Abstract
The family Anelloviridae includes human and animal torque teno viruses (TTVs) with extensive genetic diversity. The antigenic diversity among anelloviruses has never been assessed. Using torque teno sus virus (TTSuV) as a model, we describe here the first investigation of the antigenic relationships among different anelloviruses. Using a TTSuV genotype 1a (TTSuV1a) or TTSuV1b enzyme-linked immunosorbent assay (ELISA) based on the respective putative ORF1 capsid antigen and TTSuV1-specific real-time PCR, the combined serological and virological profile of TTSuV1 infection in pigs was determined and compared with that of TTSuV2. TTSuV1 is likely not associated with porcine circovirus-associated disease (PCVAD), because both the viral loads and antibody levels were not different between affected and unaffected pigs and because there was no synergistic effect of concurrent PCV2/TTSuV1 infections. We did observe a higher correlation of IgG antibody levels between anti-TTSuV1a and -TTSuV1b than between anti-TTSuV1a or -1b and anti-TTSuV2 antibodies in these sera, implying potential antigenic cross-reactivity. To confirm this, rabbit antisera against the putative capsid proteins of TTSuV1a, TTSuV1b, or TTSuV2 were generated, and the antigenic relationships among these TTSuVs were analyzed by an ELISA and by an immunofluorescence assay (IFA) using PK-15 cells transfected with one of the three TTSuV ORF1 constructs. The results demonstrate antigenic cross-reactivity between the two genotypes TTSuV1a and TTSuV1b but not between the two species TTSuV1a or -1b and TTSuV2. Furthermore, an anti-genogroup 1 human TTV antiserum did not react with any of the three TTSuV antigens. These results have important implications for an understanding of the diversity of anelloviruses as well as for the classification and vaccine development of TTSuVs.
INTRODUCTION
The first anellovirus was discovered in a Japanese patient with posttransfusion non-A to -E hepatitis in 1997 and termed torque teno virus (TTV) (26, 27). Since then, a variety of anelloviruses have been identified in numerous animal species, including nonhuman primates, tupaias, pigs, cats, and dogs (3, 28). Anelloviruses are small, single-stranded, circular DNA viruses with genome sizes ranging from 2.0 to 3.9 kb (3, 27, 28). Recently, the International Committee on Taxonomy of Viruses (ICTV) established a new family, Anelloviridae, which comprises nine genera, to include all human and animal anelloviruses (4). The viruses in this family show an extremely high degree of genetic diversity (2, 27). For example, human TTVs were previously classified into five different genogroups with approximately 50% nucleotide sequence differences, including at least 39 distinct genotypes with ∼30% nucleotide sequence differences (27). In the most recent classification system, human TTV belongs to the genus Alphatorquevirus and contains 29 different species (4).
The diversity of anelloviruses was exhibited not only in the nucleotide sequence, genomic size, and animal hosts but also in the presence of a number of intragenomic rearranged subviral molecules in a single individual according to a study of human TTV reported previously (7, 20). It was also hypothesized that human TTV should display a high level of antigenic diversity due to extensive genetic heterogeneity (21). However, experimental evidence supporting such a hypothesis is still lacking thus far.
Porcine anellovirus or Torque teno sus virus (TTSuV) resembles the genomic organization of human TTV and is classified into the genus Iotatorquevirus. TTSuV comprises two species, TTSuV1 and TTSuV2, each with a genomic size of approximately 2.8 kb and with at least four putative open reading frames (ORFs), ORF1, ORF2, ORF1/1, and ORF2/2 (15, 16, 29). ORF1 of TTSuV is believed to encode a capsid protein with the largest size relative to those of the four predicted viral proteins (5, 14, 15). TTSuV1 consists of at least two genotypes, TTSuV genotype (TTSuV1a) and TTSuV1b, with ∼30% nucleotide sequence differences, whereas TTSuV2 has only one genotype, including at least three subtypes with ∼15% nucleotide sequence differences (5, 15).
Multiple infections of human TTV with different genotypes in a single human individual or TTSuV with different genotypes or subtypes in a single pig have been documented (2, 10, 11, 15, 17, 23, 25). These findings raise the question of whether the anti-ORF1 capsid antibodies recognized by the antigen from a particular TTV or TTSuV species/genotype also comprise anti-ORF1 antibodies against other distinct TTV or TTSuV species/genotypes and whether the anti-ORF1 antibodies from one TTV or TTSuV genotype can cross-protect against infection with another genotype. To our knowledge, for human TTV or TTSuV infection, there has been no information on this topic available to date. Furthermore, the antigenic diversity and relationship of anelloviruses have never been assessed (21). It is reasonable to speculate that there is little, if any, antigenic cross-reactivity between different anellovirus species/genotypes, due to the fact that concurrent infections with multiple anelloviruses in a single individual or animal exist.
We have previously developed and validated serum Western blot (WB) assays and indirect enzyme-linked immunosorbent assays (ELISAs) for the detection of the IgG antibody against TTSuV2 in porcine sera using the purified recombinant TTSuV2-ORF1 protein expressed in Escherichia coli (14). By using TTSuV2-specific real-time quantitative PCR (qPCR) and ELISA, we further presented the combined virological and serological profile of TTSuV2 infection under natural or diseased conditions using 160 porcine sera collected from different sources (14). In the present study, we initially aimed to assess the serological profiles of the two TTSuV1 genotypes (TTSuV1a and TTSuV1b) in pigs. Subsequently, we aimed to compare the virological and serological profiles of TTSuV1a and TTSuV1b with that of TTSuV2 and to determine the degree of correlation of IgG antibody levels between anti-TTSuV1a and -TTSuV1b and between anti-TTSuV1a or -1b and anti-TTSuV2 antibodies. Finally, for the first time, we assessed the antigenic relationships between two TTSuV1 genotypes (TTSuV1a and TTSuV1b), between two species (TTSuV1 and TTSuV2), and between porcine and human genogroup 1 anelloviruses using ELISAs and immunofluorescence assays (IFAs) with antibody cross-reactions in PK-15 cells transfected with recombinant plasmids expressing the ORF1 proteins from TTSuV1a, TTSuV1b, and TTSuV2, respectively.
MATERIALS AND METHODS
Cell line.
A porcine circovirus type 1 (PCV1)-free porcine kidney cell line, PK-15, was used in this study (9). Cells were grown in modified Eagle's medium supplemented with 10% fetal bovine serum and antibiotics at 37°C.
Sources of porcine sera.
The porcine sera used in this study were described previously (14). Briefly, serum samples for WB analysis were collected from 20 conventional adult boars with no clinical symptoms from a Virginia pig farm; 7 gnotobiotic pigs from Virginia (pigs 4 to 7, 224, 229, and 230; kindly provided by Lijuan Yuan and Guohua Li from Virginia Tech) and 12 from Iowa (group D); 5 cesarean-derived, colostrum-deprived pigs; and approximately 50 conventional piglets from a Wisconsin pig farm. TTSuV2-seropositive porcine serum, which was manufactured in New Zealand and free of all known OIE (World Organization for Animal Health)-notifiable diseases, was also used in this study.
One hundred sixty porcine serum samples were used for assessing the virological and serological profiles of TTSuV1a and TTSuV1b infections and were divided into five groups (groups A to E), as described previously (14): (i) 20 group A samples were obtained from 10 specific-pathogen-free (SPF) pigs (60 to 80 days old at arrival) free of known pathogens and were collected upon arrival at the facility and 2 months after arrival; (ii) 60 group B samples were collected from 105-day-old pigs in a farm with an outbreak of porcine circovirus-associated disease (PCVAD), including 30 samples from clinically affected pigs and 30 from clinically unaffected pigs; (iii) 58 group C samples were collected from 28-day-old pigs of an unknown disease status, 28 from clinically affected and 30 from clinically unaffected pigs; (iv) 12 group D samples were obtained from 14- to 42-day-old gnotobiotic pigs located in Iowa; and (v) 10 group E sera were obtained from 21- to 30-day-old SPF pigs used for an experimental PCV2 infection study.
Construction of the TTSuV1a- and TTSuV1b-ORF1 expression plasmids.
The C-terminal part of the ORF1 proteins of two TTSuV1 strains, PTTV1a-VA (GenBank accession no. GU456383) and PTTV1b-VA (GenBank accession no. GU456384), was amplified from the available PCR fragments reported previously (15). The amplicon was expected to encode a truncated PTTV1a-VA ORF1 protein of 319 amino acids (aa) (positions 317 to 635 corresponding to PTTV1a-VA) or a truncated PTTV1b-VA ORF1 protein of 318 aa (positions 322 to 639 corresponding to PTTV1b-VA). An additional methionine codon was introduced at the N terminus of each amplified fragment. Two ORF1 expression plasmids, designated pTri-1aORF1 and pTri-1bORF1, were each constructed by cloning the respective PCR product into a bacterial/insect/mammalian-triple expression vector, pTriEx1.1-Neo (Novagen), between the NcoI and XhoI restriction sites to generate two C-terminally 8×His-tagged fusion proteins. The recombinant plasmids were confirmed by DNA sequencing. The TTSuV2 ORF1 expression construct pTri-2cORF1 was described previously (14).
Expression and purification of the recombinant TTSuV1a- and TTSuV1b-ORF1 proteins.
The two plasmids were each transformed into Rosetta 2(DE3)(pLacI) competent cells (Novagen, San Diego, CA). The bacteria were grown in 100 ml of Overnight Express TB medium (Novagen) for 16 to 18 h at 37°C, and the bacterial culture was then harvested by centrifugation at 3,400 rpm for 15 min at 4°C. The resulting bacterial pellet was treated with BugBuster and rLysozyme according to the manufacturer's protocol (Novagen). Benzonase nuclease (Novagen) was added to degrade DNA and RNA. The resulting inclusion bodies were lysed in a solution containing 6 M guanidine hydrochloride, 0.1 M sodium phosphate, 0.01 M Tris-chloride, and 0.01 M imidazole with a pH value of 8.0. The lysate supernatants were collected by centrifugation and were used for His-tagged protein purification with 50% Ni-nitrilotriacetic acid (NTA) HisBind resin (Novagen) under denaturing conditions with 8 M urea. Proteins were dialyzed as described previously (14). The recombinant His-tagged TTSuV1a- or TTSuV1b-ORF1 proteins used as the antigens for ELISAs and rabbit immunization were quantified by using NanoDrop spectrophotometry and frozen at −80°C until use.
Generation of anti-ORF1 antisera against TTSuV1a and TTSuV1b in rabbits.
The two ORF1 proteins of TTSuV1a and TTSuV1b expressed in E. coli were purified and used to immunize two New Zealand White rabbits using a custom antibody production service at Rockland Immunochemicals (Gilbertsville, PA). Antisera were harvested at 50 days postimmunization.
SDS-PAGE, anti-His-tagged MAb WB, and serum WB analyses.
The unpurified or purified recombinant TTSuV1 ORF1 proteins were resolved on a 4 to 12% Bis-Tris polyacrylamide gel (Invitrogen, Carlsbad, CA) by electrophoresis and were subsequently transferred onto a polyvinylidene difluoride (PVDF) membrane. Proteins were detected on the PVDF membrane by using an anti-6×His-tagged monoclonal antibody (MAb) at a 1:1,000 dilution at 4°C, followed by incubation with IRDye 800CW-conjugated goat anti-rabbit IgG (Li-Cor Biosciences, Lincoln, NE) at a 1:10,000 dilution at room temperature. After three washing steps using Tris-buffered saline–0.05% Tween 20 (TBS-T), the membrane was analyzed by using the Odyssey infrared imaging system (Li-Cor Biosciences).
For serum WB analysis, the purified TTSuV1a- or TTSuV1b-ORF1 proteins were incubated with individual porcine sera at a 1:200 dilution and with IRDye 800CW-conjugated rabbit F(ab′)2 anti-swine IgG (Rockland Immunochemicals, Inc.) at a 1:10,000 dilution at room temperature. The membrane was then analyzed by using the Odyssey infrared imaging system.
Indirect ELISAs.
TTSuV1a- and TTSuV1b-based ELISAs were developed. The optimal concentrations of the antigens and the optimal dilutions of sera and horseradish peroxidase (HRP) conjugates were determined by checkerboard titrations. Similar to a TTSuV2-based ELISA reported previously (14), the optimal amount of the ORF1 antigen of TTSuV1a or TTSuV1b was 68 ng per well. The optimal ELISA results were obtained by using a 1:100 dilution of serum samples and a 1:4,000 dilution of IgG conjugates.
The ELISA was initiated by diluting the purified ORF1 proteins into carbonate coating buffer (pH 9.6), which was used for coating 96-well ELISA plates (Greiner Bio-One, Monroe, NC) with 100 μl/well. After incubation at 37°C for 2 h, each well was washed 3 times with 300 μl of TBS-T and blocked with protein-free blocking buffer (Pierce, Rockford, IL) in a volume of 300 μl for 1 h at 37°C. One hundred microliters of each diluted serum sample was transferred into the corresponding well on the ELISA plates and incubated at 37°C for 2 h. After the wells were washed three times with 300 μl of TBS-T buffer, the diluted HRP-conjugated rabbit anti-swine IgG (Rockland) was added to each well in a volume of 100 μl, and the plate was incubated at 37°C for 1 h. A volume of 100 μl of Sure Blue Reserve 1-Component (KPL, Gaithersburg, MD) was added to each well and incubated for 10 min at room temperature. The reaction was stopped by the addition of 100 μl/well of 1 N HCl. The plates were then read at 450 nm by using a spectrophotometer. All serum samples were run in duplicates. Positive and negative controls run in quadruplicates were included on each plate. In general, the mean OD value of the negative control was less than 0.5, whereas the mean OD value of the positive control was greater than 1.5. The ELISA value was calculated as the S/N value, which was expressed as a ratio of the mean OD value of a sample to the mean OD value of the negative control (n = 4). A subjective cutoff S/N value of 1.2 was used to distinguish between positive and negative samples.
Real-time qPCR assay for quantitation of TTSuV1.
A SYBR green-based TTSuV1-specific real-time qPCR developed recently in our laboratory was used to measure the total TTSuV1 viral loads (both TTSuV1a and TTSuV1b) in the five groups of pig sera, as described previously (13). Since some of the samples were run with standards with a detection limit of 1.0 × 103 copies/ml, while other samples were run with standards with a detection limit of 1.0 × 104 copies/ml, to normalize the final results, the minimal detection limit was set to 1.0 × 104 copies per ml in this study. The TTSuV1 qPCR assay does not cross-amplify TTSuV2 DNA (13). The quantitation of TTSuV2 and PCV2 viral loads in group B sera was reported previously (14).
Statistical analyses.
Data were analyzed by using SAS software (version 9.2; SAS Institute, Inc., Cary, NC) and GraphPad Prism software (version 5.0; GraphPad, San Diego, CA). Antibody levels (represented by S/N values) were compared between categories of log10 viral titers by using the Kruskal-Wallis test followed by Dunn's procedure. For each group that contained clinically affected and nonaffected pigs (groups B and C), log10 virus titers of pigs with and pigs without clinical signs were compared by using a Wilcoxon two-sample test. Antibody levels of pigs with and those without disease were compared by using a two-sample t test. Using a cutoff point of 1.2, the proportions of affected and unaffected pigs with antibodies were compared by using a Fisher exact test.
Correlations between S/N values for TTSuV1a and S/N values for TTSuV1b and between S/N values for TTSuV1a or TTSuV1b (separately) and TTSuV2 were assessed by using Spearman's correlation coefficient. The correlations were separately generated for a combination of 3 groups (group A to group C).
To assess the synergistic effects of PCV2 and TTSuV1 on disease prevalence, the pigs in group B were categorized as follows: pigs positive for both PCV2 and TTSuV1, pigs positive for PCV2 only, pigs positive for TTSuV1 only, and pigs with neither PCV2 nor TTV1 infection. Subsequently, the proportions of affected pigs were compared between groups by using Fisher's exact test. Statistical significance was set to an alpha value of 0.05.
Transfection of PK-15 cells with TTSuV expression constructs.
PK-15 cells were seeded onto a 6-well plate and grown until they reached 70% to 80% confluence before transfection. Two micrograms of each of the three constructs pTri-1aORF1, pTri-1bORF1, and pTri-2cORF1, mixed with 10 μl of Lipofectamine LTX (Invitrogen), was transfected into the cells. Cells were cultured for 3 days and were subjected to IFA to detect ORF1 expression.
IFA.
Five rabbit antisera were used for IFA staining: anti-TTSuV1a, anti-TTSuV1b, anti-TTSuV2 antisera; prebleed rabbit negative-control serum; and rabbit anti-human genogroup 1 TTV ORF1 antiserum (AK47; a generous gift from Annette Mankertz at the Robert Koch Institute, Berlin, Germany) (22). The AK47 antiserum was also produced in rabbits immunized with a C-terminal fragment (aa 402 to 733) of ORF1 of strain P/1C1 (GenBank accession no. AF298585). Transfected cells were fixed with acetone. Five hundred microliters of each of the five antisera, at a 1:500 dilution in phosphate-buffered saline (PBS), was added on top of the cells in each well and incubated for 1 h at room temperature. After three washing steps with PBS, the cells were incubated with 500 μl Alexa Fluor 488-labeled goat anti-rabbit IgG (Invitrogen) at a 1:200 dilution for 1 h of incubation at room temperature. Cells were stained with 500 μl 4′,6-diamidino-2-phenylindole (DAPI) (KPL, Inc.) at a 1:1,000 dilution and visualized under a fluorescence microscope.
RESULTS
Expression and purification of the N-terminally truncated TTSuV1a and TTSuV1b ORF1 proteins.
Previously, we successfully expressed a truncated TTSuV2 ORF1 protein in E. coli (14). Using a similar strategy, the C-terminal region of the TTSuV1a ORF1 (1a-ORF1) or TTSuV1b ORF1 gene with a C-terminally engineered 8×His tag was inserted into the triple expression vector pTriEx1.1-Neo, resulting in two recombinant constructs, pTri-1aORF1 and pTri-1bORF1. We also constructed an ORF1 C-terminally truncated version of 1b-ORF1 as a control, termed pTri-1bORF1-ctruc (1bORF1-ctruc), which is 71 aa shorter than 1b-ORF1, to compare the size with that of pTri-1bORF1 by SDS-PAGE and WB analyses.
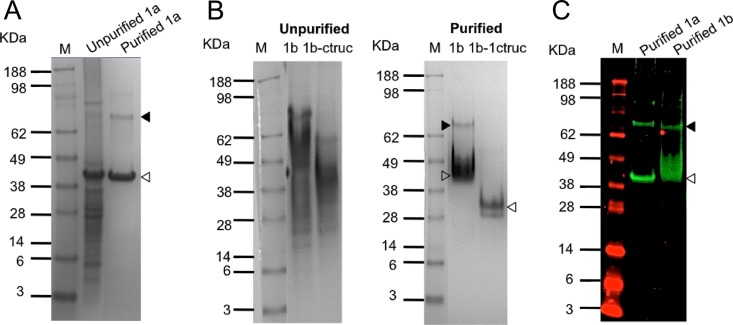
The three recombinant proteins 1a-ORF1, 1b-ORF1, and 1bORF1-ctruc were found to be insoluble and were expressed within the bacteria as inclusion bodies. The purification of the crude lysates from 1a-ORF1 products with a nickel affinity column resulted in the visualization of two bands of ∼40 kDa (Fig. 1A, white arrowheads) and ∼70 kDa (black arrowheads), as analyzed by Coomassie blue staining (Fig. 1A). The ∼40-kDa band is the expected product of the truncated 1a-ORF1 protein, whereas the ∼70-kDa polypeptide is an unknown product but should be derived from the former, since it also reacted with an anti-His-tagged MAb (see below). The expression of 1b-ORF1 or 1bORF1-ctruc showed a smear in the crude lysates (Fig. 1B). After purification, two bands of ∼40 kDa and ∼70 kDa, similar to 1a-ORF1, were also identified in the purified 1b sample, whereas only a ∼30-kDa polypeptide (Fig. 1B, white arrowheads) was detected in the purified 1b-ctruc sample (Fig. 1B). The bands of ∼40 kDa and ∼30 kDa were consistent with the expected sizes of the 1b-ORF1 and 1bORF1-ctruc protein products, respectively. All the identified polypeptides in the purified products were detected by WB using the anti-His-tagged MAb (Fig. 1C). The results indicated that both the truncated 1a-ORF1 and 1b-ORF1 proteins were successfully expressed in E. coli and thus can be used as antigens for TTSuV1a and TTSuV1b antibody detection in porcine sera.
Fig 1.
Expression and purification of the amino-terminally truncated TTSuV1a and TTSuV1b ORF1 proteins. (A) SDS-PAGE analysis of unpurified and purified TTSuV1a-ORF1 products. (B) SDS-PAGE analysis of unpurified and purified TTSuV1b-ORF1 products. An amino- and carboxyl-terminally double-truncated TTSuV1b-ORF1 (1b-ctruc) of a smaller product size served as the control. (C) Near-infrared fluorescent WB analysis of purified 1a- and 1b-ORF1 products using an anti-His-tagged MAb. Open arrowheads indicate the truncated ORF1 protein of the expected size, whereas filled arrowheads show the presumed homodimers of the expected proteins. M, protein markers.
Development of TTSuV1a- and TTSuV1b-based serum WB and indirect ELISAs.
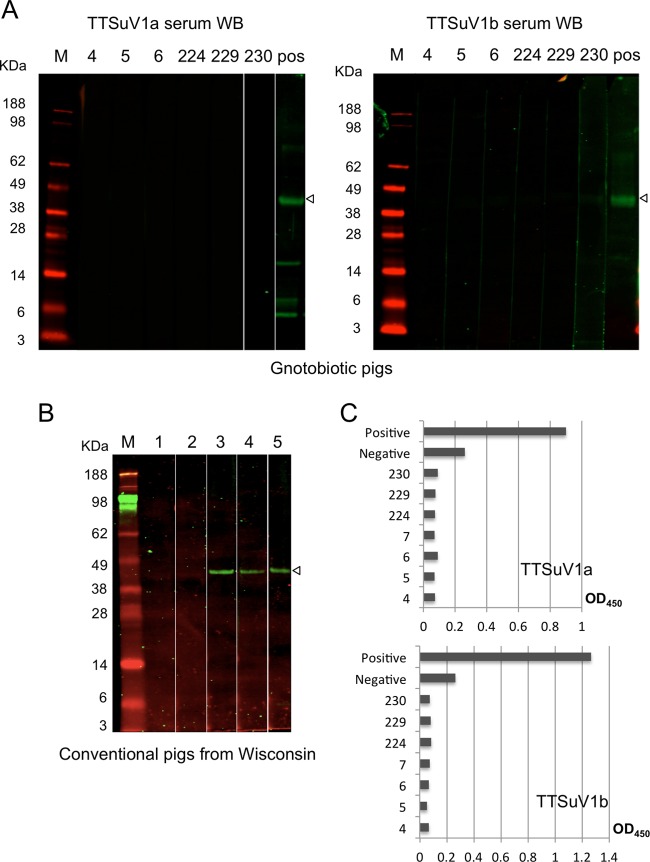
In order to identify reference positive and negative sera as controls, a total of 100 serum samples from different sources, including those from the gnotobiotic pigs, were collected. Samples were screened for anti-TTSuV1a or anti-TTSuV1b IgG seropositivity by serum WB analysis using purified 1a-ORF1 or 1b-ORF1 as the antigen, respectively. A TTSuV2-seropositive and TTSuV1/TTSuV2 DNA-positive porcine serum sample (16) showed reactivity with the 1a-ORF1 and the 1b-ORF1 antigens, as the ∼40-kDa band was present in the WB analysis (Fig. 2A, rightmost two lanes). Therefore, this serum was considered to be TTSuV1a and TTSuV1b seropositive and thus was used as a reference positive control for the ELISAs. All the 7 Virginia and 12 Iowa gnotobiotic pigs had no detectable TTSuV1a and TTSuV1b antibodies (Fig. 2A). Except for a few serum samples from conventional pigs from a Wisconsin swine farm (Fig. 2B, leftmost two lanes), the remaining samples tested positive for both TTSuV1a and TTSuV1b antibodies by WB analysis. The dual-negative serum samples from the conventional pigs from Wisconsin were pooled and used as negative-control reference serum.
Fig 2.
TTSuV1a or TTSuV1b serum WB and ELISA. (A) WB analyses using gnotobiotic pig serum samples from Virginia and commercial OIE disease-free porcine serum as the positive-control reference serum (pos). (B) Representative results of TTSuV1a WB analyses of conventional pig sera from a farm in Wisconsin. Purified 1a-ORF1 protein was used as the antigen. Sera that tested negative for both TTSuV1a and TTSuV1b antibodies by WB were pooled and used as the negative-control reference serum. Open arrowheads indicate the truncated ORF1 protein of the expected size. Only the bands in green were considered positive. M, protein markers. (C) TTSuV1a or TTSuV1b ELISA results for the seven Virginia gnotobiotic pig serum samples and positive- and negative-control reference sera.
With the available positive- and negative-control reference sera, TTSuV1a- and TTSuV1b-based ELISAs were subsequently developed and standardized, respectively. The concentrations of the purified 1a-ORF1 or 1b-ORF1 antigen, porcine sera, and the IgG conjugate were determined by a checkerboard titration assay to ensure a low background signal and to give the highest difference in OD at 450 nm (OD450) values between the positive and negative controls. WB-negative gnotobiotic porcine sera showed very low OD values (<0.1) compared to that of the negative-control reference serum (Fig. 2C), suggesting that these pig sera should not serve as a negative-control reference for the detection of porcine field samples in the ELISA.
TTSuV1 viral DNA loads and anti-TTSuV1a and anti-TTSuV1b IgG antibody levels.
A total of 160 serum samples were collected and evaluated for the prevalence and viral DNA load of TTSuV1 by real-time qPCR and for seroprevalence and antibody levels (represented by S/N values) of anti-TTSuV1a and anti-TTSuV1b IgG antibodies by the ELISAs. Among the 160 samples, 138 sera in groups A to C were collected from three herds under field conditions, whereas the remaining 22 sera from groups D (gnotobiotic pigs) and E were collected from pigs raised and housed under strictly controlled experimental conditions in research facilities.
None of the 12 TTSuV1a/TTSuV1b-seronegative gnotobiotic pigs in group D had detectable viremia. In group E pigs, only one pig was viremic, whereas six were seropositive for TTSuV1a, and among them, one pig was also seropositive for TTSuV1b.
In groups A and C, 44 of 138 pigs were viremic (31.9%), whereas 128 were TTSuV1a seropositive (92.8%) and 121 were TTSuV1b seropositive (87.7%) (Table 1). The incidence of TTSuV1 viremia was much lower than the TTSuV1a- or 1b-seropositive rate, suggesting a previous clearance of the virus by neutralizing antibodies during the post-TTSuV1 infection convalescent period.
Table 1.
Distribution of TTSuV1 viremia and anti-TTSuV1a and anti-TTSuV1b IgG antibodies among 138 serum samples from three different herds
| No. of samples with profile | Distribution of results |
||
|---|---|---|---|
| TTSuV1 viremia (31.9% positive; 44/138) | Anti-TTSuV1a IgG (92.8% positive; 128/138) | Anti-TTSuV1b IgG (87.7% positive; 121/138) | |
| 40 | + | + | + |
| 2 | + | + | − |
| 2 | + | − | + |
| 0 | + | − | − |
| 77 | − | + | + |
| 9 | − | + | − |
| 2 | − | − | + |
| 6 | − | − | − |
All three markers of TTSuV1 infection, TTSuV1 DNA and TTSuV1a/1b antibodies, were found in 40 serum samples. Notably, the number of pigs that were dually TTSuV1a/TTSuV1b seropositive but viral DNA negative (77 samples) was higher than the number of pigs with TTSuV1a or TTSuV1b seropositivity only (Table 1). In addition, the total number of porcine sera with both antibodies was 117 (40 plus 77) among the 138 serum samples, implying that (i) coinfection rates of pigs with TTSuV1a and TTSuV1b are high, which was expected, and/or (ii) a certain degree of cross-reactivity may exist between anti-TTSuV1a and anti-TTSuV1b IgG antibodies.
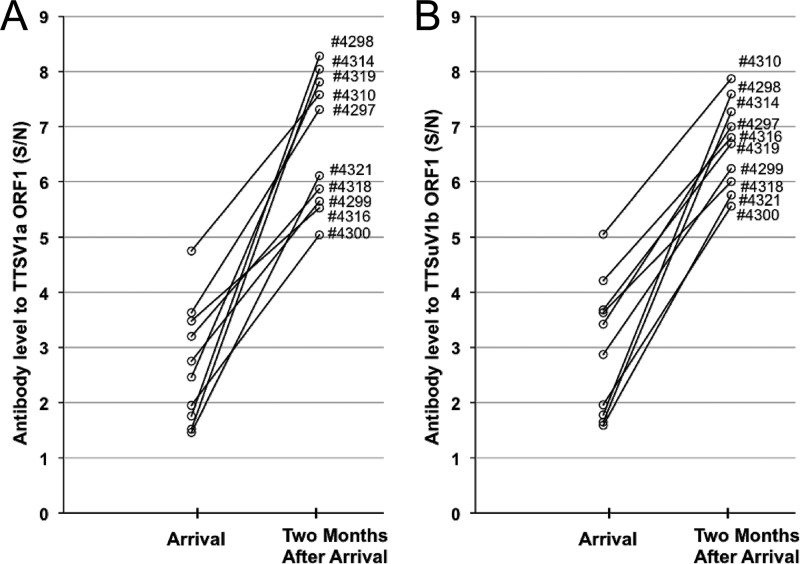
We previously demonstrated that, over a 2-month period, the 10 group A pigs had decreasing TTSuV2 viral loads that were associated with elevated anti-TTSuV2 ORF1 IgG antibody levels (14). Whether an analogous situation for TTSuV1 in these 10 pigs existed was subsequently analyzed in this study by comparing the TTSuV1 viral DNA loads and the anti-TTSuV1a or anti-TTSuV1b antibody levels in sera from the time of their arrival until 2 months later. Both the anti-TTSuV1a and anti-TTSuV1b antibody titers increased in all 10 pigs (Fig. 3). Five of the 10 pigs had undetectable TTSuV1 DNA levels during the 2 months, and in 4 pigs, the viral DNA loads decreased after 2 months, including 3 pigs with no detectable TTSuV1 DNA (data not shown). These results were consistent with those of the TTSuV2 study. Low levels of serum TTSuV DNA that are beyond the detection limit of the qPCR may explain why some pigs are negative for TTSuV DNA even though they develop antibodies.
Fig 3.
Retrospective evaluation of levels of TTSuV1 antibody to the ORF1 proteins of TTSuV1a (A) and TTSuV1b (B) in 10 pigs from group A from the time of their arrival at the research facility to 2 months after arrival.
TTSuV1 is likely not associated with PCVAD.
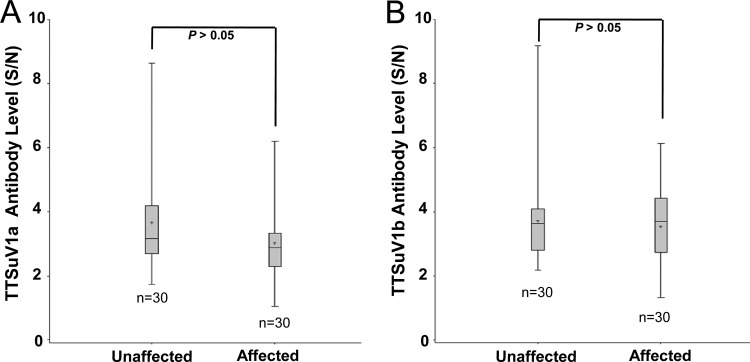
Although PCV2 is considered the primary causative agent for the induction of clinical PCVAD, it was recently shown that TTSuV partially contributed to the experimental induction of PCVAD in pigs (8). It was also observed that PCVAD-affected pigs with low or no detectable PCV2 infection had a higher TTSuV2 DNA prevalence than non-PMWS-affected pigs in Spain (19). We previously found that PCVAD-affected pigs had a significantly lower level of TTSuV2 antibody than did PCVAD-unaffected pigs in group B (14). However, determinations of the levels of anti-TTSuV1a and anti-TTSuV1b IgG antibodies in these serum samples did not reveal a difference between the PCVAD-affected and -unaffected pigs (Fig. 4). In addition, there was no statistically significant difference in TTSuV1 viral loads between the PCVAD-affected and -unaffected pigs (see Fig. S2A in the supplemental material). In contrast, the PCV2 viral load was significantly higher (P < 0.05) in PCVAD-affected pigs than in PCVAD-unaffected pigs (see Fig. S2B in the supplemental material).
Fig 4.
Box plots showing comparisons of anti-TTSuV1a (A) and anti-TTSuV1b (B) ORF1 antibody levels between PCVAD-affected and -unaffected pigs.
We further analyzed whether there existed a synergistic effect of PCV2 and TTSuV1 associated with PCVAD. Serum viral DNA prevalence rates (viremia) of PCVAD-affected pigs were as follows: 50% (16/32) for PCV2 and TTSuV1, 56% (14/25) for PCV2 only, 0% (0/1) for TTSuV1 only, and 0% (0/2) for no detectable virus. These proportions were not significantly different (P = 0.4339). The above-described results suggested that TTSuV1 is likely not associated with PCVAD.
Comparison and correlations of seroprevalence and antibody levels among anti-TTSuV1a, anti-TTSuV1b, and anti-TTSuV2 antibodies.
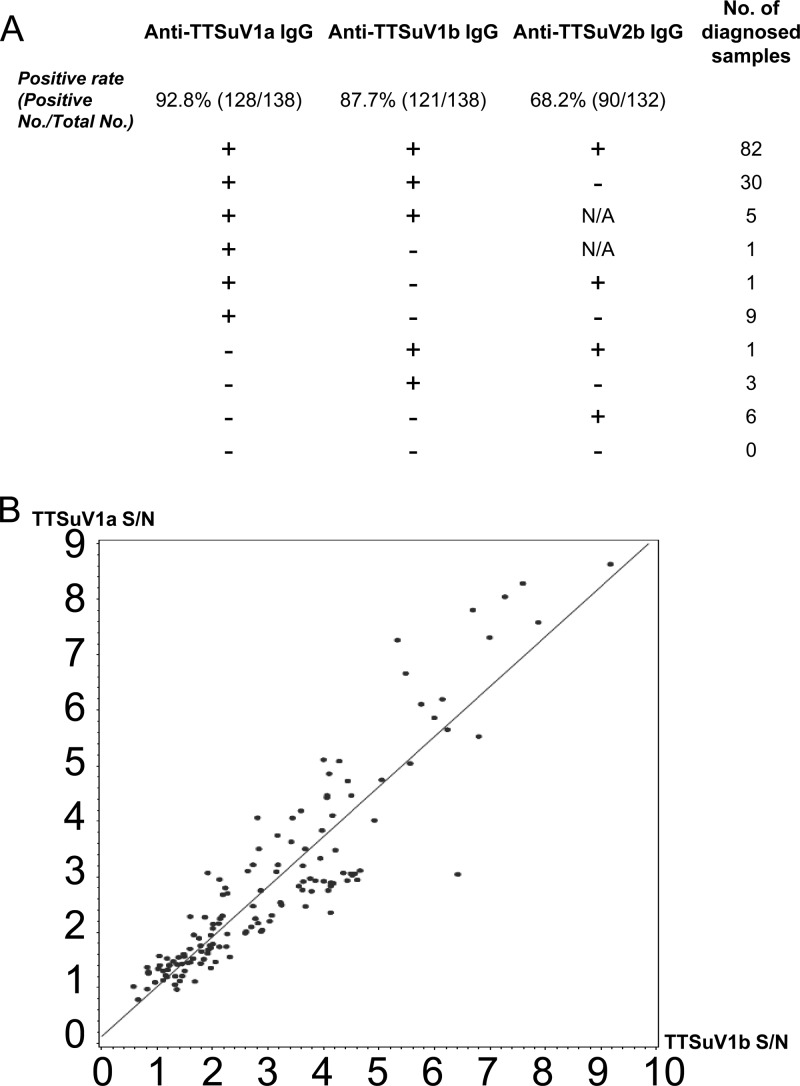
Mixed infections by TTSuV1 and TTSuV2 are common in pigs, as determined by the presence of viral DNAs of both TTSuV1 and TTSuV2 in the same pig using PCR (11, 13–15). In this study, we provided serological evidence to support this conclusion by analyzing the seroprevalence distributions of anti-TTSuV1a, -TTSuV1b, and -TTSuV2 IgG antibodies in 138 serum samples from groups A to C. As shown in Fig. 5A, 82 of 138 serum samples were triple seropositive, indicating that these pigs had been infected by TTSuV1 (TTSuV1a and/or TTSuV1b) and TTSuV2.
Fig 5.
High correlation between anti-TTSuV1a and anti-TTSuV1b IgG antibodies in 138 serum samples. (A) Distribution of anti-TTSuV1a, -TTSuV1b, and -TTSuV2 IgG antibodies. (B) Scatter plots showing a good linear relationship of antibody levels between anti-TTSuV1a and anti-TTSuV1b antibodies (P < 0.0001).
The distributions of dual-seropositive samples were significantly different. A total of 117 (82 plus 30 plus 5) porcine sera were dually seropositive for both anti-TTSuV1a and anti-TTSuV1b antibodies, which was consistent with the number calculated in Table 1. In contrast, dual seropositivity to anti-TTSuV1a and anti-TTSuV2 antibodies or to anti-TTSuV1b and anti-TTSuV2 antibodies occurred in only one sample each (Fig. 5A).
Furthermore, the correlations of antibody levels between anti-TTSuV1a and anti-TTSuV1b, between anti-TTSuV1a and anti-TTSuV2, and between anti-TTSuV1b and anti-TTSuV2 antibodies were assessed for the 138 serum samples by using Spearman's correlation coefficient. A good linear relationship between the anti-TTSuV1a and anti-TTSuV1b antibodies was observed (Spearman's rank correlation coefficient of 0.91; P < 0.0001) (Fig. 5B). When all 160 samples were included, a better agreement was obtained (Spearman's rank correlation coefficient of 0.93; P < 0.0001). A lesser degree of correlation between anti-TTSuV1a and anti-TTSuV2 or between anti-TTSuV1b and anti-TTSuV2 antibodies was found than the degree of correlation between anti-TTSuV1a and anti-TTSuV1b antibodies (data not shown). The results further revealed an association of seroprevalence and antibody levels between anti-TTSuV1a and anti-TTSuV1b antibodies, and thus, it is logical to hypothesize that there exists antigenic cross-reactivity between genotypes TTSuV1a and TTSuV1b.
Analysis of antigenic relationships among TTSuV1a, TTSuV1b, and TTSuV2 by ELISA.
Three antisera against the truncated recombinant ORF1s of TTSuV1a, TTSuV1b, or TTSuV2 were raised by the immunization of rabbits with the respective purified recombinant antigens. The specificity of these antisera was demonstrated by WB analysis using the TTSuV ORF1 antigens and the bacterial control (cell lysis product from bacteria harboring the empty expression vector) as well as by transfection experiments with PK-15 cells (see below) with the TTSuV ORF1 expression construct pTriEx1.1-Neo (see Fig. S1 in the supplemental material) (15).
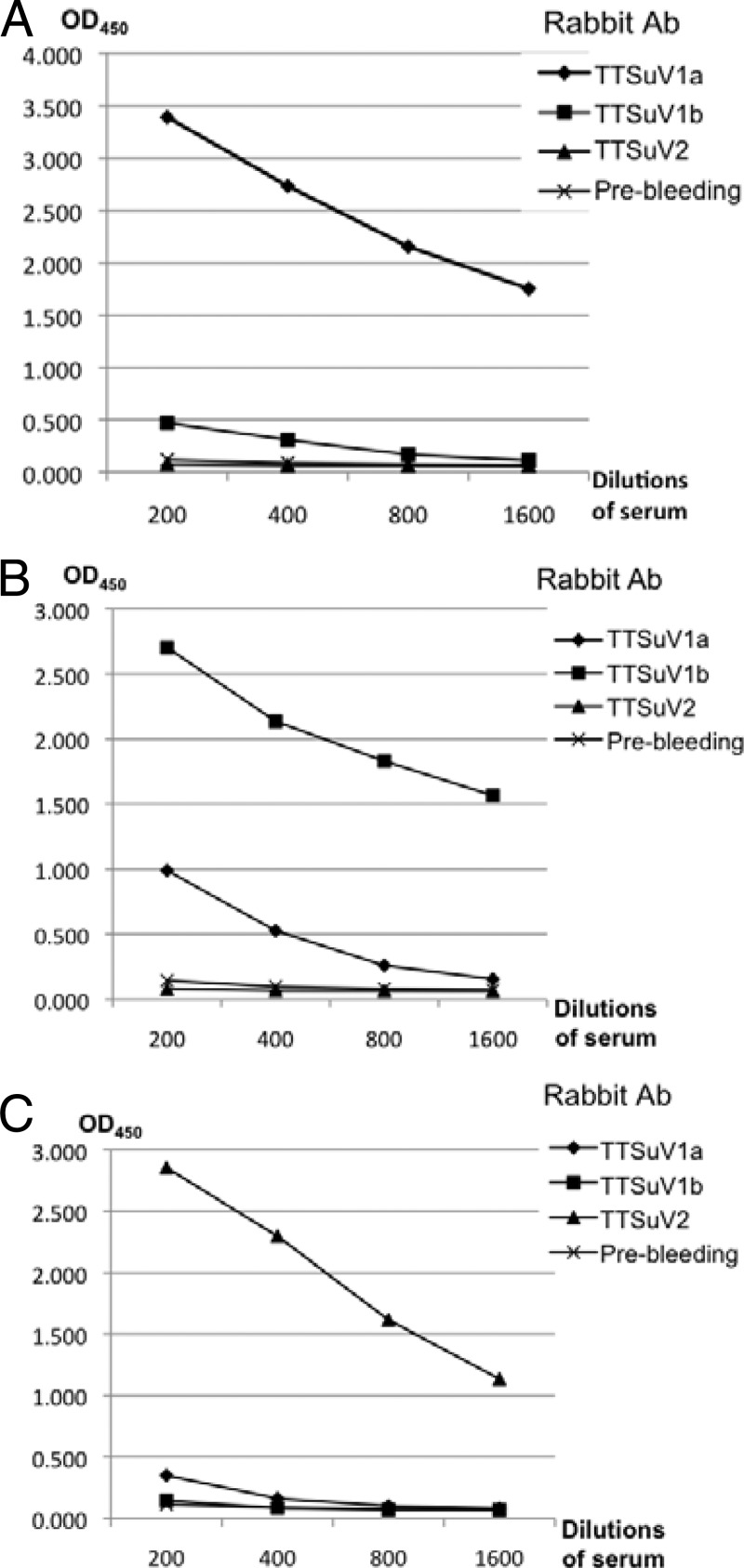
Cross-immunoreactivity studies were initially performed to assess whether one of these antigens could cross-react with antisera against the other two antigens in an ELISA format. The prebleed rabbit serum was used as the negative control. As expected, each of the three TTSuV antigens reacted with its corresponding homologous antiserum but not with the prebleed negative-control serum (OD values of <0.1) that was serially diluted 1:200 to 1:1,600 (Fig. 6).
Fig 6.
Reactivities of the three purified TTSuV ORF1 antigens of TTSuV1a (A), TTSuV1b (B), and TTSuV2 (C) with rabbit antisera against ORF1 of TTSuV1a, TTSuV1b, or TTSuV2 or with prebleed rabbit serum with 2-fold serial dilutions by ELISAs. Each antigen was tested against each serum sample in duplicate. Mean OD values are presented. Ab, antibody.
The TTSuV2 antigen did not appear to cross-react with TTSuV1a or TTSuV1b antiserum even at a 1:200 dilution, since the OD value was relatively low (Fig. 6C). In contrast, the TTSuV1b antigen did cross-react with the anti-TTSuV1a serum (as shown at 1:200 and 1:400 dilutions, both with OD values of >0.5) but not with the anti-TTSuV2 serum (Fig. 6B), whereas the TTSuV1a antigen likely cross-reacted with the anti-TTSuV1b serum (at a 1:200 dilution) but not with the anti-TTSuV2 serum (Fig. 6A). The ELISA results strongly supported our hypothesis that there is antigenic cross-reactivity between the two TTSuV1a and TTSuV1b genotypes but not between the two species TTSuV1a or -1b and TTSuV2.
Demonstration of antigenic relationships among TTSuV1a, TTSuV1b, and TTSuV2 and between TTSuVs and a genogroup 1 human TTV by IFA.
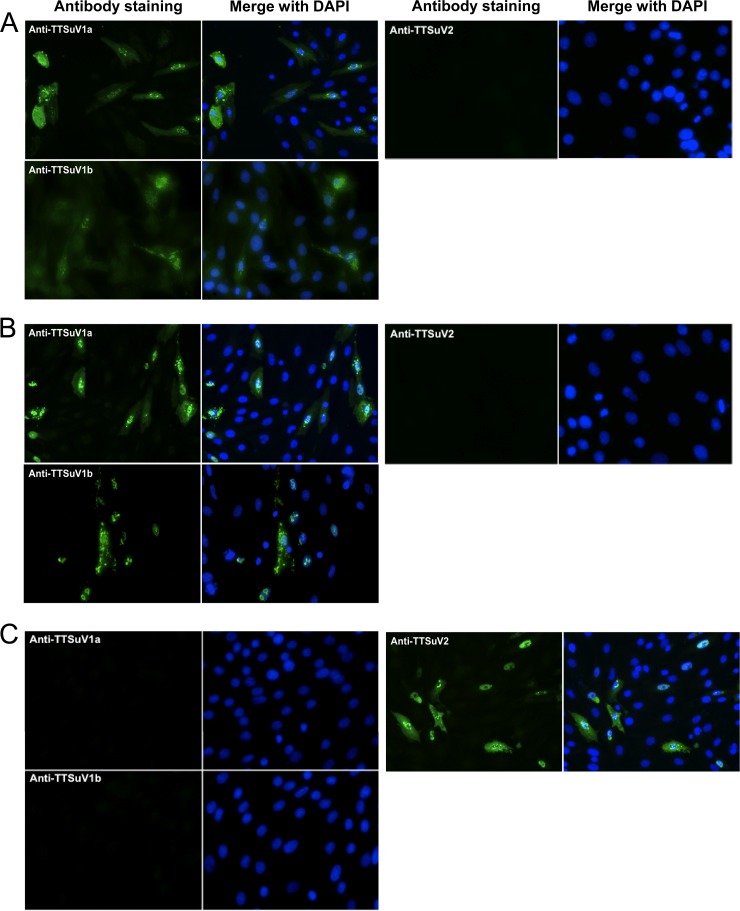
In order to analyze the antigenic cross-reactivity among these viruses more definitively, an antibody cross-reactivity experiment was performed by using IFA staining. PK-15 cells were transfected with three plasmid constructs, pTri-1aORF1, pTri-1bORF1, and pTri-2cORF1, which harbor the truncated ORF1 capsid genes from TTSuV1a, TTSuV1b, and TTSuV2, respectively. At 3 days posttransfection, cells were stained with anti-TTSuV1a, anti-TTSuV1b, anti-TTSuV2 antisera and prebleed serum. As shown in Fig. 7, cells transfected with pTri-1aORF1 (Fig. 7A) or pTri-1bORF1 (Fig. 7B) stained positive with both the anti-TTSuV1a and anti-TTSuV1b antisera but not with the anti-TTSuV2 antiserum (Fig. 7A and B) or the prebleed serum (data not shown), whereas cells transfected with pTri-2cORF1 reacted only with the anti-TTSuV2 serum (Fig. 7C). Each TTSuV1 antiserum reacted more strongly with its own homologous antigen than with the heterologous antigen based on a comparison of the positive cell numbers and fluorescence intensities (Fig. 7A and B). The truncated ORF1 proteins were expressed in both the nucleus and cytoplasm of the transfected cells (Fig. 7), which was different from what we found for cells transfected with full-length TTSuV DNA clones (16), probably due to the lack of most of the putative nuclear localization signals (NLSs) located at the N-terminal part of ORF1 in the truncated genes (by computer analysis) (data not shown). Table 2 summarizes the results of the cross-reactive immunostaining study. In addition, when transfected cells were each stained with an anti-human genogroup 1 TTV ORF1 antiserum (AK47, raised in rabbits), no fluorescent signal was detected. Mock-transfected cells did not stain with any of the five antisera (Table 2). The IFA result further confirmed the presence of antigenic cross-reactivity between TTSuV1a and TTSuV1b, as shown by the ELISA, but not between TTSuV1a or -1b and TTSuV2. The results also revealed that there was no antigenic cross-reactivity between genogroup 1 human TTV and porcine anelloviruses.
Fig 7.
Immunofluorescence assay (IFA) results for PCV1-free PK-15 cells transfected with plasmid pTri-1aORF1 (A), pTri-1bORF1 (B), or pTri-2cORF1 (C) at 3 days posttransfection. pTri-1aORF1-, pTri-1bORF1-, or pTri-2cORF1-transfected cells were stained with rabbit anti-TTSuV1a, anti-TTSuV1b, and anti-TTSuV2 ORF1 antisera, respectively. Alexa Fluor 488-conjugated goat anti-rabbit IgG (green) was used as the secondary antibody in the IFA. Antibody staining merged with nuclear staining using DAPI (blue) is also shown. Magnification, ×200.
Table 2.
Reactivities of anti-TTSuV1a, anti-TTSuV1b, anti-TTSuV2, prebleed rabbit, and anti-human TTV (AK47) sera in PCV1-free PK-15 cells transfected with plasmids encoding truncated ORF1 proteins from TTSuV1a, TTSuV1b, and TTSuV2, respectively, as determined by IFA
| Antibody transfection | Antibody reactivitya |
||||
|---|---|---|---|---|---|
| Anti-TTSuV1a | Anti-TTSuV1b | Anti-TTSuV2 | Prebleed rabbit serum | Anti-human TTV (AK47) | |
| pTri-1aORF1 | ++ | + | − | − | − |
| pTri-1bORF1 | + | ++ | − | − | − |
| pTri-2cORF1 | − | − | ++ | − | − |
| Mock | − | − | − | − | − |
The intensity of the fluorescent signal was determined visually and is expressed as ranging from − to ++.
Identification of two putative antigenic sites on ORF1 shared by TTSuV1a and TTSuV1b by sequence analyses.
The full-length ORF1 proteins of TTSuV1 and TTSuV2 shared only 22.4 to 25.8% amino acid sequence identity, with no significantly conserved regions being identified (15). The ORF1 proteins of the two TTSuV species share only 19.1 to 21.0% amino acid sequence identity with the sequence of human genogroup 1 TTV isolate P/1C1 (GenBank accession no. AF298585). The high degrees of ORF1 sequence divergences between TTSuV1 and TTSuV2 and between porcine and human anelloviruses likely account for the absence of antigenic cross-reactivity observed in this study.
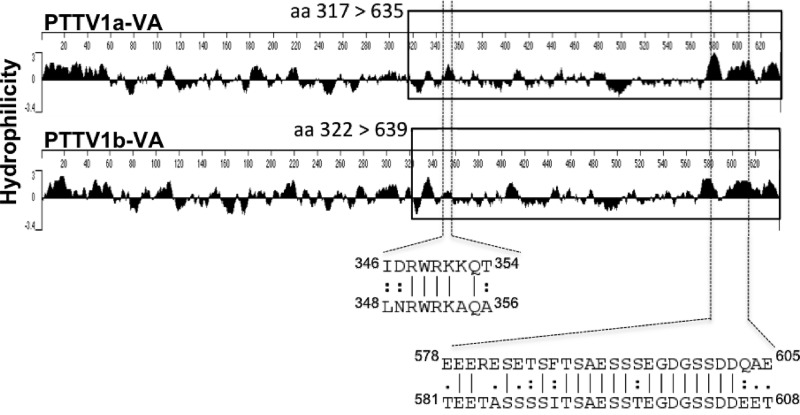
However, the amino acid sequence identity of the ORF1 proteins of genotypes TTSuV1a and TTSuV1b (six isolates available in GenBank) ranged between 49.4 and 52.4%. We previously found that conserved sites exist in the ORF1 proteins of different TTSuV1 strains, except for the four proposed variable regions (30.0 to 37.5% amino acid identity) (15). In order to identify the common antigenic sites on ORF1 between genotypes TTSuV1a and TTSuV1b, we performed a comparative analysis of the hydrophilicity profiles of the ORF1 amino acid sequences between PTTV1a-VA and PTTV1b-VA. Two conserved hydrophilic regions located at the middle and C-terminal regions were identified (Fig. 8). The C-terminal antigenic domain appeared to be more antigenic than the domain in the middle region. An alignment of the two putative antigenic regions among all published TTSuV1 sequences revealed a high degree of sequence conservation (data not shown).
Fig 8.
Comparison of hydrophilicity profiles of TTSuV1a (strain PTTV1a-VA) and TTSuV1b (strain PTTV1b-VA) ORF1 proteins and identification of two putative common antigenic domains in ORF1 of TTSuV1. The C-terminal region used for the expression of the truncated 1a- or 1b-ORF1 is indicated by a box. The corresponding alignment of amino acid sequences and amino acid positions of the two domains are also shown. Favorable mismatches of the amino acids are displayed as colons, whereas neutral mismatches are depicted as periods.
DISCUSSION
The immunology of anelloviruses is poorly understood (21). The detection of specific adaptive immune responses can provide insights into anellovirus epidemiology. By analogy to chicken anemia virus (CAV), another single-stranded circular DNA virus, the ORF1 product of anelloviruses is believed to function as the putative capsid protein and thus represents the major viral antigen (6, 21).
The detection of human TTV IgG antibodies in human populations based on human TTV ORF1 as the antigen was reported previously (21). In most cases, the successful expression of the human ORF1 antigen depends on the utilization of a strategy targeting the C-terminal region (12, 18, 22, 30). Most recently, our group successfully used the C-terminal fragment of the ORF1 protein of a U.S. strain of TTSuV2 as the antigen to detect TTSuV2-specific IgG antibodies in pig sera by an ELISA (14). Together with the present study, for serological detections of the two porcine TTV species 1 genotypes TTSuV1a and TTSuV1b, the obtained data suggest that the C-terminal portion of the ORF1 proteins of anelloviruses is an appropriate target for the development of serodiagnostic assays.
Indeed, based on the CAV virion structure determined by cryo-electron microscopic images, the C-terminal half of ORF1 was proposed to form the outer part of the capsid that is exposed to the virion surface, whereas the basic N-terminal part of CAV ORF1 was proposed to be inside the capsid to bind the viral DNA, and the middle part of ORF1 was proposed to form the inner shell of the capsid (6). The ORF1 polypeptide of anelloviruses was suggested previously to be organized in the same way as that of CAV (6). This proposed structure is consistent with a computer analysis of the ORF1 hydrophilicity profiles of TTSuV1 (Fig. 8) and TTSuV2 (14). In either case, there are two conserved major hydrophilic regions located at the middle and C-terminal regions that span the C-terminal half of ORF1 (14).
The reliability and specificity of the established ELISAs for differential TTSuV antibody detection were guaranteed by the screening of positive and negative reference sera through serum WB. This was further demonstrated by the triple seronegativity of TTSuV1a, TTSuV1b, and TTSuV2 in gnotobiotic pigs of group D (Fig. 2). However, it should be noted that the current definition of the cutoff value (S/N = 1.2) for the ELISAs may not completely normalize plate-to-plate variations. In a previous study, we utilized a more appropriate cutoff definition (the mean value of negative controls plus three times the standard deviation) for the TTSuV2 ELISA (14). Therefore, the cutoff value in this study may not cover 100% of the truly negative individuals in a given population. Future improvements of the TTSuV1 ELISA should include additional negative samples in every plate to allow for the determination of a more appropriate cutoff value. The presently reported ELISA results were checked for cross-reactivity by Western blot analyses of sera.
The viremia rate of TTSuV1 was low (31.9%) in comparison with that of TTSuV2 (59.9%) (14) in the 138 pigs. This may result from the low sensitivity of the TTSuV1 qPCR assay (10 copies/μl), the potential absence of extracted viral DNA from sera due to the cell-associated circulation of TTSuV1 in blood, or both. Therefore, by utilizing a more sensitive real-time qPCR assay such as the TaqMan probe-based approach or setting up an internal control for qPCR quantification using heparinized blood or plasma samples to test whether TTSuV is hidden in blood cells in a latent state will be needed in future studies.
A high seropositive rate for TTSuV1a (92.8%) or TTSuV1b (87.7%) was revealed in the 138 pigs of groups A to C (Table 1), which was higher than that for TTSuV2 (∼60%) (14), indicating a wider spread of actual TTSuV1 infection or the presence of long-persisting anti-TTSuV1 ORF1 antibodies in these pigs regardless of a low incidence of TTSuV1 viremia. Accordingly, these results, for the first time, provided serological evidence supporting dual infections by TTSuV1 and TTSuV2 in the same pigs. To our knowledge, this is also the first study demonstrating dual anellovirus infections in the same animals by using serological diagnosis in addition to the PCR assay. Therefore, the subsequent question raised was to determine the specificity of the seropositivity and cross-antigenic reactivity among different TTSuV species and genotypes.
In this study, we demonstrated by investigating four different aspects that, indeed, there exists antigenic cross-reactivity between genotypes TTSuV1a and TTSuV1b but not between the two TTSuV species (TTSuV1a or -1b and TTSuV2). First, compared to the serum samples with single TTSuV1a or TTSuV1b seropositivity, the number of serum samples with TTSuV1a/1b dual seropositivity was much higher (Table 1), likely implying a certain degree of cross-antigenic reactivity between the TTSuV1a and TTSuV1b antibodies. Second, the number of serum samples with dual TTSuV1a and TTSuV1b seropositivity was significantly higher than the number of samples with dual seropositivity to TTSuV1a and TTSuV2 or to TTSuV1b and TTSuV2 (Fig. 5A). In addition, a high correlation of antibody levels between anti-TTSuV1a and anti-TTSuV1b antisera, as assessed by Spearman's correlation coefficient, was observed (Fig. 5B). These analyses were conducted under the background of multiple TTSuV infections in field samples, which led us to propose a logical hypothesis regarding the presence of antigenic cross-reactivity between TTSuV1a and TTSuV1b. Third, this hypothesis was experimentally confirmed by an analysis of the antigenic relationships among TTSuV1a, TTSuV1b, and TTSuV2 through antigen-specific ELISAs (Fig. 6) and antibody cross-reactivity studies using PK-15 cells transfected with the three TTSuV ORF1 constructs, respectively (Fig. 7 and Table 2). Finally, a sequence comparison of ORF1 proteins of TTSuVs also supported the observed epidemiologic and experimental data in this study: while there was no significant sequence homology of TTSuV1a or -1b ORF1 with TTSuV2 ORF1, we identified two putative antigenic sites on ORF1 that are shared by TTSuV1a and TTSuV1b (Fig. 8).
In addition, in this study, we also demonstrated the absence of antigenic cross-reactivity between TTSuVs and a human genogroup 1 TTV by IFA. Taken together, the results from this study have important implications in predicting the antigenic cross-reactivities among different anelloviruses based on ORF1 amino acid sequence homology. Currently, anelloviruses are classified into nine genera according to the infected host species (human/ape, tamarin, douroucouli, tupaia, pig, dog, and cat), nucleotide sequence identity, and the genome size of primate anelloviruses (TTV, TTMV [torque teno minivirus], and TTMDV [torque teno midi virus]) (4). ORF1 of TTSuV (genus Iotatorquevirus) shares 15.6 to 22.3% amino acid sequence identity with the other eight genera based on multiple-sequence alignments (data not shown), which is similar to that between TTSuVs and human genogroup 1 TTV (19.1 to 21.0%). Therefore, it is reasonable to deduce that porcine anellovirus is not antigenically cross-reactive with other anelloviruses in other animal species. The ORF1 amino acid sequence homologies among the nine genera range from 15.0% to 27.3% (data not shown), thus implying that antigenic diversity between different genera does exist.
The two TTSuV species (TTSuV1 and TTSuV2) do not share antigenicity in the ORF1 antigen since they had only 22.4 to 25.8% amino acid sequence identity, whereas the two TTSuV1 genotypes (TTSuV1a and -1b) were antigenically related and cross-reactive due to their higher amino acid sequence homology (49.4 to 52.4%). It is possible that the antigenic relationship of different anelloviruses in the same genus may depend on a threshold or a range of amino acid sequence homologies. The available data using TTSuV as a model will provide insights into similar research on the antigenic diversity of human anelloviruses (TTV, TTMV, and TTMDV) in the future.
The present study of TTSuV1 together with our previous study of TTSuV2 (14) also revealed a broader picture of the nature of mixed TTSuV infections under natural or clinical disease conditions by assessing serological and virological profiles. It is not surprising to see in this study that several features of TTSuV1 infection were consistent with those of TTSuV2 (Fig. 3; see also Fig. S2 in the supplemental material) (14). More importantly, we provided new evidence to support the current opinion that TTSuV1 is likely not associated with PCVAD (1, 19, 24), by demonstrating that both viral loads and antibody levels were not significant different between PCVAD-affected and -unaffected pigs (Fig. 4; see also Fig. S2 in the supplemental material) and that there was no significant synergic PCV2/TTSuV1 effect. It is not known whether the presence of the ORF1 antibody is protective against homologous TTSuV infection (14). However, since antibodies to TTSuV1 or TTSuV2 ORF1 do not cross-react with the heterologous TTSuV antigen, it appears that TTSuV1 infection and the consequent humoral immune response do not interfere with TTSuV2 infection. Therefore, this may make the development of a single vaccine against the two recognized TTSuV species difficult. Together, the results from the present study have important implications in our understanding of the diversity of anelloviruses and in diagnosis and vaccine development for TTSuVs.
Supplementary Material
ACKNOWLEDGMENTS
This study is funded by a grant from Boehringer Ingelheim Vetmedica, Inc.
We thank Stephen Werre in the Statistical Services Laboratory of the Virginia-Maryland Regional College of Veterinary Medicine for his expert assistance with statistical analysis of the data. We thank Lijuan Yuan and Guohua Li of the Virginia-Maryland Regional College of Veterinary Medicine for generously providing us the gnotobiotic pig serum samples used in the study. We also thank Annette Mankertz at the Robert Koch Institute (Berlin, Germany) for kindly providing us the rabbit anti-human genogroup 1 TTV antiserum.
Footnotes
Published ahead of print 18 July 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Aramouni M, et al. 2011. Torque teno sus virus 1 and 2 viral loads in postweaning multisystemic wasting syndrome (PMWS) and porcine dermatitis and nephropathy syndrome (PDNS) affected pigs. Vet. Microbiol. 153:377–381 [DOI] [PubMed] [Google Scholar]
- 2. Ball JK, et al. 1999. TT virus sequence heterogeneity in vivo: evidence for co-infection with multiple genetic types. J. Gen. Virol. 80(Pt 7):1759–1768 [DOI] [PubMed] [Google Scholar]
- 3. Biagini P. 2009. Classification of TTV and related viruses (anelloviruses). Curr. Top. Microbiol. Immunol. 331:21–33 [DOI] [PubMed] [Google Scholar]
- 4. Biagini P, et al. 2011. Anelloviridae, p 331–341 In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed), Virus taxonomy. Ninth report of the International Committee on Taxonomy of Viruses Elsevier Academic Press, London, United Kingdom [Google Scholar]
- 5. Cortey M, Macera L, Segales J, Kekarainen T. 2011. Genetic variability and phylogeny of torque teno sus virus 1 (TTSuV1) and 2 (TTSuV2) based on complete genomes. Vet. Microbiol. 148:125–131 [DOI] [PubMed] [Google Scholar]
- 6. Crowther RA, Berriman JA, Curran WL, Allan GM, Todd D. 2003. Comparison of the structures of three circoviruses: chicken anemia virus, porcine circovirus type 2, and beak and feather disease virus. J. Virol. 77:13036–13041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de Villiers EM, Borkosky SS, Kimmel R, Gunst K, Fei JW. 2011. The diversity of torque teno viruses: in vitro replication leads to the formation of additional replication-competent subviral molecules. J. Virol. 85:7284–7295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ellis JA, Allan G, Krakowka S. 2008. Effect of coinfection with genogroup 1 porcine torque teno virus on porcine circovirus type 2-associated postweaning multisystemic wasting syndrome in gnotobiotic pigs. Am. J. Vet. Res. 69:1608–1614 [DOI] [PubMed] [Google Scholar]
- 9. Fenaux M, Opriessnig T, Halbur PG, Elvinger F, Meng XJ. 2004. A chimeric porcine circovirus (PCV) with the immunogenic capsid gene of the pathogenic PCV type 2 (PCV2) cloned into the genomic backbone of the nonpathogenic PCV1 induces protective immunity against PCV2 infection in pigs. J. Virol. 78:6297–6303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Forns X, et al. 1999. High prevalence of TT virus (TTV) infection in patients on maintenance hemodialysis: frequent mixed infections with different genotypes and lack of evidence of associated liver disease. J. Med. Virol. 59:313–317 [PubMed] [Google Scholar]
- 11. Gallei A, Pesch S, Esking WS, Keller C, Ohlinger VF. 2010. Porcine torque teno virus: determination of viral genomic loads by genogroup-specific multiplex RT-PCR, detection of frequent multiple infections with genogroups 1 or 2, and establishment of viral full-length sequences. Vet. Microbiol. 143:202–212 [DOI] [PubMed] [Google Scholar]
- 12. Handa A, Dickstein B, Young NS, Brown KE. 2000. Prevalence of the newly described human circovirus, TTV, in United States blood donors. Transfusion 40:245–251 [DOI] [PubMed] [Google Scholar]
- 13. Huang YW, et al. 2010. Development of SYBR green-based real-time PCR and duplex nested PCR assays for quantitation and differential detection of species- or type-specific porcine torque teno viruses. J. Virol. Methods 170:140–146 [DOI] [PubMed] [Google Scholar]
- 14. Huang YW, et al. 2011. Expression of the putative ORF1 capsid protein of torque teno sus virus 2 (TTSuV2) and development of Western blot and ELISA serodiagnostic assays: correlation between TTSuV2 viral load and IgG antibody level in pigs. Virus Res. 158:79–88 [DOI] [PubMed] [Google Scholar]
- 15. Huang YW, Ni YY, Dryman BA, Meng XJ. 2010. Multiple infection of porcine torque teno virus in a single pig and characterization of the full-length genomic sequences of four U.S. prototype PTTV strains: implication for genotyping of PTTV. Virology 396:289–297 [DOI] [PubMed] [Google Scholar]
- 16. Huang YW, et al. 2012. Rescue of a porcine anellovirus (torque teno sus virus 2) from cloned genomic DNA in pigs. J. Virol. 86:6042–6054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jelcic I, Hotz-Wagenblatt A, Hunziker A, Zur Hausen H, de Villiers EM. 2004. Isolation of multiple TT virus genotypes from spleen biopsy tissue from a Hodgkin's disease patient: genome reorganization and diversity in the hypervariable region. J. Virol. 78:7498–7507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kakkola L, et al. 2008. Expression of all six human torque teno virus (TTV) proteins in bacteria and in insect cells, and analysis of their IgG responses. Virology 382:182–189 [DOI] [PubMed] [Google Scholar]
- 19. Kekarainen T, Sibila M, Segales J. 2006. Prevalence of swine torque teno virus in post-weaning multisystemic wasting syndrome (PMWS)-affected and non-PMWS-affected pigs in Spain. J. Gen. Virol. 87:833–837 [DOI] [PubMed] [Google Scholar]
- 20. Leppik L, et al. 2007. In vivo and in vitro intragenomic rearrangement of TT viruses. J. Virol. 81:9346–9356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Maggi F, Bendinelli M. 2009. Immunobiology of the torque teno viruses and other anelloviruses. Curr. Top. Microbiol. Immunol. 331:65–90 [DOI] [PubMed] [Google Scholar]
- 22. Mueller B, Maerz A, Doberstein K, Finsterbusch T, Mankertz A. 2008. Gene expression of the human torque teno virus isolate P/1C1. Virology 381:36–45 [DOI] [PubMed] [Google Scholar]
- 23. Niel C, Saback FL, Lampe E. 2000. Coinfection with multiple TT virus strains belonging to different genotypes is a common event in healthy Brazilian adults. J. Clin. Microbiol. 38:1926–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nieto D, Aramouni M, Grau-Roma L, Segales J, Kekarainen T. 2011. Dynamics of torque teno sus virus 1 (TTSuV1) and 2 (TTSuV2) DNA loads in serum of healthy and postweaning multisystemic wasting syndrome (PMWS) affected pigs. Vet. Microbiol. 152:284–290 [DOI] [PubMed] [Google Scholar]
- 25. Ninomiya M, Takahashi M, Nishizawa T, Shimosegawa T, Okamoto H. 2008. Development of PCR assays with nested primers specific for differential detection of three human anelloviruses and early acquisition of dual or triple infection during infancy. J. Clin. Microbiol. 46:507–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nishizawa T, et al. 1997. A novel DNA virus (TTV) associated with elevated transaminase levels in posttransfusion hepatitis of unknown etiology. Biochem. Biophys. Res. Commun. 241:92–97 [DOI] [PubMed] [Google Scholar]
- 27. Okamoto H. 2009. History of discoveries and pathogenicity of TT viruses. Curr. Top. Microbiol. Immunol. 331:1–20 [DOI] [PubMed] [Google Scholar]
- 28. Okamoto H. 2009. TT viruses in animals. Curr. Top. Microbiol. Immunol. 331:35–52 [DOI] [PubMed] [Google Scholar]
- 29. Okamoto H, et al. 2002. Genomic characterization of TT viruses (TTVs) in pigs, cats and dogs and their relatedness with species-specific TTVs in primates and tupaias. J. Gen. Virol. 83:1291–1297 [DOI] [PubMed] [Google Scholar]
- 30. Ott C, et al. 2000. Use of a TT virus ORF1 recombinant protein to detect anti-TT virus antibodies in human sera. J. Gen. Virol. 81:2949–2958 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.