase leader sequence (nt 1411 to 1419) was then incorporated upstream of the AdPol via overlapping PCR to generate an enzymatically active, full-length, 140-kDa Ad5 DNA polymerase (27). A six-histidine affinity tag was then incorporated into the C-terminal region of the polymerase via overlapping PCR to generate plasmid pAcGP67A-AdPol6×His. All plasmids were sequence verified. Plasmid pAcGP67A-AdPol6×His along with linearized baculovirus AcNPV DNA (BD Bioscience) were cotransfected into Spodoptera frugiperda (SF9) insect cells using Cellfectin (Invitrogen), according to the manufacturer's protocol. Five days following transfection, the recombinant baculoviruses were harvested and amplified for several rounds. The titers of recombinant baculovirus stocks were determined by using the BacPak baculovirus rapid titer kit (Clontech), as specified by the manufacturer.
Purification of adenovirus DNA polymerase mutants.
SF9 insect cells were grown as suspension cultures in SF-900 II medium (Gibco-Invitrogen) on an orbital shaker at 27°C. Liter culture preps were infected with recombinant baculoviruses expressing either the wild-type or mutant AdPol at an MOI of 0.5. At 4 days postinfection (dpi), cells were harvested and washed in ice-cold phosphate-buffered saline (PBS). Cells were resuspended in a hypotonic lysis buffer containing 50 mM sodium phosphate (pH 7), 10% glycerol, 300 mM sodium chloride (NaCl), 5 mM 2-β-mercaptoethanol (βME), 1% Triton X-100, and complete EDTA-free protease inhibitor cocktail tablets (Roche). Cells were then placed on ice for 10 min and afterwards vortexed for 30 s. Cell debris was pelleted at 15,000 × g for 15 min at 4°C, and the resulting crude cell lysate was further clarified by centrifugation at 20,000 × g for an additional 15 min at 4°C.
For purification to near homogeneity, the clarified cell lysate was incubated with Talon resin (Clontech) for 1 h at 4°C on a nutating mixer. The Talon resin was equilibrated with 5 mM wash buffer containing 50 mM sodium phosphate (pH 7), 10% glycerol, 300 mM NaCl, 5 mM βME, 5 mM imidazole, and complete EDTA-free protease inhibitor cocktail tablets prior to incubation with the clarified cell lysate. After binding, the resin-lysate mixture was transferred into glass Econo-chromatography columns (Bio-Rad) and washed extensively with wash buffer containing an imidazole gradient that ranged from 7.5 mM to 25 mM imidazole. The protein was eluted with 150 mM elution buffer containing 50 mM sodium phosphate (pH 7), 10% glycerol, 300 mM NaCl, and 150 mM imidazole and dialyzed against three changes of dialysis buffer composed of 50 mM sodium phosphate (pH 7), 10% glycerol, and 300 mM NaCl. The resulting eluate was concentrated by using 50-kDa-molecular-mass-cutoff Amicon centrifugal filters (Millipore). Protein concentration and purity were estimated by gel electrophoresis and Coomassie brilliant blue staining; typical yields were approximately 2 mg/liter protein, with an approximate protein purity of ≥90%.
Multiple-nucleotide incorporation analysis.
A DNA 30-mer template was annealed to a 20-mer 5′-32P-end-labeled primer as described previously (20). A total of 50 nM the template-primer (T/P) complex was extended with the AdPol proteins under standard reaction conditions at 37°C for 10 min; the reaction mixture was then quenched with stop dye (10 μl of 40 mM EDTA–99% formamide). A standard polymerase assay was performed with a 20-μl reaction mixture volume containing 1 M Tris-HCl (pH 7.5), 1 mg/ml bovine serum albumin (BSA), 5 mM MgCl2, 1 M dithiothreitol (DTT), and 4% glycerol. Prior to the performance of quantitative assays of enzyme activity, serially diluted aliquots of purified polymerase preparations were analyzed in reaction mixtures containing 60 μM dNTPs; this allowed us to normalize the input activity of the AdPol proteins in subsequent quantitative assays (thereby controlling for potential differences in the conformation/quality of the polymerase preparations) (20, 26). Finally, quantitative assays of polymerase activity were conducted in multiple-nucleotide extension reactions that used serially diluted dNTPs (at concentrations of 60, 30, 6, 3, 1, and 0 μM). This allowed us to determine the efficiencies of dNTP utilization by the various mutant polymerases.
RESULTS
Site-directed mutagenesis of the (I/Y)xGG, A, and B motifs in AdPol.
To create AdPol mutants that targeted regions of the polymerase expected to alter the dNTP binding affinity and/or AdPol activity, we performed standard PCR-based site-directed mutagenesis as described in Materials and Methods. An E3-deleted adenovirus vector was used for our studies, in order to accommodate a GFP reporter transgene, which facilitated the visualization of the virus in cultured cells.
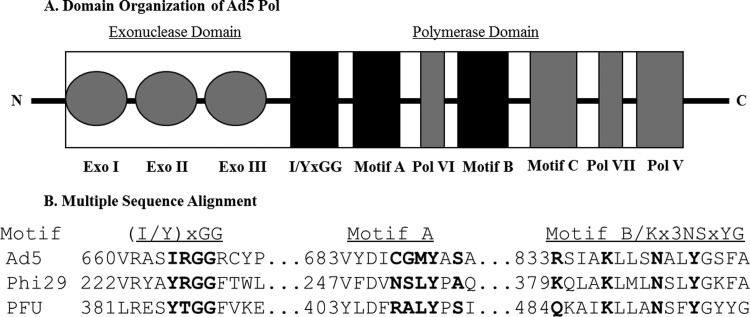
Mutations were introduced into residues C687, G688, M689, and Y690 of the conserved polymerase motif A as well as residues R833, K837, N841, and Y844 of the conserved polymerase motif B (Fig. 1 and Table 1). We also generated AdPol mutants that were expected to be impaired in their ability to stably bind template strand DNA by conducting site-directed mutagenesis of residues I664, R665, and GG666/7 of the conserved (I/Y)xGG motif. Based on structural considerations, we expected that the latter template mutants would also be likely to reduce the affinity for the incoming dNTP.
Fig 1.
Schematic representation of Ad5 polymerase domain organization and location of conserved residues. (A) Domain organization of Ad5 Pol. Shaded regions represent conserved domains; regions shaded in black (and identified in boldface type) represent the conserved motifs that underwent mutagenesis in this study ([I/Y]xGG, motif A, and motif B). (B) Multiple-sequence alignment of conserved domains in the Ad5, Phi29, and Pfu DNA polymerases.
Table 1.
Ad5 polymerase mutantsa
| Mutation | Type | Polymerase motif | Description of mutation | Result of mutation |
|---|---|---|---|---|
| I664V | C | (I/Y)xGG | Disrupts interaction of template DNA and polymerase | Viable |
| I664 M | C | (I/Y)xGG | Disrupts interaction of template DNA and polymerase | Viable (weakly) |
| R665K | C | (I/Y)xGG | Disrupts interaction of template DNA and polymerase | Viable (weakly) |
| M689V | C | A | Disrupts dNTP binding and substrate selection | Viable (weakly) |
| C687S | C | A | Disrupts dNTP binding and substrate selection | Viable (weakly) |
| I664S | NC | (I/Y)xGG | Disrupts interaction of template DNA and polymerase | Nonviable |
| I664Y | NC | (I/Y)xGG | Disrupts interaction of template DNA and polymerase | Nonviable |
| GG666/7AA | C | (I/Y)xGG | Disrupts interaction of template DNA and polymerase | Nonviable |
| M689I | C | A | Disrupts dNTP binding and substrate selection | Nonviable |
| G688S | C | A | Disrupts dNTP binding and substrate selection | Nonviable |
| Y690A | NC | A | Disrupts dNTP binding and substrate selection | Nonviable |
| Y690F | C | A | Disrupts dNTP binding and substrate selection | Nonviable |
| Y690I | NC | A | Disrupts dNTP binding and substrate selection | Nonviable |
| Y690V | NC | A | Disrupts dNTP binding and substrate selection | Nonviable |
| S692Y | NC | A | Disrupts dNTP binding and substrate selection | Nonviable |
| R833T | NC | B | Disrupts dNTP binding affinity | Nonviable |
| Y844S | NC | B | Disrupts dNTP binding affinity | Nonviable |
| K837N | NC | B | Disrupts dNTP binding affinity | Nonviable |
| N841E | NC | B | Disrupts dNTP binding affinity | Nonviable |
| N841Y | C | B | Disrupts dNTP binding affinity | Nonviable |
The table shows each mutant viral molecular clone that was constructed and analyzed. The position of each mutation is shown, along with the type of mutation (C, conservative amino acid substitution; NC, nonconservative substitution), the motif targeted ([I/Y]xGG, motif A, or motif B), the expected effect of the mutation, and the replication capacity of the viral molecular clone. Clones that replicated in Hek 293A host cells are indicated in boldface type. Clones were either nonviable (failed to replicate in any cell type, even with the addition of exogenous dNs), viable (replicated similarly to the wild-type virus in all cell types tested), or semiviable (replicated moderately well in Hek 293A cells but poorly in other cell types tested).
AdPol mutations abrogate virus replication.
To assess the viability (replication competency) of each viral molecular clone, the mutant AdPol genome was transfected into virus-permissive Hek 293A cells and monitored daily for virus production. A virus was considered viable if it demonstrated any of the following properties: plaque formation and/or a progressively increasing CPE. Hek 293A cells were chosen to propagate virus because they were previously shown to contain high levels of endogenous dNTPs (16). Of the 20 AdPol mutants constructed, only 5 were found to be viable in 293 cells, while the rest were found to be replication defective. The five replication-competent virus mutations were I664V, I664M, and R665K ([I/Y]xGG motif mutants) and C687S and M689V (motif A mutants); all of these viable mutants contained conservative substitutions at the target residues. All mutations in motif B were lethal (Table 1), including the conservative N841Y mutation.
In order to ensure that the availability of dNTPs was not the rate-limiting factor for virus replication, the nonviable virus mutants were further tested in Hek 293A cells treated with 0.5 mM exogenous deoxynucleosides (dNs). The addition of exogenous dNs was previously shown to increase cellular dNTP levels by several hundredfold (16) but failed to rescue any of the nonviable virus mutants.
AdPol mutants were impaired in their ability to replicate in Wi38 normal lung fibroblast cells.
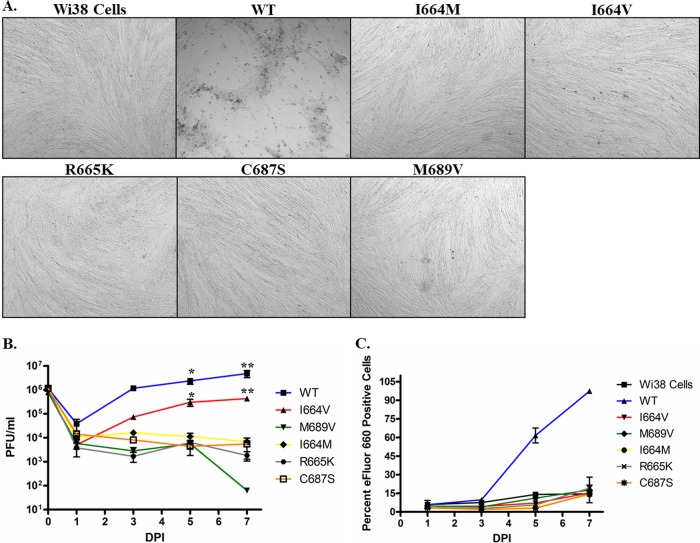
Wi38 normal lung fibroblast cells were used to assess the kinetics of virus replication in a low-dNTP environment over a period of 7 days. Wi38 normal lung fibroblast cells contain substantially lower dNTP concentrations (0.1 μM to 0.2 μM) than cancer cells (16). As expected, all AdPol mutants exhibited impaired viral replication in these cells (Fig. 2). As such, infections with these mutants did not result in CPE or virus-mediated cell death (Fig. 2A and C). Viral titers were also assessed by harvesting cells and culture supernatants at days 1, 3, 5, and 7 postinfection and performing a TCID50 assay (Fig. 2B). This analysis confirmed that all of the AdPol mutants, except for the I664V mutant, were unable to productively replicate in Wi38 cells. In contrast, wild-type Ad5 caused a productive, cytolytic infection in Wi38 cells.
Fig 2.
Replication kinetics of Ad5 polymerase mutants in Wi38 cells. Primary human lung fibroblasts (Wi38 cells) were infected with each indicated Ad5 mutant virus. Infections were allowed to proceed for 7 days. (A) Bright-field images were recorded at day 7 postinfection for each AdPol mutant virus. Wi38 cells represent mock-infected cells at day 7. Images were taken at a ×5 magnification. (B) Titers of infectious virus were determined at days 1, 3, 5, and 7 postinfection by TCID50 analysis of infected-cell lysates. (C) Cell death was assessed at 1, 3, 5 and 7 days postinfection (DPI) by flow cytometric quantitation of the percentage of cells that were positively stained with the viability dye eFluor 660, which irreversibly labels dead cells. In panels B and C, data represent mean values of 3 technical replicates; error bars denote the standard deviations. The data shown are representative of 3 independent experiments that yielded similar results. A statistical comparison of the virus titers was performed at each time point. * (P < 0.05) and ** (P < 0.001) denote statistically significant differences in titers of the indicated viruses compared to the all other virus mutants analyzed.
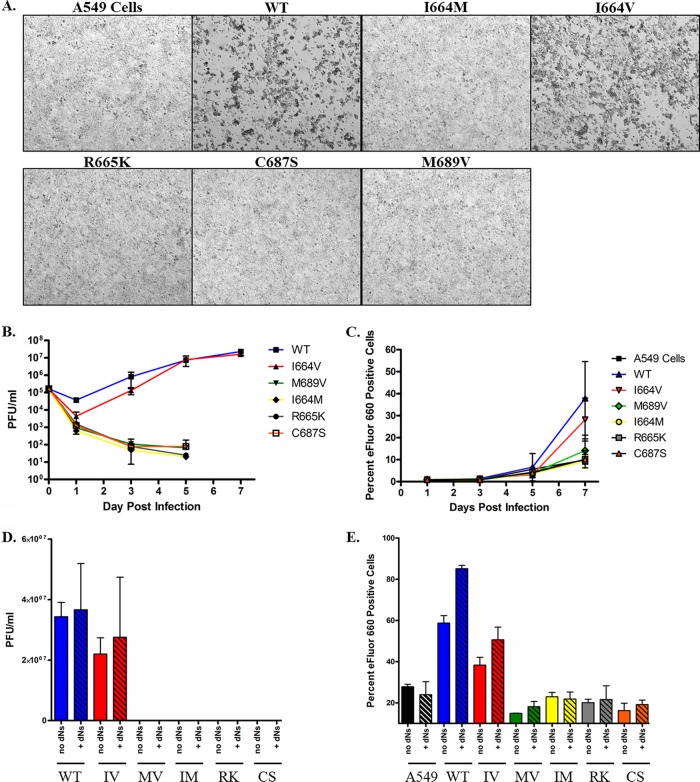
Only the I664V Ad5 mutant replicates efficiently in A549 lung carcinoma cells.
To assess the kinetics of virus replication in a high-dNTP environment, A549 lung carcinoma cells (1.7 to 3.3 μM dNTPs) (16) were used (Fig. 3). The I664V Ad5 mutant replicated with kinetics very similar to those of wild-type Ad5 in these cells (Fig. 3B). The I664V mutant virus also caused progressive CPE and the eventual lysis of A549 cells (Fig. 3A and C). In contrast, the I664M, R665K, C687S, and M689V Ad5 mutants were unable to productively replicate in this cell line (Fig. 3B).
Fig 3.
Replication kinetics of Ad5 polymerase mutants in A549 cells. A549 lung carcinoma cells were infected with each indicated Ad5 mutant virus. Infections were allowed to proceed for 7 days. (A) Bright-field images were recorded as described in the legend of Fig. 2. A549 cells represent mock-infected cells at day 7. (B) Titers of infectious virus were determined at days 1, 3, 5, and 7 postinfection by TCID50 analysis of infected-cell lysates in the absence of exogenous dNs. (C) Cell death, in the absence of exogenous dNs, was assessed as described in the legend of Fig. 2. (D) Titers of infectious virus were determined at 7 days postinfection, in the presence (+dNs) or absence (no dNs) of exogenous dNs. (E) Cell death was also assessed at 7 days postinfection in the presence (0.25 mM) (+dNs) or absence (no dNs) of exogenous dNs. In panels B to E, data represent mean values of 3 technical replicates; error bars denote the standard deviations. The data shown are representative of 3 independent experiments that yielded similar results. IV, I664V mutation.
In an attempt to rescue virus replication for the impaired AdPol mutants, exogenous dNs were added to the A549 cells in order to raise intracellular dNTP levels, and virus replication was then assessed at 7 days postinfection. The addition of exogenous dNs to culture supernatants failed to rescue any of the impaired AdPol mutants (Fig. 3D and E). Because the I664V mutant Ad5 was the only AdPol mutant to replicate efficiently in A549 cancer cells, regardless of the presence of exogenous dNs, it was selected for further evaluation.
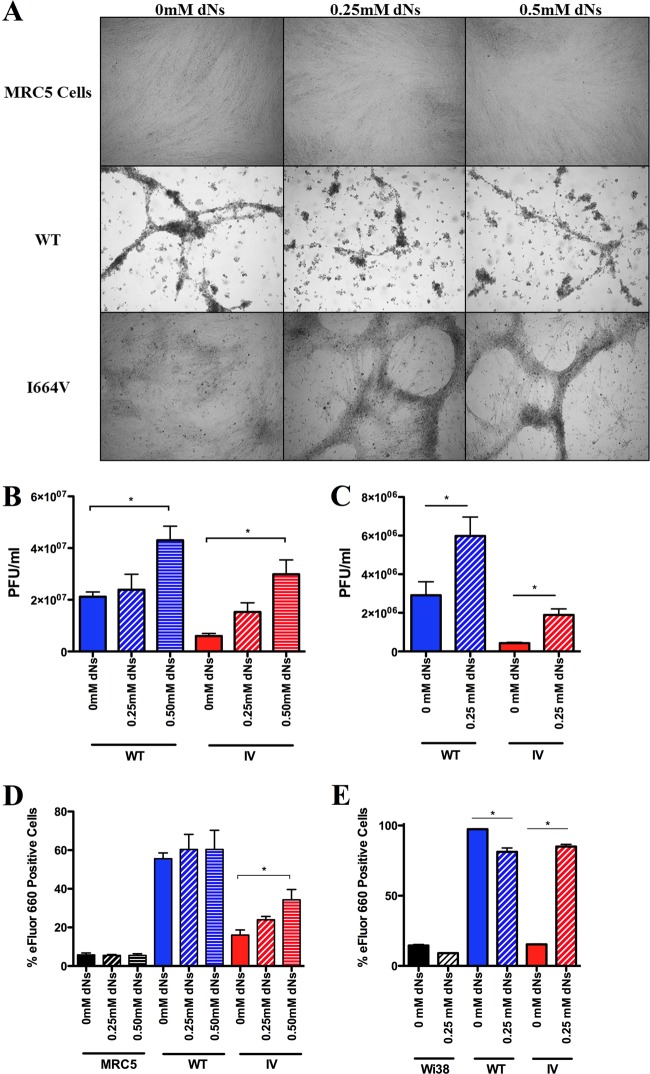
I664V Ad5 replication was rescued in normal lung fibroblasts (MRC5 and Wi38 cells) treated with exogenous dNs.
Because the I664V Ad5 mutant was found to be impaired in its ability to replicate in primary Wi38 cells, we next evaluated the ability of exogenous dNs to rescue virus replication and virus-mediated cell death in primary cells. For this analysis, we used primary lung fibroblasts (both MRC5 and Wi38 cells) (Fig. 4). Infections were allowed to proceed for 7 days, with culture medium being supplemented with either 0.25 mM or 0.5 mM exogenous dNs every other day for the duration of the experiment. The addition of 0.5 mM exogenous dNs to the culture medium of the MRC5 cells resulted in a statistically significant, 5-fold increase in the replication of the I664V Ad5 mutant, while the addition of 0.25 mM exogenous dNs elicited a 2.5-fold increase in the replication of this virus (Fig. 4B). In contrast, the addition of 0.25 mM exogenous dNs to cells infected with WT Ad5 resulted in only a minimal (1.1-fold) rise in the WT virus replication level, and even the addition of 0.5 mM exogenous dNs elicited only a 2-fold rise in the WT virus replication level (Fig. 4B). We observed very similar findings with Wi38 cells, where the addition of 0.25 mM exogenous dNs elicited a statistically significant, 4.4-fold increase in the replication of the I664V Ad5 mutant, versus a more modest 2.1-fold increase in the replication of the wild-type virus (Fig. 4C). Consistent with the observed effect on virus replication, the addition of 0.5 mM exogenous dNs to MRC5 cells also resulted in a statistically significant, 2.1-fold increase in cell killing by the I664V Ad5 mutant but had no statistically significant effect on cell killing by WT Ad5 (Fig. 4D). Similar results were again observed for Wi38 cells, where the addition of 0.25 mM exogenous dNs resulted in a statistically significant, 5.7-fold increase in cell killing by the I664V Ad5 mutant, versus a modest, but statistically significant, reduction in cell killing by WT Ad5 (Fig. 4E).
Fig 4.
Replication of the I664V Ad5 mutant virus is rescued by addition of exogenous dNs to primary fibroblasts. Primary human lung fibroblasts (MRC5 cells or Wi38 cells) were infected with either wild-type Ad5 or the I664V Ad5 mutant. Culture supernatants were supplemented with 0, 0.25, or 0.5 mM each dN (for MRC5) or 0 or 0.25 mM each dN (for Wi38). Infections were allowed to proceed for 7 days. (A) Bright-field images of MRC5 cell cultures were recorded at day 7 postinfection. (B and C) Titers of infectious virus were determined at 7 days postinfection for MRC5 (B) and Wi38 (C) cells. (D and E) Cell death was determined at 7 days postinfection for MRC5 (D) and Wi38 (E) cells. In panels B to E, data represent mean values of either six (MRC5) or three (Wi38) technical replicates; error bars denote the standard deviations. The data shown are representative of 2 independent experiments that yielded similar results. * indicates a P value of <0.05, as determined by a one-way analysis of variance with the Bonferroni post hoc test.
The I664V Ad5 mutant preferentially lyses cancer cells.
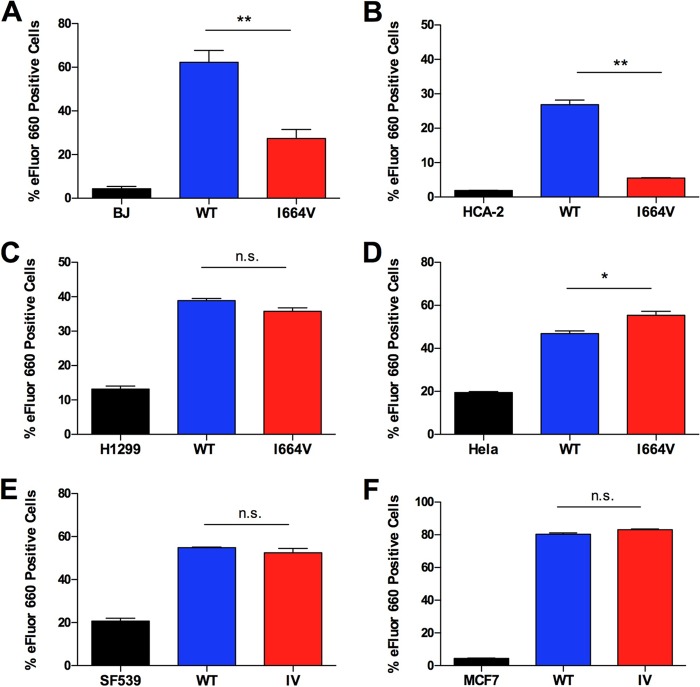
We next evaluated the ability of the I664V Ad5 mutant to induce cell killing in a small panel of normal (primary) cells and cancer cells. BJ and HCA-2 primary foreskin fibroblast cells were chosen as representative normal cells. The I664V Ad5 mutant had statistically significant, 2.5-fold-reduced (BJ cells) and 6.8-fold-reduced (HCA-2 cells) abilities to induce the killing of these primary cells compared to the wild-type virus (Fig. 5A and B). HeLa (cervical adenocarcinoma), H1299 (non-small-cell lung carcinoma), SF539 (glioma), and MCF-7 (breast cancer) cells were selected as representative tumor cells. The I664V Ad5 mutant efficiently lysed all of the tumor cell types to a level that was either statistically indistinguishable from that of the wild-type virus (H1299, SF539, and MCF-7 cells) or slightly elevated compared to that of the WT virus (HeLa cells) (Fig. 5C to E).
Fig 5.
The I664V Ad5 mutant is selectively cytotoxic for cancer cells but not normal cells. (A and B) BJ (A) and HCA-2 (B) normal foreskin fibroblast cells were infected with either the WT or the I664V Ad5 mutant at an MOI of 20, and cell killing was measured 7 days later. (C to E) Various cancer cell lines (H1299 [C], HeLa [D], SF539 [E], and MCF-7 [F]) were infected with the WT or the I664V Ad5 mutant at an MOI of 5, and cell killing was measured 7 days later. Data represent mean values of 6 technical replicates; error bars denote the standard deviations. * indicates a P value of <0.01 and ** indicates a P value of <0.001; n.s. indicates values that were not statistically significantly different, as determined by a one-way analysis of variance with the Bonferroni post hoc test.
The I664V Ad5 mutant preferentially replicates in cancer cells.
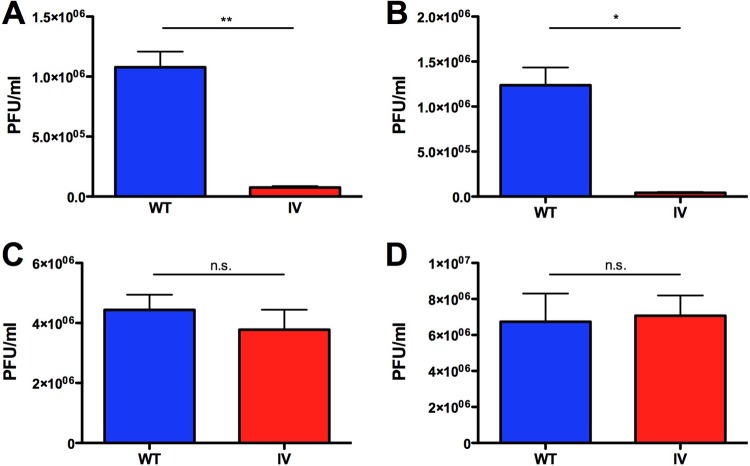
In light of the ability of the I664V Ad5 mutant to selectively induce cell killing in cancer cells compared to normal cells, we conducted follow-up experiments to examine virus replication in a subset of the cell lines examined in Fig. 5. This analysis revealed that the I664V virus replicated much less efficiently in normal cells (BJ and HCA-2 cells) than did wild-type Ad5 (Fig. 6A and B). The difference in virus titers between WT Ad5 and the I664V Ad5 mutant was statistically significant in both cases (14-fold in BJ cells and 29-fold in HCA-2 cells). In contrast, the wild-type virus and the I664V mutant replicated to statistically indistinguishable titers in cancer cells (H1299 and HeLa cells) (Fig. 6C and D).
Fig 6.
The I664V Ad5 mutant virus replicates preferentially in cancer cells but not normal cells. (A and B) BJ (A) and HCA-2 (B) normal foreskin fibroblast cells were infected with either WT or I664V Ad5, at an MOI of 20, and titers of infectious virus were measured 7 days later. (C and D) H1299 and HeLa cancer cell lines were infected with WT or I664V Ad5 at an MOI of 5, and titers of infectious virus were measured 7 days later. Data represent mean values of 6 technical replicates; error bars denote the standard deviations. * indicates a P value of <0.001, and ** indicates a P value of <0.0001; n.s. indicates values that were not statistically significantly different, as determined by an unpaired t test.
AdPol mutants are impaired in their dNTP utilization efficiencies.
To further ascertain if the altered replication capacity of the AdPol mutants was associated with a dNTP-binding defect, the DNA polymerase proteins of the five viable virus mutants were analyzed biochemically. A multiple-nucleotide incorporation analysis was employed to assess the efficiencies of dNTP utilization by the various purified polymerases (10, 16, 20). This assay evaluates the ability of the purified polymerase to incorporate dNTPs (in a concentration-dependent fashion) onto a 5′-32P-labeled primer (20-mer) that has been annealed to a 30-mer template.
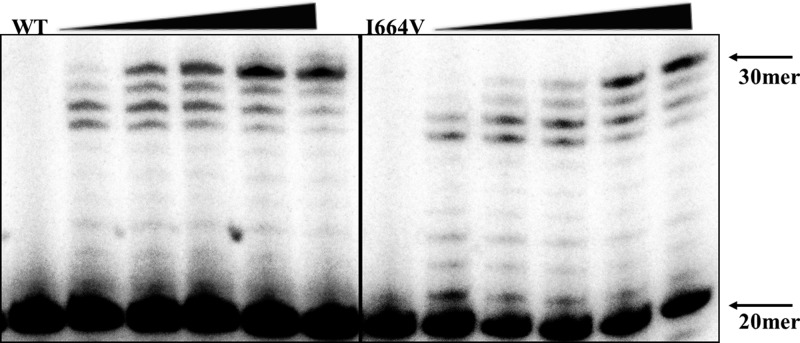
For this experiment, dNTP incorporation was evaluated at concentrations ranging from 0 to 60 μM dNTPs. Typical analysis results are shown in Fig. 7 for the wild-type and the I664V Ad5 DNA polymerases. The dNTP concentration necessary for 50% of the maximal extension of the template-primer (T/P) was determined by using this assay for each of the mutant polymerases (Table 2). A total of 7.6 μM dNTPs was sufficient to permit a 50% maximal extension of the T/P by wild-type Ad5 Pol. This is consistent with the efficient replication of wild-type Ad5 in A549 cells (which contain a dNTP concentration of ∼1.7 to 3.3 μM) (Table 3). The I664M polymerase was only modestly affected in dNTP utilization efficiency, requiring 13.2 μM dNTPs for a 50% maximal T/P extension (a 1.7-fold increase compared to the wild-type Ad5 Pol). Despite the modest change in dNTP utilization, the I664M Ad5 mutant was significantly impaired in its ability to replicate in both cancer and primary cells. This suggests that the I664M AdPol mutation likely influences other aspects of polymerase function (such as the ability to carry out initiation or elongation activities), thus accounting for its incompatibility with virus replication.
Fig 7.
Adenovirus DNA polymerase dNTP utilization efficiency is reduced by the I664V mutation. Multinucleotide extension assay reactions were performed by using a 5′-32P-labeled 20-mer primer annealed to a 30-mer template with serially diluted dNTPs at final concentrations of 0, 1, 3, 6, 30, and 60 μM. A representative image is presented for the wild-type and I664V Ad5 DNA polymerases. Arrows indicate the positions of the 20-mer (nonelongated primer) and the 30-mer (elongated primer) products. The data shown are representative of 3 independent experiments that yielded similar results.
Table 2.
dNTP utilization by mutant Ad5 DNA polymerases
| Mutation | Mean dNTP concn (μM) required for 50% maximal extension ± SDa | Fold changeb |
|---|---|---|
| Wild type | 7.60 ± 1.05 | |
| I664M | 13.2 ± 1.14 | 1.7 |
| I664V | 19.2 ± 1.18 | 2.5 |
| R665K | 29.5 ± 1.34 | 4 |
| C687S | 30.6 ± 1.25 | 4 |
| M689V | 29.0 ± 2.39 | 4 |
Mean values and standard deviations are shown for 3 different experiments.
Fold change values compared to the value for the wild-type polymerase protein.
Table 3.
dNTP contents of selected cell lines
| Cell line | Cell type | Intracellular dNTP concn (μM) | Description (reference) | Reference |
|---|---|---|---|---|
| A549 | Cancer | 1.7–3.3 | 16 | |
| HeLa | Cancer | 3.7–31 | HeLa cells contain 9.8–82 pmol each dNTP per 106 cells (25) and have a vol of 2.6 × 103 μm3 (34) | 25 |
| 293FT | Cancer | 1.0–2.3 | The present study used 293A cells; 293A and 293FT cells are both subclones of the parental 293 cell line | 16 |
| PANC1 | Cancer | 1.3–2.4 | 16 | |
| Wi38 | Primary | 0.15–0.34 | 18 | |
| MRC5 | Primary | 0.38 | This is the measured value for dTTP (31) | 31 |
The I664V polymerase mutant was also only modestly impaired in its dNTP utilization efficiency, requiring 19.2 μM dNTPs for a 50% maximal T/P extension (a 2.5-fold increase). This is consistent with the fact that the I664V Ad5 mutant replicated efficiently in dN-supplemented cells and tumor cells but not in primary cells. In contrast, all of the other polymerase mutants (R665K, C687S, and M689V) had a more profound (4-fold) defect in dNTP utilization and failed to support virus replication in Wi38 and A549 cells, regardless of dN supplementation.
DISCUSSION
Oncolytic virotherapy utilizing adenoviruses engineered to selectively replicate in and lyse cancer cells is being actively pursued as a cancer treatment modality (9, 11, 19, 24). Most previous approaches to the development of conditionally replicating adenoviruses have focused on the transcriptional regulation of the essential Ad5 E1 genes or have targeted the adenovirus replication proteins located in the E2 region (8, 14, 22). Our study aimed at regulating adenovirus replication by the rational mutagenesis of its DNA polymerase. We based our approach on studies which showed that one can create DNA polymerase mutants that are catalytically active only at high dNTP concentrations, such as those found in cancer cells, by selectively mutagenizing regions of the protein that affect dNTP substrate utilization (7, 10, 16, 21, 32).
We focused our efforts on Ad5 DNA polymerase motifs known to interact with the dNTP substrate either directly (motif A or B), through the polymerase active site, or indirectly ([I/Y]xGG), through the template DNA (2–4, 7, 28, 33). The mutagenesis of the selected residues had a dramatic effect on the replicative capacity of Ad5. None of the nonconservative mutations gave rise to viable virus, nor did any of the motif B mutants. This included mutations of motif B that targeted amino acid residues R833 and K837, which have been shown to directly bind and stabilize the incoming dNTP within the polymerase active site via their basic side chains (1, 29, 33). The loss of these basic side chains (R833T and K837N mutants) was sufficient to abrogate virus replication (Table 1). Additional mutations in motif B, N841Y and Y844S, which were expected to impair the formation of the proper nascent base pair binding pocket, required for the stabilization of the nucleotide in the templating position, were also not tolerated (1, 4, 23).
Mutations of the (I/Y)xGG motif gave rise to three of the five viable virus mutants that were obtained. This motif has been shown to be involved in stably binding the template DNA at the polymerization active site, in shuttling the primer terminus between the polymerase and exonuclease active sites, and in playing an essential role in the transition between initiation and elongation (7, 28, 30). In particular, residue I664 has been implicated in having a direct interaction with the ribose functional group of the 3′ nucleotide that precedes the templating nucleotide, directly influencing template strand stability in the polymerase active site. Similar to previous studies, different site-directed mutations made at I664 gave rise to viruses with distinct phenotypes (i.e., I664V [which was replication competent], I664M [which was grossly attenuated], and I664S and I664Y [which were completely replication defective]). This is likely due to the fact that such mutants can lead to alterations in the polymerase/exonuclease equilibrium, with potentially deleterious effects on virus replication (7, 15, 28, 30). Interestingly, mutations that favor exonucleolysis have been found to require high dNTP concentrations to carry out efficient polymerization and exhibited altered dNTP binding affinities compared to the wild-type polymerase (7). This is consistent with the phenotype exhibited by the I664V Ad5 mutant, which replicated to wild-type levels in all cancer cells in which it was tested but was significantly impaired in all normal cells (which are expected to have approximately 10-fold-lower levels of endogenous dNTPs than cancer cells [Table 3]). Moreover, the replication of the I664V Ad5 mutant was significantly enhanced by the addition of exogenous dNs to primary MRC5 and Wi38 cells (Fig. 4).
Mutagenesis of the (I/Y)xGG region of the Ad5 DNA polymerase has also been shown to produce mutants that are impaired in their ability to transition from initiation to elongation. Such mutations reduce elongation activity even in the presence of high dNTP concentrations (7). This may be the case for the I664M mutation, which showed only a modest defect in dNTP utilization (a 1.7-fold change compared to the wild-type polymerase) as well as impaired replication in all cell lines tested, regardless of the concentration of dNTPs present during infection.
The severe phenotype of the R665K mutant virus was unexpected, since this is a highly conservative substitution that has a minimal effect in the context of the closely related Phi29 DNA polymerase (28, 30). In Phi29, the residues of the (I/Y)xGG motif have been shown to interact with the templating nucleotide through a series of Van der Waals interactions and hydrogen bonds that are facilitated by a network of water molecules that mediate a large portion of the protein-nucleic acid interactions (1). It is possible that while the lysine residue maintains the basic charge and size of the side-chain group, the removal of amine groups that are present in arginine may impair the hydrogen bonding that is critical for template DNA binding or I664 orientation.
The mutagenesis of motif A gave rise to two viable virus mutants. Residues in this motif have been shown to interact with both incoming dNTPs and two highly conserved aspartate residues important for the proper catalysis of the polymerase reaction. None of the mutations carried out on the “steric gate” tyrosine (Y690) gave rise to replication-competent virus mutants. Even the highly conservative Y690F mutant was nonviable, suggesting that both the phenolic ring and the hydroxyl group are essential for proper polymerase function.
In conclusion, our data show that even modest changes in polymerase activity, associated with dNTP substrate utilization, are sufficient to completely abrogate adenovirus replication. Despite this, one mutation (I664V) was found to replicate in a dNTP-dependent manner and to selectively induce the killing of tumor cells rather than primary cells. This mutant provides encouraging proof-of-concept support for the notion that conditionally replicating, tumor-selective adenovirus vectors can be created by modifying the efficiency with which the viral DNA polymerase utilizes dNTP substrates.
ACKNOWLEDGMENTS
We thank Casey Maguire for the construction of plasmid pAdTrack-E1SV40/eGFP and Edward Kennedy for helpful discussions.
This work was supported by Department of Defense grant W81XWH-07-10376 (to S.D. and B.K.), NIH grant F31 GM095190 (to W.D.), NIH grant T32DE007202 (to C.C.), NIH grant T32GM068411, and NIH grant T32GM007356 (to V.F.).
Footnotes
Published ahead of print 18 July 2012
REFERENCES
- 1. Berman A, et al. 2007. Structures of phi29 DNA polymerase complexed with substrate—the mechanism of translocation in B-family polymerases. EMBO J. 26:3494–3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Blasco M, Lazaro J, Bernad A, Blanco L, Salas M. 1992. Phi29 DNA polymerase active site: mutants in conserved residues TYR254 and TYR390 are affected in dNTP binding. J. Biol. Chem. 267:19427–19434 [PubMed] [Google Scholar]
- 3. Blasco M, Lazaro J, Blanco L, Salas M. 1993. Phi29 DNA polymerase active site: residue ASP249 of conserved amino acid motif “DX2SLYP” is critical for synthetic activities. J. Biol. Chem. 268:24106–24113 [PubMed] [Google Scholar]
- 4. Blasco M, Lazaro J, Blanco L, Salas M. 1993. Phi29 DNA polymerase active site: the conserved amino acid motif “Kx3NSxYG” is involved in template-primer binding and dNTP selection. J. Biol. Chem. 268:16763–16770 [PubMed] [Google Scholar]
- 5. Bonnin A, Lazaro JM, Blanco L, Salas M. 1999. A single tyrosine prevents insertion of ribonucleotides in the eukaryotic-type phi29 DNA polymerase. J. Mol. Biol. 290:241–251 [DOI] [PubMed] [Google Scholar]
- 6. Brenkman AB, Breure EC, van der Vliet PC. 2002. Molecular architecture of adenovirus DNA polymerase and location of the protein primer. J. Virol. 76:8200–8207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brenkman AB, Heideman MR, Truniger V, Salas M, van der Vliet PC. 2001. The (I/Y)XGG motif of adenovirus DNA polymerase affects template DNA binding and the transition from initiation to elongation. J. Biol. Chem. 276:29846–29853 [DOI] [PubMed] [Google Scholar]
- 8. Brunori M, Malerba M, Kashiwazaki H, Iggo R. 2001. Replicating adenoviruses that target tumors with constitutive activation of the wnt signaling pathway. J. Virol. 75:2857–2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cody JJ, Douglas JT. 2009. Armed replicating adenoviruses for cancer virotherapy. Cancer Gene Ther. 16:473–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Diamond TL, et al. 2004. Macrophage tropism of HIV-1 depends on efficient cellular dNTP utilization by reverse transcriptase. J. Biol. Chem. 279:51545–51553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dobbelstein M. 2004. Replicating adenoviruses in cancer therapy. Curr. Top. Microbiol. Immunol. 273:291–334 [DOI] [PubMed] [Google Scholar]
- 12. Field J, Gronostajski RM, Hurwitz J. 1984. Properties of the adenovirus DNA polymerase. J. Biol. Chem. 259:9487–9495 [PubMed] [Google Scholar]
- 13. Gao L, et al. 2008. Apparent defects in processive DNA synthesis, strand transfer, and primer elongation of Met-184 mutants of HIV-1 reverse transcriptase derive solely from a dNTP utilization defect. J. Biol. Chem. 283:9196–9205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hardcastle J, Kurozumi K, Chiocca EA, Kaur B. 2007. Oncolytic viruses driven by tumor-specific promoters. Curr. Cancer Drug Targets 7:181–189 [DOI] [PubMed] [Google Scholar]
- 15. Jacewicz A, Makiela K, Kierzek A, Drake JW, Bebenek A. 2007. The roles of Tyr391 and Tyr619 in RB69 DNA polymerase replication fidelity. J. Mol. Biol. 368:18–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jamburuthugoda VK, Chugh P, Kim B. 2006. Modification of human immunodeficiency virus type 1 reverse transcriptase to target cells with elevated cellular dNTP concentrations. J. Biol. Chem. 281:13388–13395 [DOI] [PubMed] [Google Scholar]
- 17. Jamburuthugoda VK, Guo D, Wedekind JE, Kim B. 2005. Kinetic evidence for interaction of human immunodeficiency virus type 1 reverse transcriptase with the 3′-OH of the incoming dTTP substrate. Biochemistry 44:10635–10643 [DOI] [PubMed] [Google Scholar]
- 18. Jamburuthugoda VK, et al. 2008. Reduced dNTP binding affinity of 3TC-resistant M184I HIV-1 reverse transcriptase variants responsible for viral infection failure in macrophage. J. Biol. Chem. 283:9206–9216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kanerva A, Hemminki A. 2005. Adenoviruses for treatment of cancer. Ann. Med. 37:33–43 [DOI] [PubMed] [Google Scholar]
- 20. Kennedy EM, Hergott C, Dewhurst S, Kim B. 2009. The mechanistic architecture of thermostable Pyrococcus furiosus family B DNA polymerase motif A and its interaction with the dNTP substrate. Biochemistry 48:11161–11168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim B, Loeb LA. 1995. Human immunodeficiency virus reverse transcriptase substitutes for DNA polymerase I in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 92:684–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kirn D. 2001. Clinical research results with dl1520 (Onyx-015), a replication-selective adenovirus for the treatment of cancer—what have we learned? Gene Ther. 8:89–98 [DOI] [PubMed] [Google Scholar]
- 23. Liu H, Naismith JH, Hay RT. 2000. Identification of conserved residues contributing to the activities of adenovirus DNA polymerase. J. Virol. 74:11681–11689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu T-C, Galanis E, Kirn D. 2007. Clinical trial results with oncolytic virotherapy: a century of promise, a decade of progress. Nat. Clin. Pract. Oncol. 4:101–117 [DOI] [PubMed] [Google Scholar]
- 25. North TW, Bestwick RK, Mathews CK. 1980. Detection of activities that interfere with the enzymatic assay of deoxyribonucleoside 5′-triphosphates. J. Biol. Chem. 255:6640–6645 [PubMed] [Google Scholar]
- 26. Saturno J, Blanco L, Salas M, Esteban JA. 1995. A novel kinetic analysis to calculate nucleotide affinity of proofreading DNA polymerases. Application to phi 29 DNA polymerase fidelity mutants. J. Biol. Chem. 270:31235–31243 [DOI] [PubMed] [Google Scholar]
- 27. Stunnenberg HG, Lange H, Philipson L, van Miltenburg RT, van der Vliet PC. 1988. High expression of functional adenovirus DNA polymerase and precursor terminal protein using recombinant vaccinia virus. Nucleic Acids Res. 16:2431–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Truniger V, Blanco L, Salas M. 1999. Role of the “YxGG/A” motif of Phi29 DNA polymerase in protein-primed replication. J. Mol. Biol. 286:57–69 [DOI] [PubMed] [Google Scholar]
- 29. Truniger V, Lázaro JM, Salas M. 2004. Two positively charged residues of ϕ29 DNA polymerase, conserved in protein-primed DNA polymerases, are involved in stabilisation of the incoming nucleotide. J. Mol. Biol. 335:481–494 [DOI] [PubMed] [Google Scholar]
- 30. Truniger V, Lazaro JM, Salas M, Blanco L. 1996. A DNA binding motif coordinating synthesis and degradation in proofreading DNA polymerases. EMBO J. 15:3430–3441 [PMC free article] [PubMed] [Google Scholar]
- 31. Wachsman M, Hamzeh FM, Saito H, Lietman PS. 1996. Anticytomegaloviral activity of methotrexate associated with preferential accumulation of drug by cytomegalovirus-infected cells. Antimicrob. Agents Chemother. 40:433–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Weiss KK, et al. 2004. A role for dNTP binding of human immunodeficiency virus type 1 reverse transcriptase in viral mutagenesis. Biochemistry 43:4490–4500 [DOI] [PubMed] [Google Scholar]
- 33. Yang G, Lin TC, Karam J, Konigsberg WH. 1999. Steady-state kinetic characterization of RB69 DNA polymerase mutants that affect dNTP incorporation. Biochemistry 38:8094–8101 [DOI] [PubMed] [Google Scholar]
- 34. Zhao L, et al. 2008. Intracellular water-specific MR of microbead-adherent cells: the HeLa cell intracellular water exchange lifetime. NMR Biomed. 21:159–164 [DOI] [PMC free article] [PubMed] [Google Scholar]