Abstract
Limited antiviral compounds are available for the control of influenza, and the emergence of resistant variants would further narrow the options for defense. The H275Y neuraminidase (NA) mutation, which confers resistance to oseltamivir carboxylate, has been identified among the seasonal H1N1 and 2009 pandemic influenza viruses; however, those H275Y resistant variants demonstrated distinct epidemiological outcomes in humans. Specifically, dominance of the H275Y variant over the oseltamivir-sensitive viruses was only reported for a seasonal H1N1 variant during 2008-2009. Here, we systematically analyze the effect of the H275Y NA mutation on viral fitness and transmissibility of A(H1N1)pdm09 and seasonal H1N1 influenza viruses. The NA genes from A(H1N1)pdm09 A/California/04/09 (CA04), seasonal H1N1 A/New Caledonia/20/1999 (NewCal), and A/Brisbane/59/2007 (Brisbane) were individually introduced into the genetic background of CA04. The H275Y mutation led to reduced NA enzyme activity, an increased Km for 3′-sialylactose or 6′-sialylactose, and decreased infectivity in mucin-secreting human airway epithelial cells compared to the oseltamivir-sensitive wild-type counterparts. Attenuated pathogenicity in both RG-CA04NA-H275Y and RG-CA04 × BrisbaneNA-H275Y viruses was observed in ferrets compared to RG-CA04 virus, although the transmissibility was minimally affected. In parallel experiments using recombinant Brisbane viruses differing by hemagglutinin and NA, comparable direct contact and respiratory droplet transmissibilities were observed among RG-NewCalHA,NA, RG-NewCalHA,NA-H275Y, RG-BrisbaneHA,NA-H275Y, and RG-NewCalHA × BrisbaneNA-H275Y viruses. Our results demonstrate that, despite the H275Y mutation leading to a minor reduction in viral fitness, the transmission potentials of three different antigenic strains carrying this mutation were comparable in the naïve ferret model.
INTRODUCTION
Effective antiviral drugs provide immediate control during influenza epidemics or pandemics. Neuraminidase (NA) inhibitors are recommended by WHO for the pharmacological management of the 2009 pandemic H1N1 viruses [A(H1N1)pdm09], which carry the S31N mutation in the M2 protein that are known to confer resistance to adamantanes (ion channel blockers for influenza A viruses) (3). A total of 350 million treatment courses of oseltamivir were distributed to countries worldwide for pandemic preparedness between 2004 and December 2009 (56). Emergence of resistant variants has been a concern with increased NA inhibitor usage during the H1N1 pandemic, especially if such variants may spread efficiently among humans. Structure-based-designed NA inhibitors target the enzyme site (66) to block the NA activity that hydrolyzes the α-ketosidic linkage between sialic acid and the adjacent oligosaccharide. Such activity is important for establishing infections (44) and for the release of viral progeny (1). NA mutations within the enzyme site that confer resistance to NA inhibitors would also reduce NA enzyme activity and thereby compromise viral fitness (19). As different NA mutations result in different levels of NA functional loss, NA inhibitor-resistant variants have been shown to differ in their fitness in vitro and in vivo (27, 71). The fitness of the NA inhibitor-resistant variants can also be confounded by the genetic composition of the virus, including the hemagglutinin (HA) that binds to sialic acid-containing receptors and possesses a counteracting activity to that of NA (19, 45).
The H275Y NA mutation (using N1 numbering) is the most commonly reported mutation arising from oseltamivir selection pressure within the N1 subtype. The mutation was first isolated from 2 out of 54 human subjects post-oseltamivir treatment experimentally infected with seasonal H1N1 influenza virus (A/Texas/36/91) (20). The H275Y mutation has also been detected in H5N1 patients after oseltamivir treatment (10, 41). Surveillance monitoring of influenza virus susceptibility to NA inhibitors showed limited detection rates for this mutation among the N1 subtype prior to October 2007 (14, 49, 61, 64), but since then, a globally spreading H275Y variant has emerged (11, 31, 40, 67), leading to the dominance of an oseltamivir-resistant variant over the sensitive H1N1 strains. Epidemiological studies could not provide evidence to link this finding to oseltamivir usage, suggesting that the transmissibility of the H275Y variant was at least comparable to that of the circulating oseltamivir-sensitive strains (11, 26, 47). The sudden increase of the H1N1 oseltamivir-resistant viruses occurred concurrently with a major change of HA antigenicity, specifically, the change from A/New Caledonia/20/1999-like virus (NewCal; 2000/2001 to 2006/2007) to A/Solomon Island/3/2006-like virus (2007/2008) and A/Brisbane/59/2007-like virus (Brisbane; 2008/2009). In addition to the change in HA antigenicity, Sandbulte et al. recently reported a major change in NA antigenicity that was associated with the emergence of the Brisbane-like virus (58). Studies have shown that the presence of secondary mutations (4) or epistatic mutations (39) within NA may restore the NA function upon acquisition of the H275Y mutation. Specifically, the V234M and R222Q amino acid changes have been shown to increase surface NA expression while accommodating the H275Y NA mutation (4). Furthermore, mutations occurring in the HA have been reported to promote the replication of seasonal H1N1 viruses with the H275Y NA mutation (16). Collectively, these studies indicate that the fitness outcome of viruses carrying the H275Y mutation may be varied across antigenetic strains, and therefore the emergence of new variants merits independent investigations.
The A(H1N1)pdm09 virus, which emerged through genetic reassortment, contains gene segments derived from human, swine, and avian origins (15, 62), with the HA from the classical Swine influenza virus (descendant of the 1918 pandemic virus) and the NA from the Eurasian swine influenza virus (which originated in an avian reservoir) (59). While an increased usage of NA inhibitors was expected during the pandemic period, the detection rate of the H275Y NA mutation among A(H1N1)pdm09 viruses was reported as only <2% in the general population (6, 7, 46, 68). A case-controlled study identified immunosuppression as the major risk factor associated with detection of an oseltamivir-resistant H275Y variant among hospitalized patients (6). The transmission of the A(H1N1)pdm09 variant carrying the H275Y NA mutation was found to be limited (9, 18, 42, 50), although more recent studies have suggested the presence of community transmission of oseltamivir-resistant H275Y variants in the United States and Australia during 2010-2011 (32, 63). The fitness of clinically derived or recombinant A(H1N1)pdm09 H275Y variants compared to their respective wild-type counterparts has been evaluated using in vitro and animal models (5, 13, 23, 37, 48, 52, 60). The majority of these reports found that A(H1N1)pdm09 virus carrying the H275Y mutation had comparable replication efficiency and transmissibility as that of their wild-type counterparts (23, 37, 48, 60), while some investigators detected a slightly compromised viral fitness (5, 13, 24). Overall, there was little correlation between the epidemiological data and the laboratory-based in vitro or in vivo experimental findings, and there is a need to reevaluate the role of the H275Y NA mutation in viral fitness by using selectively different virus strains to serve as control groups.
To achieve a comprehensive understanding of the various degrees of NA functional loss that have been implicated by studies of the H275Y NA mutation, here we have adopted a systematic approach by generating three pairs of recombinant H1N1 viruses that differ in their NA genes. The first pair is A(H1N1)pdm09 A/California/04/09 (CA04) viruses with or without the H275Y NA mutation. Two other pairs of oseltamivir-sensitive (wild-type) and resistant recombinant CA04 viruses were generated for comparison, with their NA gene derived from the NewCal and Brisbane human seasonal H1N1 influenza virus strains known to possess different transmissibilities in human populations based on epidemiological data. While previous studies focused primarily on the A(H1N1)pdm09 viruses alone, we have included two previous human seasonal H1N1 viruses into the study design. This approach allowed us to directly compare the effect of the H275Y NA mutation in three different NA backgrounds. The NA enzyme properties were characterized, and the viral replication efficiencies in conventional cell lines and in differentiated human normal bronchial epithelial (NHBE) cells were evaluated. Lastly, the transmissibilities of the CA04 recombinant viruses via direct contact or respiratory droplet routes were further compared to those of the seasonal NewCal and Brisbane viruses. This study provides the first in vitro and in vivo comparison of the effect of the H275Y mutation across three antigenic strains exhibiting different epidemiological outcomes.
MATERIALS AND METHODS
Compounds.
NA inhibitors oseltamivir carboxylate [GS4071; 4-N-acetyl-5-amino-3-(1-ethylpropoxy)-1-cyclohexane-1 carboxylic acid] was provided by Hoffmann-La Roche (Basel, Switzerland), and zanamivir (GG167; 4-guanidino-Neu5Ac2en) was provided by GlaxoSmithKline (Stevenage, United Kingdom). The compounds were dissolved in sterile DNase/RNase-free water, and 10 mM aliquots were stored at −20°C until used.
Viruses and cells.
A/New Caledonia/20/99 (NewCal) and A/Brisbane/59/07 (Brisbane) seasonal H1N1 influenza viruses were kindly provided from Ian Barr (WHO Collaborating Centre for Reference and Research on Influenza, Australia); the A(H1N1)pdm09 A/California/04/09 (CA04) virus was provided by Robert G. Webster at St. Jude Children's Research Hospital. Madin-Darby canine kidney (MDCK) cells and human embryonic kidney 293T cells were obtained from the American Type Culture Collection and were maintained in minimum essential medium (MEM) with 10% fetal calf serum (FCS) and Opti-MEM (Invitrogen) supplemented with 5% FCS, respectively. MDCK-SIAT1 cells, which overexpress human α-2,6-sialyltransferase, were kindly provided by Mikhail N. Matrosovich (Philipps University, Marburg, Germany) and were maintained as previously described (43). Differentiated NHBE cells derived from human donors were generated as described previously (8). Briefly, NHBE cells (Lonza) were expanded in tissue culture flasks no more than three times before seeding onto human collagen IV-coated transwell inserts in 24-well plates to allow differentiation in an air-liquid interface for at least 28 days in an incubator (5% CO2, 37°C) with BEBM medium (Lonza) supplemented with retinoic acid as previously described (8). The BEBM medium from the basolateral compartment was changed every other day. Human bronchial epithelial cells (HBEC) were isolated from patients who had undergone an operation at Queen Mary Hospital, Hong Kong; HBEC were isolated as described previously (17) and differentiated under the same conditions as NHBE cells.
Generation of recombinant viruses.
The full genome of CA04 and Brisbane viruses as well as the HA and NA genes of NewCal virus were amplified by one-step reverse transcription-PCR (RT-PCR; Qiagen) and cloned into the pHW2000 plasmid as described previously (28). The H275Y mutation was introduced into the plasmid carrying the NA gene for all three viruses by site-directed mutagenesis (Agilent Technologies). Recombinant viruses in the CA04 genetic background were generated by transfecting human embryonic kidney 293T cells (TransIT-LT1; Mirus) with eight plasmids. Cocultured MDCK and 293T cells were used to rescue the Brisbane and NewCal viruses due to low viral yields when we used 293T cells alone. The rescued viruses were then propagated in MDCK cells for two or three passages at a multiplicity of infection (MOI) of 0.001 PFU/cell to prepare stocks. The HA and NA of the recombinant viruses were verified.
Infectivity and replication kinetics.
The 50% tissue culture infectious dose (TCID50) was determined by incubating serial half-log dilutions of virus in MDCK cells at 37°C for 72 h. A hemagglutination assay was performed to determine the endpoint of infection, and the TCID50 was calculated by the Reed-Muench method (57). A plaque assay was performed by incubating serial dilutions of virus in confluent MDCK cells seeded onto 6-well plates. After 1 h of incubation at 37°C, inocula were removed and overlaid with infection medium (MEM with 0.3% bovine serum albumin) containing 0.4% agarose. Plaques were visualized after incubation at 37°C for 3 days by overnight fixation with 4% formaldehyde in phosphate-buffered saline (PBS) followed by staining with 0.2% crystal violet. Replication kinetics in MDCK or MDCK-SIAT1 cells was determined in triplicate wells at an MOI of 0.001 PFU/cell. After incubation, the cells were washed and overlaid with infection medium. Supernatants were collected at 8, 12, 24, 36, 48, and 60 h postinfection and stored at −80°C prior to virus titration. Replication kinetics in differentiated NHBE and HBEC was determined at an MOI of 0.01 PFU/cell. To test the potential effect of mucin at the initial stage of infection, NHBE cells were divided into two groups, either with or without extensive PBS washings prior to infection. Cells were infected via the apical side with 200 μl of diluted virus. After 1 h of incubation at 37°C, viral inoculum was removed; both apical and basolateral surfaces of the transwell were washed once with PBS before replenishment with fresh medium and incubation at 37°C. Virus was collected at 24, 48, 72, and 96 h postinfection by adding 200 μl medium onto the apical surface followed by equilibration for 15 to 20 min at 37°C, samples were stored at −80°C for later titration.
Competitive growth of RG-CA04 and RG-CA04NA-H275Y in differentiated NHBE and in MDCK-SIAT1 cells.
RG-CA04 and its mutant counterpart RG-CA04NA-H275Y were mixed into four different ratios to infect prewashed differentiated NHBE cells in duplicate transwells at an MOI of 0.01 PFU/cell. Total RNA was extracted from the inoculum (0 h postinfection) and viruses were collected from differentiated NHBE cells at 24, 48, and 72 h postinfection (RNeasy; Qiagen). A 475-bp PCR product was amplified (One-step RT-PCR; Qiagen) using specific primers (forward, 5′-AGAACTCAAGAGTCTGAATGTG-3′; reverse, 5′-TCCCAGTCCATCCGTTCG-3′). The RT-PCR product was cloned into the pCR4-TOPO vector (Invitrogen). Plasmid DNA was isolated (Miniprep; Qiagen), and a total of 20 to 40 clones were sequenced using the T3 primer, covering the nucleotide encoding residue 275.
To test the inhibitory effects of oseltamivir in a coinfection environment, competitive growth was repeated in the presence or absence of oseltamivir carboxylate by using MDCK-SIAT1 cells at an MOI of 0.001 PFU/cell. Sensitivity of RG-CA04 virus to oseltamivir carboxylate was predetermined in MDCK-SIAT1 by using serial 3.16-fold dilutions of oseltamivir carboxylate (the final concentration ranged from 0 to 100 μM in triplicate samples). The half-maximal effective concentration (EC50) was determined by plotting virus titer as a function of compound concentration, followed by variable-slope dose-response curve fitting using GraphPad Prism software. We performed the competition assay in MDCK-SIAT1 cells under two conditions: in the absence or presence of 0.2 μM oseltamivir carboxylate, which is approximately at the EC40, according to the dose-response curve for the RG-CA04 virus. Prior to infection, MDCK-SIAT1 cells were preincubated with medium or with medium containing 0.2 μM oseltamivir carboxylate for 1 h at 37°C. After 1 h, cells were infected using RG-CA04 and RG-CA04NA-H275Y viruses that were mixed into five different ratios at an MOI of 0.001 PFU/cell. Total RNA was extracted from the inoculum (0 h postinfection) and from viruses collected at 24 and 48 h postinfection. TOPO clonal sequencing was repeated using the same methods described above.
NA inhibition assay and NA kinetics.
NA inhibition was assayed with viruses standardized to an equivalent NA enzyme activity. Viruses were first incubated with oseltamivir carboxylate or zanamivir at concentrations of 0 to 100 μM at 37°C for 30 min, after which the mixtures were further incubated with the fluorogenic substrate 2′-(4-methylumbelliferryl)-α-d-N-acetylneuraminic acid (MUNANA) at a final concentration of 167 μM (22, 53) at 37°C for 30 min. The reaction was stopped by adding 150 μl stop solution (0.014 M NaCl, 0.1 M glycine in 25% ethanol; pH 10.7). The half-maximal inhibitory concentration (IC50) for each virus was determined by plotting fluorescence as a function of compound concentration, followed by variable-slope dose-response curve fitting using GraphPad Prism software. NA kinetics with MUNANA were determined at final concentrations of 0 to 600 μM at 37°C in a total volume of 50 μl with viruses diluted to approximately 5,000 PFU/ml. The fluorescence of the released 4-methylumbelliferone was measured every 68 s for 68 min by using FLUOstar Optima (BMG Labtech), with excitation and emission wavelengths of 355 and 460 nm, respectively. NA kinetics using sialosides 3′-sialyllactose (3′SL) and 6′-sialyllactose (6′SL) as the substrates (Carbosynth, United Kingdom) were determined in a coupled enzyme assay. Viruses (standardized to approximately 1.74 × 106 PFU/ml) were incubated with 3′SL (final concentration, 0 to 4,500 μM) or 6′SL (final concentration, 0 to 7,500 μM) at pH 6.0 with 0.2 M phosphate buffer, excess galactose oxidase (Sigma-Aldrich), horseradish peroxidase (Invitrogen), and Amplex UltraRed reagent (Invitrogen). The reaction was conducted at 37°C, and the fluorogenic product was measured every 5 min for 4 to 6 h using FLUOstar Optima (BMG Labtech), with excitation and emission wavelengths of 530 and 590 nm, respectively. The enzyme kinetics data were fitted to the Michaelis-Menten equation by using nonlinear regression (GraphPad Prism) to determine the Michaelis constant (Km) and maximum velocity (Vmax) of substrate conversion.
Transmission experiments in ferrets.
Transmissibility was tested in 4- to 6-month-old male ferrets obtained from Southland (New Zealand) or Sangosho (China). All studies were conducted in a biosafety level 3+ facility at the LKS Faculty of Medicine, the University of Hong Kong, in compliance with all applicable laws and with ethics approval from the Committee on the Use of Live Animals in Teaching and Research (CULATR) at the University of Hong Kong. Ferrets used in the study were confirmed to be seronegative against influenza virus A NP protein by using the ID Screen influenza virus A antibody competition enzyme-linked immunosorbent assay kit (ID.vet) and had a hemagglutination inhibition (HI) titer of ≤20 against pandemic influenza H1N1 virus (A/CA/07/2009) and influenza B virus (B/Brisbane/60/08). Virus transmission experiments used groups of two inoculated ferrets, two naïve direct contact ferrets, and two naïve respiratory droplet contact ferrets. Donors were inoculated intranasally with 104 PFU of recombinant virus in 0.5 ml medium under isoflurane anesthesia. All recombinant viruses used were stock viruses after two passages in MDCK cells, except for RG-CA04, which were passaged three times in MDCK cells. After 1 day, a naïve direct contact ferret was placed in the same cage with each inoculated ferret, and one naïve respiratory droplet contact ferret was placed in an adjacent compartment of the cage, separated by a single-layered perforated divider. To monitor virus shedding, nasal washes were collected from all ferrets every other day for 14 days and titrated in MDCK cells. The area under the curve (AUC) was calculated for each ferret as an approximation of the cumulative amount of virus shed in the nasal washes over time by using GraphPad Prism software. Ferret body temperatures and clinical signs were monitored daily, and weights were recorded every other day. Naïve respiratory droplet contact ferrets were handled before naïve direct contact ferrets, and inoculated ferrets were handled last; hands and equipment were disinfected before and after handling each ferret. Separate gloves and tools were used for respiratory droplet contact ferrets.
RESULTS
Generation and characterization of A(H1N1)pdm09 H1N1 recombinant viruses encoding different NA genes.
Three pairs of recombinant CA04 viruses that contain NA genes from three selected strains were generated, with each pair differing by the H275Y NA mutation alone: RG-CA04, RG-CA04NA-H275Y, RG-CA04 × BrisbaneNA, RG-CA04 × BrisbaneNA-H275Y, RG-CA04 × NewCalNA, and RG-CA04 × NewCalNA-H275Y. Recombinant viruses were generated via the eight-plasmid system in 293T cells as described previously (28); virus stocks were prepared in MDCK cells after two to three passages at an MOI of 0.001 PFU/cell. The susceptibilities of the recombinant viruses to zanamivir and oseltamivir carboxylate were determined in enzyme-based NA inhibition assays. We observed that the H275Y NA mutation led to an over-1,000-fold increase in oseltamivir carboxylate IC50s, with RG-CA04 × BrisbaneNA-H275Y exhibiting the greatest with respect to its wild-type counterpart. All viruses remained sensitive toward zanamivir (Table 1).
Table 1.
Growth and sensitivities of the recombinant viruses to NA inhibitors
| Virus | Virus yielda (log10 PFU/ml) | Sensitivity to NA inhibitorb |
|||
|---|---|---|---|---|---|
| Oseltamivir carboxylate |
Zanamivir |
||||
| IC50 (nM) | Fold change | IC50 (nM) | Fold change | ||
| RG-CA04 | 6.7 ± 0.2 | 0.8 ± 0.2 | 1.0 | 0.9 ± 0.2 | 1.0 |
| RG-CA04NA-H275Y | 7.1 ± 0.4 | 1,205.5 ± 16.3* | 1,506.9 | 1.1 ± 0.2 | 1.2 |
| RG-CA04 × NewCalNA | 7.3 ± 0.4 | 1.7 ± 0.2 | 1.0 | 1.3 ± 0.3 | 1.0 |
| RG-CA04 × NewCalNA-H275Y | 7.3 ± 0.3 | 2,365.5 ± 143.5* | 1,391.5 | 2.5 ± 0.5 | 1.9 |
| RG-CA04 × BrisbaneNA | 7.3 ± 0.4 | 1.1 ± 0.2 | 1.0 | 0.9 ± 0.2 | 1.0 |
| RG-CA04 × BrisbaneNA-H275Y | 7.3 ± 0.3 | 2,060.0 ± 216.4* | 1,872.7 | 1.3 ± 0.1 | 1.4 |
Viral titers are means ± standard deviations derived from 2 to 3 independent experiments.
Viral sensitivities to NA inhibitors were determined in NA inhibition assays. The values were fit to nonlinear regression curves with the variable slope model to determine the IC50, using GraphPad Prism. Results are means ± standard deviations derived from 2 to 3 independent experiments. IC50 fold changes were calculated relative to the counterpart wild-type virus.
, P < 0.05 compared to the counterpart wild-type virus.
All recombinant viruses grew to comparable titers, suggesting that the H275Y NA mutation did not significantly affect the growth of the virus in MDCK cells (Table 1). Furthermore, the comparable growth between RG-CA04 and RG-CA04 × BrisbaneNA or RG-CA04 × NewCalNA suggested that the HA/NA balance was not significantly impaired when we used these particular HA/NA combinations.
The HAs and NAs of the stock recombinant viruses were sequenced to ensure their identities to the field strains; the introduced H275Y NA mutation was confirmed to be maintained in the recombinant viruses after two to three passages in MDCK cells. Heterogeneous plaques were noticed with increased passage of the recombinant viruses in MDCK cells. To explore whether these heterogeneous plaque phenotypes were related to possible mutations occurring in HA and NA gene sequences, the RG-CA04 and RG-CA04NA-H275Y viruses were subjected to three rounds of plaque purification in triplicates. The clinically derived isolate A/California/04/2009, which was grown in MDCK for five passages, also exhibited heterogeneous plaque morphology and was included for analysis. Viral RNA was extracted from the third round of plaque expansion, and the HA and NA genes were amplified using one-step RT-PCR and analyzed by Sanger sequencing. After three rounds of plaque isolation and expansion in MDCK cells, 8 of the 18 viruses did not retain homogeneity in plaque size (Table 2). Assessment of the 10 homogeneous phenotypes showed amino acid changes in the HA or NA, although we did not observe any common mutation that could be associated with the observed large or small plaque morphology for the recombinant RG-CA04 or RG-CA04NA-H275Y viruses (Table 2). In contrast, the G155E (H1 numbering) HA mutation was observed consistently from the large plaques isolated from the clinically derived CA04 virus in all triplicates (Table 2). This HA mutation has been previously reported for the A(H1N1)pdm09 virus as one of the mutations contributing to increased virulence after adaptation in mice (34). The S183P mutation was detected in both small and large plaques for the clinically derived isolate and has also been linked with increased virulence in mice (70). Overall, we observed heterogeneity in plaque morphology of the recombinant A(H1N1)pdm09 virus after passaging in MDCK cells; while sporadic mutations were found in HA or NA after three rounds of plaque purification in MDCK cells, the consensus sequence remained identical to the field strain.
Table 2.
Amino acid changes identified in heterogeneous plaques
| Virus | MDCK passage history | Replicate | Plaque population and phenotype after expansion | Mean plaque diama | Amino acid changeb |
|
|---|---|---|---|---|---|---|
| HA | NA | |||||
| A/California/04/2009 | C5 | Small-1 | Medium | 1.04 ± 0.26 | T197A | Nil |
| Small-2 | Small | 0.83 ± 0.21 | S183P | Nil | ||
| Small-3 | Small | 0.68 ± 0.20 | L191I | Nil | ||
| Large-1 | Mixed, medium-large | — | G155E, L191I | K390R | ||
| Large-2 | Large | 2.81 ± 0.46 | G155E, S183P | Nil | ||
| Large-3 | Large | 2.94 ± 0.18 | G155E, S183P | Nil | ||
| RG-CA04 | C2 | Small-1 | Small | 0.89 ± 0.20 | Nil | V94I |
| Small-2 | Small | 0.84 ± 0.31 | Nil | Nil | ||
| Small-3 | Small | 0.64 ± 0.21 | Nil | Nil | ||
| Large-1 | Large | 2.44 ± 0.50 | K153Q | Nil | ||
| Large-2 | Medium | 1.70 ± 0.28 | D127E | Nil | ||
| Large-3 | Mixed, small-large | — | G155E | Nil | ||
| RG-CA04NA-H275Y | C2 | Small-1 | Mixed, small-large | — | Nil | Nil |
| Small-2 | Small | 0.84 ± 0.17 | Nil | Nil | ||
| Small-3 | Mixed, small-large | — | Nil | S31L | ||
| Large-1 | Mixed, medium-large | — | F390L | Nil | ||
| Large-2 | Mixed, medium-large | — | F390L | Nil | ||
| Large-3 | Large | 2.35 ± 0.47 | N156D | Nil | ||
Plaque diameters (means ± standard deviations, in mm) were determined from 5 randomly selected plaques. —, for mixed populations, mean plaque diameters were not determined.
Amino acid changes identified in HA and NA of heterogeneous plaques were obtained after three rounds of plaque purification in MDCK cells. After the third round of plaque purification, RNA was isolated for RT-PCR and sequence analysis; Nil, identical to the field strain.
NA enzyme kinetics revealed that the H275Y NA mutation leads to differential NA functional loss in different NA backgrounds.
To evaluate the effect of the H275Y framework mutation on NA enzyme activity, kinetics assays were performed using the fluorogenic substrate MUNANA or 3′SL and 6′SL with standardized viral doses based on the PFU. Viruses carrying the H275Y mutation showed lower Vmax and higher Km values (except RG-CA04NA-H275Y in catalyzing 3′SL) compared to their wild-type counterparts (Table 3), suggesting that the H275Y mutation generally led to decreased NA activity and a reduced affinity for substrates.
Table 3.
NA enzyme kinetics with MUNANA, 3′SL, and 6′SL substrates
| Virus | NA enzyme kinetics (95% CI) with substratea |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| MUNANA |
3′SL |
6′SL |
|||||||
| Km (μM) | Vmax [(excitation/emission at 355/460 nm)/s] | Vmax ratiob | Km (μM) | Vmax [(excitation/emission at 530/590 nm)/min] | Vmax ratiob | Km (μM) | Vmax [(excitation/emission at 530/590 nm)/min] | Vmax ratiob | |
| RG-CA04 | 26.5 (8.2–44.7) | 4.0 (3.3–4.6) | 1.0 | 622.7 (428.1–817.4) | 101.0 (91.5–110.5) | 1.0 | 3,469 (458.5–6,480) | 68.2 (40.0–96.6) | 1.0 |
| RG-CA04NA-H275Y | 61.9 (35.4–88.5) | 3.5 (3.1–3.9) | 0.9 | 905.3 (657.1–1,154) | 105.8 (96.8–115.8) | 1.1 | 4,224 (2,176–6,272) | 57.8 (43.5–72.2) | 0.8 |
| RG-CA04 × NewCalNA | 42.3 (25.3–59.3) | 4.3 (3.8–4.7) | 1.0 | 741.5 (434.2–1,049) | 111.3 (96.5–126.1) | 1.0 | 2,355 (1,529–3,181) | 84.5 (72.2–96.9) | 1.0 |
| RG-CA04 × NewCalNA-H275Y | 84.5 (41.2–127.9) | 2.4 (2.1–2.8)* | 0.6 | 1,370.0 (178.5–2,561) | 69.5 (45.1–93.95)* | 0.6 | 3,987 (1,586–6,389) | 57.2 (39.9–74.6) | 0.7 |
| RG-CA04 × BrisbaneNA | 14.0 (3.6–24.4)§ | 6.8 (5.7–7.9) | 1 | 454.2 (143.6–764.8) | 152.0 (124.0–180.1) | 1.0 | 2,108 (1,425–2,790) | 105.6 (92.0–119.3) | 1.0 |
| RG-CA04 × BrisbaneNA-H275Y | 26.4 (13.5–39.3) | 4.3 (3.8–4.8)* | 0.6 | 698.9 (400.0–997.8) | 97.5 (84.4–110.6)* | 0.6 | 2,462 (1,646–3,278) | 67.8 (58.3–77.3)* | 0.6 |
Standardized virus doses were used for the NA kinetics assay. The enzyme kinetics data were fit to the Michaelis-Menten equation by nonlinear regression to determine the Michaelis constant (Km) and maximum velocity (Vmax) of substrate conversion by using GraphPad Prism. Results are means from 2 independent experiments. *, P < 0.05 compared to the counterpart wild-type virus; §, P < 0.05 compared to the RG-CA04 × NewCalNA virus.
The Vmax ratio of the value for the H275Y variant relative to that of its cognate wild-type NA is shown.
When we applied the fluorogenic MUNANA substrate for enzyme kinetics, we observed that the H275Y mutation significantly reduced the NewCal and Brisbane NA activities by 40% (P < 0.05), while only a minor reduction (10%) was observed for CA04 NA activity (Table 3, Vmax and Vmax ratio). In addition, the H275Y mutation increased the Km for CA04, NewCal, and Brisbane NAs (Table 3). It was noted that the Km of RG-CA04 × BrisbaneNA was significantly lower (14 μM) than that of RG-CA04 × NewCalNA (42.3 μM; P < 0.05) or RG-CA04 (26.5 μM). The Km for RG-CA04 × BrisbaneNA-H275Y (26.4 μM) was more comparable to that of RG-CA04 × NewCalNA or RG-CA04 viruses. This observation is consistent with a previous report by Rameix-Welti et al., who compared the enzymatic properties of seasonal H1N1 influenza viruses (54).
A similar trend was observed when applying 3′SL or 6′SL as the substrates for enzyme kinetics; the H275Y mutation significantly reduced NA activity only for NewCal NA (3′SL) and Brisbane NA (3′SL and 6′SL) (P < 0.05). Interestingly, the RG-CA04NA-H275Y virus did not show a decreased Vmax compared to RG-CA04 while catalyzing 3′SL, although an ∼20% reduction in activity was observed for 6′SL. Using 3′SL as the substrate, we observed again that the Km values of RG-CA04 × NewCalNA and RG-CA04 closely resembled that of RG-CA04 × BrisbaneNA-H275Y and were higher than that of RG-CA04 × BrisbaneNA (Table 3). Using 6′SL as the substrate, the Km values for seasonal N1 derived from NewCal or Brisbane (2,355 and 2,108 μM, respectively) were observed to be lower than that of the A(H1N1)pdm09 N1 (3,469 μM), suggesting that the NAs of seasonal H1N1 viruses were more efficient in catalyzing 6′SL than that of the A(H1N1)pdm09 NA. Overall, we observed that the H275Y mutation may have led to decreased NA activity and consistently led to increased Km values for NAs derived from seasonal or A(H1N1)pdm09 H1N1 influenza viruses.
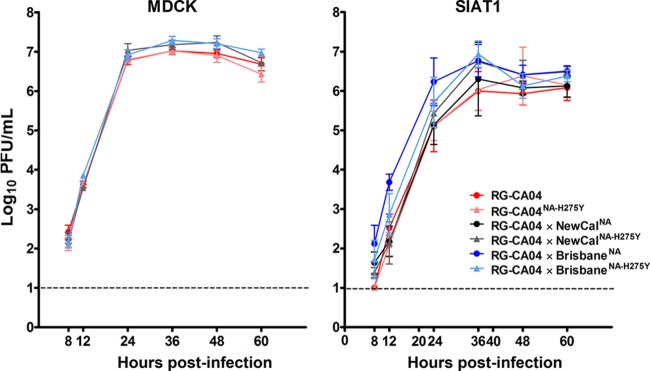
Replication kinetics in MDCK cells and the MDCK-SIAT1 cell line yielded comparable virus replication rates between wild-type and corresponding H275Y mutants.
To assess if the H275Y NA mutation would compromise viral growth in vitro, multicycle replication kinetics of the recombinant viruses were analyzed in MDCK and MDCK-SIAT1 cells (stably transfected with α-2,6-sialyl transferase) (43) at an MOI of 0.001 PFU/cell. Our data suggest no detectable growth advantage for RG-CA04 over RG-CA04NA-H275Y in either MDCK or MDCK-SIAT1 cells (P < 0.05) (Fig. 1). Furthermore, comparable growth was observed between RG-CA04 × NewCalNA and RG-CA04 × NewCalNA-H275Y viruses or between RG-CA04 × BrisbaneNA and RG-CA04 × BrisbaneNA-H275Y viruses in SIAT1 cells (Fig. 1). RG-CA04 × BrisbaneNA showed a marginally higher titer at 12 h postinfection in MDCK-SIAT1 cells to other viruses, except for RG-CA04 × BrisbaneNA-H275Y. Overall, the H275Y mutation in different N1-derived NAs did not lead to compromised growth efficiencies in conventional cell lines.
Fig 1.
Replication kinetics of the recombinant CA04 viruses in MDCK and MDCK-SIAT1 cells. Multicycle replication curves were determined at an MOI of 0.001 PFU/cell in MDCK and MDCK-SIAT1 cells with triplicated wells. Viruses were collected at the specified time points, and mean (± standard deviation) log10 values are shown. The detection limit was 10 PFU/ml. *, P < 0.05.
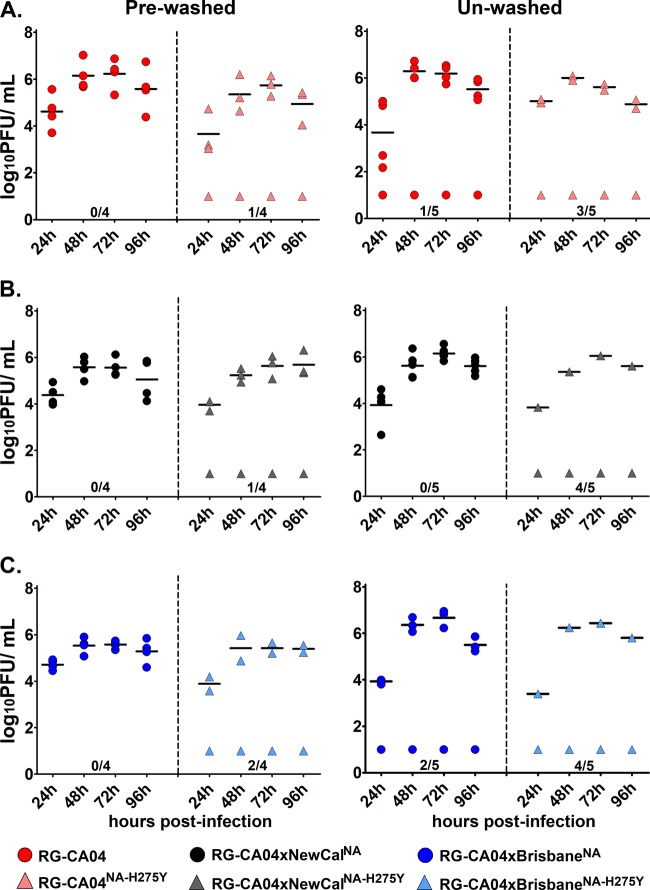
Recombinant viruses carrying the H275Y mutation were compromised in establishing infection in mucin-secreting differentiated NHBE cells.
We continued to examine the replication efficiency of the recombinant viruses in differentiated NHBE cells, which were previously shown to express high levels of α-2,6-linked sialic acids, human airway trypsin-like protease, as well as ciliated and nonciliated mucin-secreting goblet cells (8). Mucin is known to contain high level of O-linked glycans and plays an important role in host innate defense (25, 29, 36, 65). Collectively, these properties may provide a more physiological relevant model in assessing NA inhibitor-resistant viruses.
To evaluate the infectivity of the recombinant viruses with or without the H275Y mutation in differentiated NHBE cells, the cells derived from two different donors (Lonza, Switzerland) were subjected to extensive washing with PBS (cells derived from donor 1) or no washing (donors 1 and 2) prior to infection. Using cells from the prewashed protocol, all three wild-type viruses (RG-CA04, RG-CA04 × NewCalNA, and RG-CA04 × BrisbaneNA) were able to establish infection in 4/4 replicates, with viral titers peaking at approximately 72 h postinfection (Fig. 2). The recombinant viruses carrying the H275Y mutation, RG-CA04NA-H275Y, RG-CA04 × NewCalNA-H275Y, and RG-CA04 × BrisbaneNA-H275Y, successfully established infection in prewashed cells in 3/4, 3/4, and 2/4 replicates, respectively (Fig. 2). Comparable viral titers was observed between the replicates successfully infected by the wild-type and the H275Y counterparts, suggesting comparable replication kinetics once the infection was established. Among the transwells that were not successfully infected, no detectable plaques were found across all time points (detection limit, 10 PFU). In differentiated NHBE cells that were left unwashed in order to maintain the mucin layer intact prior to infection, we observed that recombinant viruses carrying the NA H275Y mutation showed a reduced ability to successfully initiate infection. For wild-type viruses, we observed that RG-CA04, RG-CA04 × NewCalNA, and RG-CA04 × BrisbaneNA could establish infection in 4/5, 5/5, and 3/5 replicates of the unwashed differentiated NHBE cells, respectively (Fig. 2). For the viruses carrying the H275Y NA mutation, RG-CA04NA-H275Y, RG-CA04 × NewCaledoniaNA-H275Y, and RG-CA04 × BrisbaneNA-H275Y could only establish infection in 2/5, 1/5, and 1/5 replicates of the unwashed differentiated NHBE cells, respectively. Due to the small sample size, a statistically significant difference was only observed between the RG-CA04 × NewCalNA and CA04 × NewCaledoniaNA-H275Y viruses (Fig. 2C) (P = 0.0476; Fisher's exact test). Overall, by using mucin-secreting differentiated NHBE cells (derived from two different donors) as an alternative in vitro model, we observed that recombinant CA04 viruses carrying the H275Y mutation led to a consistently reduced ability to establish infection in the unwashed cells, regardless if the NA originated from A(H1N1)pdm09 or seasonal H1N1 viruses.
Fig 2.
Replication efficiency of the recombinant CA04 viruses in differentiated NHBE cells. The abilities of the recombinant viruses to replicate in prewashed or unwashed differentiated NHBE cells (derived from two donors) prior to infection were evaluated. Replication efficiencies are shown between RG-CA04 and RG-CA04NA-H275Y (A), RG-CA04 × NewCalNA and RG-CA04 × NewCalNA-H275Y (B), and RG-CA04 × BrisbaneNA and RG-CA04 × BrisbaneNA-H275Y. Each data point represents the titer (in log10 PFU/ml) from one transwell collected at the specified time points. Mean titer values at each time point from the transwells with positive viral replication are denoted by horizontal lines.
Well-differentiated HBEC derived from patients who had undergone lung resections were also applied to compare the infectivities of the recombinant seasonal influenza viruses RG-Brisbane and RG-BrisbaneNA-H275Y. In parallel, we included the infection of RG-CA04 × BrisbaneNA and RG-CA04 × BrisbaneNA-H275Y viruses for comparison. These cells have been characterized to exhibit properties similar to differentiated NHBE cells (R. W. Y. Chan and M. C. W. Chan, unpublished data) and also secrete mucin. Infection experiments were performed on unwashed differentiated HBEC derived from one donor. Results showed that all viruses could establish infection in HBEC: RG-Brisbane (2/2 replicates), RG-BrisbaneNA-H275Y (3/3 replicates), RG-CA04 × BrisbaneNA (2/2 replicates), and RG-CA04 × BrisbaneNA-H275Y (3/3 replicates). It is not known if the amounts of mucin or the compositions of the mucin secreted from differentiated NHBE and HBEC were comparable. These results highlight the existence of donor-to-donor variability when using primary cells.
TOPO cloning for sequencing of coinfected RG-CA04 and RG-CA04H275Y viruses in vitro.
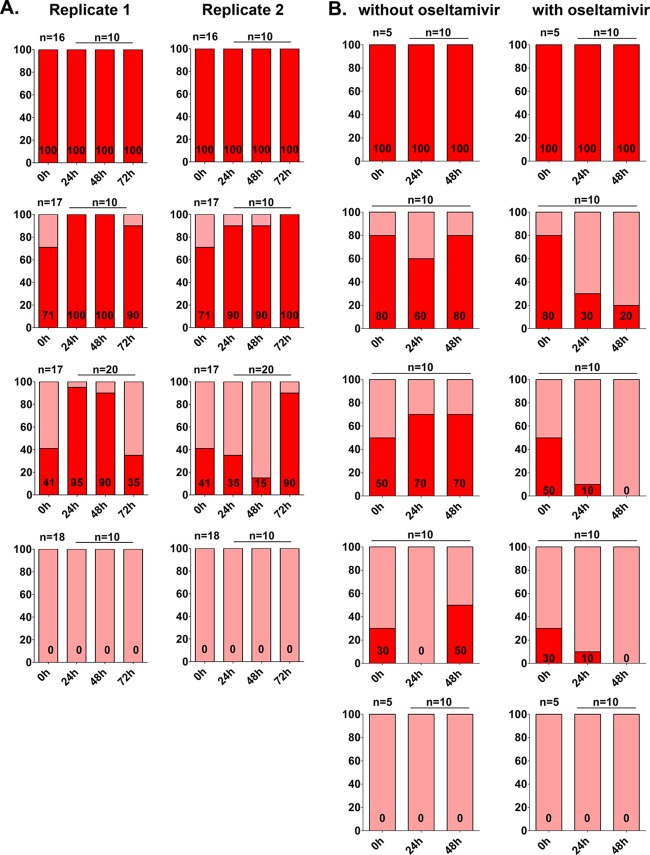
To understand if the A(H1N1)pdm09 virus possessed a survival advantage over its H275Y NA counterpart, RG-CA04 and RG-CA04NA-H275Y viruses were mixed at 100:0, 71:29, 41:59, and 0:100 ratios (verified by TOPO clonal sequencing) to coinfect the differentiated NHBE cells, which were washed extensively with PBS prior to infection. For each mixture, duplicate transwells were infected using an MOI of 0.01 PFU/cell, and viruses were collected from the apical surface every 24 h until peak titers were reached, at approximately 72 h postinfection.
At ratios of 100:0 and 0:100, we observed that both the RG-CA04 and RG-CA04NA-H275Y genotypes were stably maintained at 24, 48, and 72 h postinfection, suggesting the genetic stability of the H275Y NA mutation. With coinfection at a ratio of 71:29 (H275:Y275), we observed that RG-CA04 became dominant (100%) at 24 h (replicate 1) or 72 h (replicate 2) postinfection. For cells infected using the ratio of 41:59 (H275:Y275), we observed that RG-CA04 became dominant at 24 h (95%; replicate 1) or 72 h (90%; replicate 2) postinfection (Fig. 3A); however, we noticed an increase of RG-CA04NA-H275Y in replicate 1 at 72 h postinfection. Overall, our results suggest the genetic stability of the H275Y mutation in A(H1N1)pdm09 virus and a slightly higher survival advantage of RG-CA04 over RG-CA04NA-H275Y virus when coinfected at different ratios in prewashed differentiated NHBE cells.
Fig 3.
Competitive replication of RG-CA04 and RG-CA04NA-H275Y in vitro. RG-CA04 and RG-CA04NA-H275Y viruses were premixed at different ratios prior to infection of differentiated NHBE (A) or MDCK-SIAT1 (B) cells in the absence or presence of 0.2 μM oseltamivir carboxylate. TOPO clonal sequencing was performed to determine the ratio of RG-CA04 and RG-CA04NA-H275Y viruses at different times postinfection. NHBE cells in two separate transwells (replicates 1 and 2; both with no oseltamivir treatment) were washed with PBS prior to infection with mixtures of RG-CA04 and RG-CA04NA-H275Y viruses. The percentage of the oseltamivir-sensitive H275 genotype is shown in red, while the percentage of the oseltamivir-resistant Y275 genotype is shown in pink. Numbers of clones (n) sequenced at each time point are indicated in each panel.
We extended the competition experiment between the RG-CA04 and RG-CA04NA-H275Y in MDCK-SIAT1 cells and further evaluated the changes of the oseltamivir-sensitive and oseltamivir-resistant populations in the absence or presence of oseltamivir carboxylate. Susceptibility of RG-CA04 was first determined using increasing concentration of oseltamivir carboxylate (0 to 100 μM) in MDCK-SIAT1 cells. The EC50 of CA04 virus determined in MDCK-SIAT1 cells (0.76 μM, 95% confidence interval [CI], 0.28 to 2.05 μM) was found slightly higher than the EC50 previously reported in MDCK cells (0.18 μM) (51). Five different ratios (100:0, 80:20, 50:50, 30:70, and 0:100) of RG-CA04 and RG-CA04NA-H275Y viruses were then used for the coinfection experiment in the absence or presence of 0.2 μM oseltamivir carboxylate, which approximated the EC40 for RG-CA04 in MDCK-SIAT1 cells. For cells infected in the absence of oseltamivir carboxylate, a general trend of increasing RG-CA04 genotype over time was observed (with the exception of cells infected at 80:20 and 30:70 ratios at 24 h postinfection) (Fig. 3B); however, the H275 did not completely dominate over the Y275, even with a premixed ratio of 80:20 (H:Y) at 48 h postinfection. For cells infected in the presence of oseltamivir carboxylate, a progressive decrease in the proportion of RG-CA04 was observed across all three mixtures (80:20, 50:50, and 30:70) over time; RG-CA04 was not detectable at 48 h postinfection for cells initially infected with 50:50 and 30:70 ratios (Fig. 3B). In summary, our results suggest that RG-CA04 possesses a minor survival advantage over RG-CA04NA-H275Y in the absence of oseltamivir carboxylate in both NHBE and MDCK-SIAT1 cells. Coinfection with H275 and Y275 genotypes in the presence of oseltamivir carboxylate resulted in a gradual depletion of the oseltamivir-sensitive H275 genotype over time.
Transmissibility of the recombinant A(H1N1)pdm09 viruses in ferrets.
The transmissibility of the recombinant CA04 viruses was evaluated in the ferret model. At an inoculation dose of 104 PFU, ferrets infected with RG-CA04 virus exhibited greater levels of weight loss (Fig. 4A) and were less active and playful compared to the ferrets inoculated with RG-CA04NA-H275Y (Fig. 4B) or RG-CA04 × BrisbaneNA-H275Y (Fig. 4C) viruses. One of the two ferrets inoculated with RG-CA04 virus died on day 8 postinoculation, while all ferrets survived in the RG-CA04NA-H275Y and RG-CA04 × BrisbaneNA-H275Y groups. Transmission from inoculated ferrets to naïve direct contact and respiratory droplet contact ferrets was observed for all three recombinant CA04 viruses at day 4 postinoculation (day 3 postcontact) (Fig. 4). The AUC approximating the total amount of virus shed during the course of infection was calculated for each animal; while the donor or the direct contact ferrets infected by RG-CA04NA-H275Y consistently showed lower AUC values, the differences were not statistically significant, due to small sample sizes (Table 4). Overall, with a small sample size, we observed that RG-CA04NA-H275Y or RG-CA04 × BrisbaneNA-H275Y viruses possessed attenuated pathogenicities but retained comparable transmission efficiencies to the RG-CA04 virus.
Fig 4.
Transmission of RG-CA04, RG-CA04NA-H275Y, and RG-CA04 × BrisbaneNA-275Y viruses in ferrets. The direct contact and respiratory droplet contact transmissibility of RG-CA04 (A), RG-RG-CA04NA-H275Y (B), and RG-CA04 × BrisbaneNA-H275Y (C) viruses are shown. Ferret weight changes (% of weight change compared to the weight at day 0; mean ± standard deviation) are also shown. Viral titers (in log10 TCID50/ml) were obtained from the nasal washes of individual donor ferrets (yellow), direct contacts (green), and respiratory droplet contacts (blue). The detection limit was 102.2 TCID50/ml. One donor animal inoculated with RG-CA04 virus was found dead in the cage at day 8 postinoculation.
Table 4.
AUCs for ferret nasal washes
| Virus | AUCa (mean ± SD) |
||
|---|---|---|---|
| Donor | Direct contact | Respiratory droplet contact | |
| RG-CA04 C3 | 6.9 ± 2.4 | 8.6 ± 5.3 | 4.9 ± 1.0 |
| RG-CA04NA-H275Y | 4.9 ± 0.8 | 2.9 ± 1.9 | 3.8 ± 1.1 |
| RG-CA04 × BrisbaneNA-H275Y | 6.1 ± 1.9 | 5.2 ± 1.7 | 5.4 ± 1.9 |
| RG-NewCalHA,NA | 9.3 ± 0.9 | 7.4 ± 2.3 | 8.7 ± 0.7 |
| RG-NewCalHA,NA-H275Y | 8.2 ± 1.0 | 8.4 ± 1.2 | 7.6 ± 3.1 |
| RG-NewCalHA × BrisbaneNA-H275Y | 10.1 ± 0.1 | 7.0 ± 2.7 | 8.9 ± 0.5 |
| RG-BrisbaneHA,NA-H275Y | 8.8 ± 1.2 | 8.8 ± 0.9 | 7.1 ± 2.9 |
The AUC represents total virus shedding (in log10 TCID50/ml) at 2, 4, 6, 8, 10, and 12 days postinoculation (1, 3, 5, 7, 9, and 11 days postcontact), as calculated from the nasal washes of each ferret. Means ± standard deviations of results from two donors, two direct contacts, or two respiratory droplet contact ferrets are shown. The detection limit was 102.2 TCID50/ml.
Transmissibility of the recombinant seasonal H1N1 viruses in ferrets.
The transmissibility of human seasonal H1N1 virus carrying the H275Y NA mutation has been previously studied in ferrets using strains circulating prior to 2007 (A/Texas/36/91 and A/New Caledonia/20/99) (27, 35) or after 2007 (A/Brisbane/59/2007) (33). While the H275Y mutation was shown to reduce the infectivity of seasonal H1N1 viruses isolated before 2007 in ferrets (27, 35), donor ferrets successfully infected with NewCal-like virus carrying the H275Y mutation were able to transmit the virus to naïve direct contact ferrets (27). The transmission potentials of the Brisbane-like viruses carrying the H275Y NA mutation were previously evaluated using a coinfection model in ferrets; the H275Y mutant was reported to possess comparable transmissibility, albeit being marginally outgrown by the wild-type virus (33). Given that these studies were performed under different experimental conditions, our ability to clearly compare results obtained previously is limited. Here, we aimed to perform a side-by-side comparison among seasonal H1N1 influenza viruses that circulated prior to and after 2007 under identical experimental settings. Since the HA and NA may confer to NA inhibitor resistance, and a balanced activity between the two surface glycoproteins is important for viral fitness, we focused on HA and NA derived from the NewCal and Brisbane viruses, of which the H275Y variants have shown different epidemiological outcomes. Recombinant viruses with identical internal genes derived from the Brisbane virus but differing in the surface glycoproteins were generated: RG-BrisbaneHA,NA, RG-BrisbaneHA,NA-H275Y, RG-NewCalHA,NA, RG-NewCalHA,NA-H275Y, RG-BrisbaneHA × NewCalNA-H275Y, and RG-NewCalHA × BrisbaneNA-H275Y. These viruses replicated to comparable titers in MDCK cells except for RG-BrisbaneHA,NA-H275Y, which replicated to a significantly lower titer compared to RG-BrisbaneHA,NA at 12 h postinfection in MDCK-SIAT1 cells (P < 0.05) (Fig. 5). The RG-BrisbaneHA × NewCal NA-H275Y virus that was observed to possess an additional N125Y HA mutation replicated to a lower titer in MDCK-SIAT1 cells (data not shown).
Fig 5.
Replication kinetics of the recombinant seasonal H1N1 viruses in MDCK and MDCK-SIAT1 cells. Multicycle replication curves were determined at an MOI of 0.001 PFU/cell in MDCK and MDCK-SIAT1 cells in triplicate wells. Viruses were collected at the specified time points, and the mean (± standard deviation) log10 values are shown. The detection limit was 10 PFU/ml. *, P < 0.05.
We continued to evaluate the transmissibility of four selected recombinant seasonal H1N1 viruses (RG-BrisbaneHA,NA-H275Y, RG-NewCalHA,NA, RG-NewCalHA,NA-H275Y, and RG-NewCalHA × BrisbaneNA-H275Y) in the ferret model. At the inoculation dose of 104 PFU, peak titers were detected at 2 days postinoculation from donor ferrets inoculated with RG-BrisbaneHA,NA-H275Y, RG-NewCalHA,NA, RG-NewCalHA,NA-H275Y, or RG-NewCalHA × BrisbaneNA-H275Y, with titers ranging from 6.4 to 8.3 log10TCID50/ml (Fig. 6). Transmission of all four viruses to naïve ferrets cohoused with donor ferrets was detected at 4 days postinoculation, with peak titers ranging from 5.7 to 7.3 log10TCID50/ml (Fig. 6). At 4 days postinoculation, transmission of RG-NewCalHA,NA and RG-NewCalHA × BrisbaneNA-H275Y to 2/2 naïve respiratory droplet contacts was detected, whereas for RG-NewCalHA,NA-H275Y and RG-BrisbaneHA,NA-H275Y, only 1/2 and 0/2 naïve contacts became infected, respectively. Transmission of RG-NewCalHA,NA-H275Y and RG-BrisbaneHA,NA-H275Y was then detected from all naïve respiratory droplet contacts at 6 days postinoculation (Fig. 6). By comparing the RG-NewCalHA,NA and RG-NewCalHA,NA-H275Y viruses, we observed that the H275Y NA mutation slightly decreased the respiratory droplet transmissibility but not the direct contact transmissibility; however, statistical analysis could not be performed due to the small sample size. The transmissibilities of the RG-NewCalHA,NA and RG-NewCalHA × BrisbaneNA-H275Y viruses were more comparable. Overall, we observed at the inoculation dose of 104 PFU/ferret, all seasonal H1N1 viruses carrying the H275Y NA mutation could transmit to ferrets under cohoused or respiratory droplet contact settings.
Fig 6.
Transmission of RG-NewCalHA,NA, RG-NewCalHA,NA-H275Y, RG-NewCalHA × BrisbaneNA-H275Y, and RG-BrisbaneHA,NA-H275Y viruses in ferrets. The direct contact and respiratory droplet contact transmissibilities of RG-NewCalHA,NA (A), RG-NewCalHA,NA-H275Y (B), RG-NewCalHA × BrisbaneNA-H275Y (C), and RG-BrisbaneHA,NA-H275Y (D) viruses are shown. Viral titers (in log10 TCID50/ml) were obtained from the nasal washes of individual donor ferrets (yellow), direct contacts (green), and respiratory droplet contacts (blue). The detection limit was 102.2 TCID50/ml.
DISCUSSION
The H275Y mutation that confers resistance to oseltamivir carboxylate has been detected from human infections of different influenza N1 subtypes, including seasonal H1N1 (11, 20, 31, 40, 49, 61, 67), highly pathogenic H5N1 (10, 41), and more recently, in A(H1N1)pdm09 viruses (18, 21, 30, 32, 63). Epidemiological data suggest that the fitness and the transmission potential of resistant variants carrying the H275Y NA mutation may vary between strains, with the observation of the unprecedented global spread of the Brisbane-like seasonal H1N1 virus during the 2008-2009 influenza season. While different in vitro or in vivo models (5, 13, 23, 27, 33, 35, 37, 48, 52, 60) were used to evaluate the fitness of viruses carrying the H275Y mutation, its effect on different N1 NA cannot be directly compared. The effect of this mutation in vitro has been previously addressed in a systematic manner by using different seasonal H1N1 influenza viruses (2), while in the present study we used three antigenically representative human seasonal strains, including the pandemic 2009 virus. Our results showed that the H275Y mutation consistently led to reduced NA enzyme function and a decreased ability to establish infection in mucin-secreting primary airway epithelial cells. Under coinfection conditions in NHBE and MDCK-SIAT1 cells, we also observed a minor survival advantage of RG-CA04 over RG-CA04NA-H275Y in the absence of oseltamivir carboxylate. Despite the detections of reduced viral competencies, both seasonal and A(H1N1)pdm09 viruses carrying the H275Y mutation were efficiently transmitted to naïve direct contact or respiratory droplet contact ferrets under the same experimental condition that a control swine influenza virus could only transmit to the naïve direct contact ferrets but not to the respiratory droplet contacts (data not shown). Our results suggest that the H275Y mutation in H1N1 influenza virus leads to a minor reduction in viral fitness, with its transmission potential being minimally affected in the ferret model.
Previous studies on NA inhibitor-resistant variants have shown that mutations within or surrounding the enzyme active site can impose different levels of NA functional loss (27, 71, 72). Our data are in accord with studies demonstrating that H275Y-carrying viruses have higher Km and lower Vmax values than their wild-type counterparts (5, 13, 52, 54, 73). When comparing the enzyme activities among the three different NA, we observed, using MUNANA or 3′SL substrates, that the Km value of RG-CA04 × BrisbaneNA-H275Y remained within close range to RG-CA04 and RG-CA04 × NewCalNA viruses, while the Km of RG-CA04 × BrisbaneNA consistently remained the lowest among all. Although we did not directly compare the HA binding efficiency in the presence of different NA, it is hypothesized that NA with a low Km for sialosides may interfere with the HA binding and may not be necessarily advantageous for a virus, especially during the initiation of infection. This finding has been previously reported in a study investigating the NA enzymatic properties of seasonal H1N1 virus (54) and also identified the D344N substitution among H1N1 viruses circulating in 2007-2008 as the major determinant for increased affinity to sialic acids (55). The uniqueness of Brisbane-like NA has been described in other studies, including the identification of secondary mutations (4) or epistatic mutations (39) that may restore the NA function upon acquisition of the H275Y NA mutation.
We observed that the Km values for NA derived from seasonal H1N1 viruses were lower than that of the A(H1N1)pdm09 virus when applying 6′SL substrate, suggesting the NA derived from seasonal viruses is more efficient in catalyzing 6′SL. The adaptation of the avian origin N2 NA in humans has been previously investigated, with the observation of increased activity in catalyzing 6′SL over time (38). As the A(H1N1)pdm09 virus acquired its NA gene from the “Eurasian” swine lineage (15, 62), which originally came from avian species and has been established in swine since the 1970s (12, 59), it is not clear if adaptation of avian origin viruses in swine may also lead to increased NA activity for 6′SL.
The use of mucin-secreting differentiated NHBE cells provides a physiologically relevant model to assess viruses with impaired NA activity, which viruses show little or no compromised growth in cell lines such as MDCK or MDCK-SIAT cells. Our results suggest mucin plays a protective role in innate defense; by using cells of the same donor, we showed that extensive PBS washings prior to infection rendered the cells more permissive to infection. Furthermore, we observed that recombinant viruses carrying the H275Y NA mutation, which have reduced NA activity, were less efficient in initiating infection than their wild-type counterparts in the unwashed NHBE cells, which presumably had a more intact mucin layer. It was also noted that once the infection was established, H275Y-carrying recombinant viruses could replicate to comparable titers as their wild-type counterparts, and this recapitulates the situation observed in conventional MDCK and MDCK-SIAT1 cell lines. Although we were not able to quantify the amount of mucin or other contents being secreted by the cells, these results support previous studies that influenza virus NA is functionally important for initiating infection in human airway epithelium (44).
We addressed the relative fitness of RG-CA04 and RG-CA04NA-H275Y by infecting prewashed differentiated NHBE cells and MDCK-SIAT1 cells with the two viruses mixed at different ratios. We observed that the H275Y mutation may have slightly reduced the survival advantage of the virus in the absence of oseltamivir carboxylate, albeit being genetically stable over time. A previous study performing coinfection of recombinant A(H1N1)pdm09 viruses carrying H275 or Y275 NA residues in human airway epithelial cells also observed enrichment of the H275 genotype after 72 h postinfection; however, no growth advantage was observed for either virus when the coinfection was performed in MDCK cells (5). In addition to testing the survival advantage of RG-CA04 and RG-CA04NA-H275Y in a competitive mixtures assay, the influence of oseltamivir carboxylate was also tested in MDCK-SIAT1 cells. As expected, the TOPO clonal sequencing showed that the proportion of the oseltamivir-sensitive H275 genotype progressively declined over time. More importantly, our results demonstrated the viable coexistence of oseltamivir-sensitive and -resistant populations while under partial oseltamivir pressure (exposure to the EC40 for sensitive virus).
The transmission results showed a robust data set, which all point toward the ability for H275Y-carrying viruses to transmit efficiently to naïve ferrets either under cohousing or respiratory droplet contact settings. It has been well established through epidemiological observations that the Brisbane-like viruses carrying the H275Y mutation are easily transmissible among humans, while only low-level transmission of the H275Y variants was reported prior to 2007-2008 (14, 64). However, our data and a previous report (33) did not find a unique transmission potential for the BrisbaneNA-H275Y virus in this animal model. In fact, we showed that RG-NewCalHA,NA and RG-NewCalHA,NA-H275Y could both transmit via direct contact and respiratory droplet routes, with RG-NewCalHA,NA-H275Y showing a slight delay in respiratory droplet transmissibility. While ferrets provide a naïve animal model for us to evaluate the transmission potential of the influenza viruses, the effect of exposure to prior circulating strains cannot be fully addressed. Oseltamivir-resistant Y275 variants circulating during the 2008-2009 season showed reduced antigenicity compared to the oseltamivir-sensitive strains (69). More recent research has also shown that the NA of Brisbane-like viruses has undergone a major drift in antigenicity from previously circulating seasonal H1N1 viruses (58).
In conclusion, we have shown that the H275Y NA mutation conferring oseltamivir resistance can lead to NA functional loss in A(H1N1)pdm09 and seasonal H1N1 viruses. The reduced NA activity may affect the ability of the viruses to establish infection in the human airway, in the presence of mucin. However, all viruses carrying the H275Y mutation were found to be able to transmit to naïve direct contact and respiratory droplet contact ferrets. Our results support the urgent need for the development of novel antiviral compounds or alternative treatment options for the control of influenza virus infections.
ACKNOWLEDGMENTS
This study was supported by the Research Fund for the Control of Infectious Disease (10090142), Food and Health Bureau, Hong Kong SAR; contract HHSN266200700005C from the U.S. National Institute of Allergy and Infectious Diseases, National Institutes of Health; Seed Funding for Basic Research, The University of Hong Kong; and the Area of Excellence Scheme of the University Grants Committee (AoE/M-12/06), Hong Kong SAR.
We thank Ian Barr for providing the seasonal H1N1 viruses; the Laboratory Animal Unit, the University of Hong Kong, for the support in animal experiments; Olive T. W. Li for providing the PB2, PA, and HA plasmids for the CA04 virus; and Edward S. K. Ma, Iris Ng, Kit-Man Yuen, Joanne Fong, Alan Li, Paul Wong, and Michael Liman for technical assistance.
Footnotes
Published ahead of print 18 July 2012
REFERENCES
- 1. Air GM, Laver WG. 1989. The neuraminidase of influenza virus. Proteins 6:341–356 [DOI] [PubMed] [Google Scholar]
- 2. Baz M, Abed Y, Simon P, Hamelin ME, Boivin G. 2010. Effect of the neuraminidase mutation H274Y conferring resistance to oseltamivir on the replicative capacity and virulence of old and recent human influenza A(H1N1) viruses. J. Infect. Dis. 201:740–745 [DOI] [PubMed] [Google Scholar]
- 3. Belshe RB, Smith MH, Hall CB, Betts R, Hay AJ. 1988. Genetic basis of resistance to rimantadine emerging during treatment of influenza virus infection. J. Virol. 62:1508–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bloom JD, Gong LI, Baltimore D. 2010. Permissive secondary mutations enable the evolution of influenza oseltamivir resistance. Science 328:1272–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brookes DW, Miah S, Lackenby A, Hartgroves L, Barclay WS. 2011. Pandemic H1N1 2009 influenza virus with the H275Y oseltamivir resistance neuraminidase mutation shows a small compromise in enzyme activity and viral fitness. J. Antimicrob. Chemother. 66:466–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Calatayud L, et al. 2011. Oseltamivir-resistant pandemic (H1N1) 2009 virus infection in England and Scotland, 2009–2010. Emerg. Infect. Dis. 17:1807–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. CDC 2010. Update: influenza activity—United States, 2009–10 season. Morb. Mortal. Wkly. Rep. 59:8. [PubMed] [Google Scholar]
- 8. Chan RW, et al. 2010. Influenza H5N1 and H1N1 virus replication and innate immune responses in bronchial epithelial cells are influenced by the state of differentiation. PLoS One 5:e8713 doi:10.1371/journal.pone.0008713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen LF, et al. 2011. Cluster of oseltamivir-resistant 2009 pandemic influenza A (H1N1) virus infections on a hospital ward among immunocompromised patients, North Carolina, 2009. J. Infect. Dis. 203:838–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de Jong MD, et al. 2005. Oseltamivir resistance during treatment of influenza A (H5N1) infection. N. Engl. J. Med. 353:2667–2672 [DOI] [PubMed] [Google Scholar]
- 11. Dharan NJ, et al. 2009. Infections with oseltamivir-resistant influenza A(H1N1) virus in the United States. JAMA 301:1034–1041 [DOI] [PubMed] [Google Scholar]
- 12. Donatelli I, et al. 1991. Detection of two antigenic subpopulations of A(H1N1) influenza viruses from pigs: antigenic drift or interspecies transmission? J. Med. Virol. 34:248–257 [DOI] [PubMed] [Google Scholar]
- 13. Duan S, et al. 2010. Oseltamivir-resistant pandemic H1N1/2009 influenza virus possesses lower transmissibility and fitness in ferrets. PLoS Pathog. 6:e1001022 doi:10.1371/journal.ppat.1001022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Escuret V, et al. 2008. Detection of human influenza A (H1N1) and B strains with reduced sensitivity to neuraminidase inhibitors. J. Clin. Virol. 41:25–28 [DOI] [PubMed] [Google Scholar]
- 15. Garten RJ, et al. 2009. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325:197–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ginting TE, et al. 2012. Amino acid changes in hemagglutinin contribute to the replication of oseltamivir-resistant H1N1 influenza viruses. J. Virol. 86:121–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Godding V, et al. 1998. Secretory component production by human bronchial epithelial cells is upregulated by interferon gamma. Eur. Respir. J. 11:1043–1052 [DOI] [PubMed] [Google Scholar]
- 18. Graitcer SB, et al. 2011. Characteristics of patients with oseltamivir-resistant pandemic (H1N1) 2009, United States. Emerg. Infect. Dis. 17:255–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gubareva LV. 2004. Molecular mechanisms of influenza virus resistance to neuraminidase inhibitors. Virus Res. 103:199–203 [DOI] [PubMed] [Google Scholar]
- 20. Gubareva LV, Kaiser L, Matrosovich MN, Soo-Hoo Y, Hayden FG. 2001. Selection of influenza virus mutants in experimentally infected volunteers treated with oseltamivir. J. Infect. Dis. 183:523–531 [DOI] [PubMed] [Google Scholar]
- 21. Gubareva LV, et al. 2010. Comprehensive assessment of 2009 pandemic influenza A (H1N1) virus drug susceptibility in vitro. Antivir. Ther. 15:1151–1159 [DOI] [PubMed] [Google Scholar]
- 22. Gubareva LV, Webster RG, Hayden FG. 2002. Detection of influenza virus resistance to neuraminidase inhibitors by an enzyme inhibition assay. Antiviral Res. 53:47–61 [DOI] [PubMed] [Google Scholar]
- 23. Hamelin ME, et al. 2010. Oseltamivir-resistant pandemic A/H1N1 virus is as virulent as its wild-type counterpart in mice and ferrets. PLoS Pathog. 6:e1001015 doi:10.1371/journal.ppat.1001015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hamelin ME, et al. 2011. Reduced airbourne transmission of oseltamivir-resistant pandemic A/H1N1 virus in ferrets. Antivir. Ther. 16:775–779 [DOI] [PubMed] [Google Scholar]
- 25. Hattrup CL, Gendler SJ. 2008. Structure and function of the cell surface (tethered) mucins. Annu. Rev. Physiol. 70:431–457 [DOI] [PubMed] [Google Scholar]
- 26. Hauge SH, Dudman S, Borgen K, Lackenby A, Hungnes O. 2009. Oseltamivir-resistant influenza viruses A (H1N1), Norway, 2007–08. Emerg. Infect. Dis. 15:155–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Herlocher ML, et al. 2004. Influenza viruses resistant to the antiviral drug oseltamivir: transmission studies in ferrets. J. Infect. Dis. 190:1627–1630 [DOI] [PubMed] [Google Scholar]
- 28. Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. U. S. A. 97:6108–6113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Holmen JM, et al. 2004. Mucins and their O-glycans from human bronchial epithelial cell cultures. Am. J. Physiol. Lung Cell Mol. Physiol. 287:L824–L834 [DOI] [PubMed] [Google Scholar]
- 30. Hurt AC, et al. 2011. Oseltamivir-resistant influenza viruses circulating during the first year of the influenza A(H1N1) 2009 pandemic in the Asia-Pacific region, March 2009 to March 2010. Euro Surveill. 16:19770 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19770 [PubMed] [Google Scholar]
- 31. Hurt AC, et al. 2009. Emergence and spread of oseltamivir-resistant A(H1N1) influenza viruses in Oceania, South East Asia and South Africa. Antiviral Res. 83:90–93 [DOI] [PubMed] [Google Scholar]
- 32. Hurt AC, et al. 2011. Community transmission of oseltamivir-resistant A(H1N1)pdm09 influenza. N. Engl. J. Med. 365:2541–2542 [DOI] [PubMed] [Google Scholar]
- 33. Hurt AC, et al. 2010. Assessing the viral fitness of oseltamivir-resistant influenza viruses in ferrets, using a competitive-mixtures model. J. Virol. 84:9427–9438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ilyushina NA, et al. 2010. Adaptation of pandemic H1N1 influenza viruses in mice. J. Virol. 84:8607–8616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ives JAL, et al. 2002. The H274Y mutation in the influenza A/H1N1 neuraminidase active site following oseltamivir phosphate treatment leave virus severely compromised both in vitro and in vivo. Antiviral Res. 55:307–317 [DOI] [PubMed] [Google Scholar]
- 36. Kesimer M, et al. 2009. Tracheobronchial air-liquid interface cell culture: a model for innate mucosal defense of the upper airways? Am. J. Physiol. Lung Cell Mol. Physiol. 296:L92–L100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kiso M, et al. 2010. Characterization of oseltamivir-resistant 2009 H1N1 pandemic influenza A viruses. PLoS Pathog. 6:e1001079 doi:10.1371/journal.ppat.1001079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kobasa D, et al. 1999. Amino acid residues contributing to the substrate specificity of the influenza A virus neuraminidase. J. Virol. 73:6743–6751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kryazhimskiy S, Dushoff J, Bazykin GA, Plotkin JB. 2011. Prevalence of epistasis in the evolution of influenza A surface proteins. PLoS Genet. 7:e1001301 doi:10.1371/journal.pgen.1001301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lackenby A, et al. 2008. Emergence of resistance to oseltamivir among influenza A(H1N1) viruses in Europe. Euro Surveill. 13:8026 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=8026 [DOI] [PubMed] [Google Scholar]
- 41. Le QM, et al. 2005. Avian flu: isolation of drug-resistant H5N1 virus. Nature 437:1108. [DOI] [PubMed] [Google Scholar]
- 42. Le QM, et al. 2010. A community cluster of oseltamivir-resistant cases of 2009 H1N1 influenza. N. Engl. J. Med. 362:86–87 [DOI] [PubMed] [Google Scholar]
- 43. Matrosovich M, Matrosovich T, Carr J, Roberts NA, Klenk H-D. 2003. Overexpression of the alpha-2,6-sialyltransferase in MDCK cells increases influenza virus sensitivity to neuraminidase inhibitors. J. Virol. 77:8418–8425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk HD. 2004. Neuraminidase is important for the initiation of influenza virus infection in human airway epithelium. J. Virol. 78:12665–12667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. McKimm-Breschkin JL. 2000. Resistance of influenza viruses to neuraminidase inhibitors: a review. Antiviral Res. 47:1–17 [DOI] [PubMed] [Google Scholar]
- 46. Meijer A, et al. 2011. Oseltamivir-resistant pandemic A(H1N1) 2009 influenza viruses detected through enhanced surveillance in the Netherlands, 2009–2010. Antiviral Res. 92:81–89 [DOI] [PubMed] [Google Scholar]
- 47. Meijer A, et al. 2009. Oseltamivir-resistant influenza virus A (H1N1), Europe, 2007–08 season. Emerg. Infect. Dis. 15:552–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Memoli MJ, et al. 2011. Multidrug-resistant 2009 pandemic influenza A(H1N1) viruses maintain fitness and transmissibility in ferrets. J. Infect. Dis. 203:348–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Monto AS, et al. 2006. Detection of influenza viruses resistant to neuraminidase inhibitors in global surveillance during the first 3 years of their use. Antimicrob. Agents Chemother. 50:2395–2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Moore C, et al. 2011. Evidence of person-to-person transmission of oseltamivir-resistant pandemic influenza A(H1N1) 2009 virus in a hematology unit. J. Infect. Dis. 203:18–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nguyen JT, et al. 2010. Triple combination of amantadine, ribavirin, and oseltamivir is highly active and synergistic against drug resistant influenza virus strains in vitro. PLoS One 5:e9332 doi:10.1371/journal.pone.0009332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pizzorno A, Bouhy X, Abed Y, Boivin G. 2011. Generation and characterization of recombinant pandemic influenza A(H1N1) viruses resistant to neuraminidase inhibitors. J. Infect. Dis. 203:25–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Potier M, Mameli L, Belisle M, Dallaire L, Melancon SB. 1979. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-alpha-d-N-acetylneuraminate) substrate. Anal. Biochem. 94:287–296 [DOI] [PubMed] [Google Scholar]
- 54. Rameix-Welti MA, Enouf V, Cuvelier F, Jeannin P, van der Werf S. 2008. Enzymatic properties of the neuraminidase of seasonal H1N1 influenza viruses provide insights for the emergence of natural resistance to oseltamivir. PLoS Pathog. 4:e1000103 doi:10.1371/journal.ppat.e1000103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rameix-Welti MA, et al. 2011. Neuraminidase of 2007–2008 influenza A(H1N1) viruses shows increased affinity for sialic acids due to the D344N substitution. Antivir. Ther. 16:597–603 [DOI] [PubMed] [Google Scholar]
- 56. Reddy D. 2010. Responding to pandemic (H1N1) 2009 influenza: the role of oseltamivir. J. Antimicrob. Chemother. 65(Suppl 2):ii35–ii40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Reed LJ, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. (Lond.) 27:493–497 [Google Scholar]
- 58. Sandbulte MR, et al. 2011. Discordant antigenic drift of neuraminidase and hemagglutinin in H1N1 and H3N2 influenza viruses. Proc. Natl. Acad. Sci. U. S. A. 108:20748–20753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Scholtissek C, Burger H, Bachmann PA, Hannoun C. 1983. Genetic relatedness of hemagglutinins of the H1 subtype of influenza A viruses isolated from swine and birds. Virology 129:521–523 [DOI] [PubMed] [Google Scholar]
- 60. Seibert CW, et al. 2010. Oseltamivir-resistant variants of the 2009 pandemic H1N1 influenza A virus are not attenuated in the guinea pig and ferret transmission models. J. Virol. 84:11219–11226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sheu TG, et al. 2008. Surveillance for neuraminidase inhibitor resistance among human influenza A and B viruses circulating worldwide from 2004 to 2008. Antimicrob. Agents Chemother. 52:3284–3292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Smith GJ, et al. 2009. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459:1122–1125 [DOI] [PubMed] [Google Scholar]
- 63. Storms AD, et al. 2012. Oseltamivir-resistant pandemic (H1N1) 2009 virus infections, United States, 2010–11. Emerg. Infect. Dis. 18:308–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tashiro M, et al. 2009. Surveillance for neuraminidase-inhibitor-resistant influenza viruses in Japan, 1996–2007. Antivir. Ther. 14:751–761 [DOI] [PubMed] [Google Scholar]
- 65. Thornton DJ, Rousseau K, McGuckin MA. 2008. Structure and function of the polymeric mucins in airways mucus. Annu. Rev. Physiol. 70:459–486 [DOI] [PubMed] [Google Scholar]
- 66. von Itzstein M, et al. 1993. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature 363:418–423 [DOI] [PubMed] [Google Scholar]
- 67. WHO 2009. Influenza A(H1N1) virus resistance to oseltamivir, 2008/2009 influenza season, Northern Hemisphere. WHO, Geneva, Switzerland: http://www.who.int/influenza/resources/ documents/H1N1webupdate20090318_ed_ns.pdf [Google Scholar]
- 68. WHO 2010. Update on oseltamivir-resistant pandemic A (H1N1) 2009 influenza virus: January 2010. Wkly. Epidemiol. Rec. 85:37–40 [PubMed] [Google Scholar]
- 69. Wu WL, et al. 2012. The 2008–2009 H1N1 influenza virus exhibits reduced susceptibility to antibody inhibition: implications for the prevalence of oseltamivir resistant variant viruses. Antiviral Res. 93:144–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ye J, et al. 2010. Variations in the hemagglutinin of the 2009 H1N1 pandemic virus: potential for strains with altered virulence phenotype? PLoS Pathog. 6:e1001145 doi:10.1371/journal.ppat.1001145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yen HL, et al. 2005. Neuraminidase inhibitor-resistant influenza viruses may differ substantially in fitness and transmissibility. Antimicrob. Agents Chemother. 49:4075–4084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yen HL, et al. 2006. Importance of neuraminidase active-site residues to the neuraminidase inhibitor resistance of influenza viruses. J. Virol. 80:8787–8795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Yen HL, et al. 2007. Inefficient transmission of H5N1 influenza viruses in a ferret contact model. J. Virol. 81:6890–6898 [DOI] [PMC free article] [PubMed] [Google Scholar]