Abstract
Chimpanzees in west central Africa (Pan troglodytes troglodytes) are endemically infected with simian immunodeficiency viruses (SIVcpzPtt) that have crossed the species barrier to humans and gorillas on at least five occasions, generating pandemic and nonpandemic forms of human immunodeficiency virus type 1 (HIV-1) as well as gorilla SIV (SIVgor). Chimpanzees in east Africa (Pan troglodytes schweinfurthii) are also infected with SIVcpz; however, their viruses (SIVcpzPts) have never been found in humans. To examine whether this is due to a paucity of natural infections, we used noninvasive methods to screen wild-living eastern chimpanzees in the Democratic Republic of the Congo (DRC), Uganda, and Rwanda. We also screened bonobos (Pan paniscus) in the DRC, a species not previously tested for SIV in the wild. Fecal samples (n = 3,108) were collected at 50 field sites, tested for species and subspecies origin, and screened for SIVcpz antibodies and nucleic acids. Of 2,565 samples from eastern chimpanzees, 323 were antibody positive and 92 contained viral RNA. The antibody-positive samples represented 76 individuals from 19 field sites, all sampled north of the Congo River in an area spanning 250,000 km2. In this region, SIVcpzPts was common and widespread, with seven field sites exhibiting infection rates of 30% or greater. The overall prevalence of SIVcpzPts infection was 13.4% (95% confidence interval, 10.7% to 16.5%). In contrast, none of the 543 bonobo samples from six sites was antibody positive. All newly identified SIVcpzPts strains clustered in strict accordance to their subspecies origin; however, they exhibited considerable genetic diversity, especially in protein domains known to be under strong host selection pressure. Thus, the absence of SIVcpzPts zoonoses cannot be explained by an insufficient primate reservoir. Instead, greater adaptive hurdles may have prevented the successful colonization of humans by P. t. schweinfurthii viruses.
INTRODUCTION
Of over 40 African primate species naturally infected with simian immunodeficiency viruses (SIVs), chimpanzees (Pan troglodytes) are unique because they harbor the virus (SIVcpz) that spawned the human AIDS pandemic (20, 30, 78). It is now well established that chimpanzees are the original source of viruses currently found in chimpanzees, gorillas, and humans and that ape viruses have been transmitted to humans on at least four occasions, generating human immunodeficiency virus type 1 (HIV-1) groups M, N, O, and P (24, 64, 65). Chimpanzees also differ from most other primate species by having acquired their infection relatively more recently, as a consequence of cross-species transmission and recombination of viruses infecting monkeys on which they prey (3). Importantly, natural history studies have shown that SIVcpz is quite pathogenic and has a substantial negative impact on the health, reproduction, and life span of its natural host (17, 29, 54, 74). Thus, in addition to representing a potential source for human infection, SIVcpz also comprises a serious threat to chimpanzee populations living in the wild (54).
Chimpanzees are highly endangered, thus requiring noninvasive approaches to study SIVcpz infection in wild populations (59, 60). To address this, we have—over the past decade—developed approaches that permit the detection of virus-specific antibodies and nucleic acids in fecal and urine samples (29, 30, 35a, 41, 54, 55, 58, 59, 72, 73, 78, 79, 84). Examining the molecular epidemiology of SIVcpz in the wild, we found that only two of the four currently recognized subspecies (9), i.e., central (Pan troglodytes troglodytes) and eastern (Pan troglodytes schweinfurthii) chimpanzees but not western (Pan troglodytes verus) and Nigeria-Cameroonian (Pan troglodytes ellioti) chimpanzees, are naturally infected by SIVcpz (30, 35, 55, 58, 59, 78). We also found that SIVcpz is unevenly distributed among wild apes, with high prevalence rates (up to 50%) in some communities and rare or absent infection in others (29, 30, 54, 55, 59, 78). Molecular studies of existing SIVcpz strains revealed that they cluster in accordance to their subspecies origin in two highly divergent lineages, termed SIVcpzPtt and SIVcpzPts (65). Interestingly, all groups of HIV-1 as well as viruses (SIVgor) from western gorillas (Gorilla gorilla gorilla) fall into just one of these lineages, clustering with SIVcpzPtt from P. t. troglodytes, which implicates the central subspecies as the source of both human and gorilla infections (30, 65, 72, 79). In contrast, evidence for transmission of SIVcpzPts strains to either humans or sympatric eastern gorillas (Gorilla gorilla beringei) is lacking (41), raising questions as to the relative abundance and distribution of SIVcpz in wild-living P. t. schweinfurthii populations (45).
Eastern chimpanzees live in central and east Africa (Fig. 1A) in an area that ranges from the southeastern parts of the Central African Republic (CAR) through the northern parts of the Democratic Republic of Congo (DRC) to southwestern Sudan and the western parts of Uganda, Rwanda, Burundi, and Tanzania (50). The first SIVcpzPts strain (ANT) was identified in an ape captured at an unknown location in the DRC and exported to Belgium (20, 44, 76). Subsequent testing of fecal samples from a limited number of chimpanzees in the vicinity of Kisangani indicated that SIVcpzPts was also present in wild-living P. t. schweinfurthii communities in the DRC (84). However, extensive field studies in east Africa revealed a surprising paucity of SIVcpzPts infections. Although infected chimpanzees were identified in Gombe National Park (GM) (29, 54, 59, 60) and the Ugalla region (UG) of western Tanzania (55), communities in the Budongo Forest (BG) in northern Uganda, the Kibale National Park (KB) in western Uganda, the Bwindi Impenetrable Forest (BW) in southern Uganda, and Mahale Mountains National Park (MH) in Tanzania seemed free of SIVcpz infection (55, 59, 66). Moreover, none of over 300 fecal samples from the Nyungwe Forest Reserve (NY) in western Rwanda was virus positive (66). Thus, in contrast to the high infection rates observed for P. t. troglodytes apes in southern Cameroon (30, 41, 78) and northern Gabon (35a) field studies at the eastern limits of the P. t. schweinfurthii range identified only isolated foci of infection.
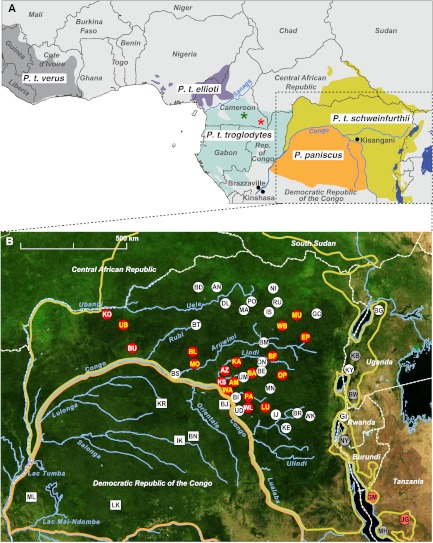
Fig 1.
Location of ape study sites. (A) Geographic ranges of chimpanzees (Pan troglodytes) and bonobos (Pan paniscus) in Saharan Africa. The four recognized chimpanzee subspecies are color coded (P. t. verus, gray; P. t. ellioti, purple; P. t. troglodytes, blue; P. t. schweinfurthii, yellow). International borders, major rivers and lakes, and select cities are shown. Asterisks indicate where the closest SIVcpz relatives of HIV-1 groups M (red) and N (green) were identified in wild-living P. t. troglodytes communities (30). A box outlines the study area, which is magnified in panel B. (B) Location of chimpanzee (circles) and bonobo (squares) study sites in the DRC, Uganda, and Rwanda. The ranges of eastern chimpanzees (yellow) and bonobos (orange) are shown as in panel A. Sites where SIVcpz was detected are indicated in red, with white and yellow lettering denoting the recovery of antibody-positive and antibody- and nucleic acid-positive samples, respectively. Previously published SIVcpz-positive and -negative sites in Uganda, Rwanda, and Tanzania are shown in dark red and gray, respectively. Forested areas are shown in green, while arid and semiarid areas are in yellow and brown. Major lakes are shown in black, with major rivers depicted in blue. Dashed white lines indicate national boundaries.
The habitat of eastern chimpanzees in Uganda, Rwanda, and Tanzania is severely fragmented (Fig. 1B) due to extensive deforestation, expanding agriculture, and human encroachment (50). To test whether this habitat loss might have contributed to the paucity of SIVcpzPts infections in east Africa, we targeted wild-living apes in the Democratic Republic of the Congo, a country that is home to half of the world's remaining chimpanzees (10, 50). There are two different chimpanzee species in the DRC, the eastern chimpanzee (Pan troglodytes schweinfurthii) and the bonobo (Pan paniscus), who live in nonoverlapping ranges north and south of the Congo River, respectively (Fig. 1A). Current population estimates suggest that there may be as many as 180,000 to 200,000 eastern chimpanzees and 30,000 to 50,000 bonobos still remaining in largely intact forest blocks (10, 19, 50). With the exception of one study (84), wild-living apes in the DRC have not previously been surveyed for SIV. Here, we show that eastern chimpanzees, but not bonobos, are widely and commonly infected with SIVcpzPts and thus represent a substantial virus reservoir.
MATERIALS AND METHODS
Study sites and sample collection.
The vast majority of ape fecal samples were collected in the DRC from nonhabituated eastern chimpanzees (n = 2,480) and bonobos (n = 543) by teams of local trackers (Table 1). Samples were collected in the vicinity of chimpanzee night nests or when encountered during forest walks, placed into 50-ml conical tubes, and preserved in an equal volume of RNAlater (Life Technologies) as described previously (30, 59, 78). Tubes were labeled with a sample number, the field site code, and GPS coordinates when available. Because local trackers were participating in the collection effort, the quality of samples varied between field sites and individual specimens were frequently divided into multiple aliquots without this being indicated. In four instances, samples were collected from pet chimpanzees kept by local villagers (BU203, KS133, KS134, and KS135). Samples were also collected from habituated chimpanzee communities in the Budongo Reserve (BG) in northern Uganda (n = 20) (52) and the Kyambura Gorge (KY) in western Uganda (n = 16) (32). At the latter two sites, fecal samples were collected from individually known apes under direct observation by resident primatologists. Finally, samples were obtained from nonhabituated chimpanzees in the Gishwati Forest (GI) in northwestern Rwanda (n = 49) (12, 49). Because of a lack of refrigeration at most field sites, especially in the DRC, samples were kept at ambient temperature for various periods of time (usually several weeks, but in some instances several months) before they could be stored at −20°C. In the DRC, this was done at a central laboratory in Kisangani, where samples were then batched and shipped to the United States. Bonobo samples from the Malebo (ML) field site were stored at −80°C at the Institut National de Recherche Biomédicale in Kinshasa and then shipped directly to the University of Montpellier.
Table 1.
Prevalence of SIVcpz infection in wild-living eastern chimpanzees and bonobosa
| Species and subspecies | Field siteb | No. of fecal samples collected | No. of fecal samples positive for mtDNA | Proportion of degraded/mixed samples | No. of individuals sampledc | No. of mtDNA haplotypesd | No. of SIVcpz antibody-positive samples | No. of infected individualse | No. of vRNA-positive samples | SIVcpz prevalence (%)f | 95% confidence interval of SIVcpz prevalence |
|---|---|---|---|---|---|---|---|---|---|---|---|
| P. t. schweinfurthii | Amunyala (AM) | 44 | 37 | 0.16 | NA | 5 | 15 | 3 | 3 | 33 | 2–43 |
| Ango (AN) | 16 | 15 | 0.06 | NA | 1 | 0 | 0 | 0 | 0 | 0–60 | |
| Azunu (AZ) | 6 | 5 | 0.17 | 3 | 2 | 4 | 2 | 0 | 67 | 9–99 | |
| Bafwaboli (BA) | 256 | 229 | 0.11 | NA | 22 | 39 | 5 | 1 | 9 | 3–20 | |
| Bondo (BD) | 15 | 15 | 0.00 | NA | 9 | 0 | 0 | 0 | 0 | 0–34 | |
| Baego (BE) | 134 | 87 | 0.35 | NA | 18 | 0 | 0 | 0 | 0 | 0–17 | |
| Bafwasende (BF) | 42 | 37 | 0.12 | NA | 7 | 12 | 1 | 12 | 11 | 0–48 | |
| Babingi (BI) | 124 | 103 | 0.17 | NA | 14 | 0 | 0 | 0 | 0 | 0–14 | |
| Bongbola (BL) | 50 | 48 | 0.04 | NA | 21 | 3 | 2 | 2 | 10 | 1–30 | |
| Bomili (BM) | 47 | 43 | 0.09 | NA | 2 | 0 | 0 | 0 | 0 | 0–31 | |
| Biruwe (BR) | 59 | 59 | 0.00 | NA | 1 | 0 | 0 | 0 | 0 | 0–23 | |
| Basoco (BS) | 20 | 10 | 0.50 | NA | 8 | 0 | 0 | 0 | 0 | 0–37 | |
| Buta (BT) | 5 | 4 | 0.20 | NA | 4 | 0 | 0 | 0 | 0 | 0–60 | |
| Bumba (BU) | 1 | 1 | 0.00 | NA | 1 | 1 | 1 | 0 | 100 | 3–100 | |
| Dingila (DL) | 5 | 5 | 0.00 | NA | 1 | 0 | 0 | 0 | 0 | 0–98 | |
| Epulu (EP) | 160 | 126 | 0.21 | NA | 19 | 7 | 2 | 4 | 7 | 1–22 | |
| Gombari (GO) | 5 | 2 | 0.60 | NA | 2 | 0 | 0 | 0 | 0 | 0–84 | |
| Ijela (IJ) | 27 | 7 | 0.74 | NA | 2 | 0 | 0 | 0 | 0 | 0–84 | |
| Isiro (IS) | 5 | 4 | 0.20 | NA | 3 | 0 | 0 | 0 | 0 | 0–71 | |
| Kabuka (KA) | 164 | 126 | 0.23 | NA | 23 | 33 | 13 | 10 | 43 | 25–63 | |
| Kasese (KE) | 15 | 7 | 0.53 | NA | 3 | 0 | 0 | 0 | 0 | 0–71 | |
| Kotakoli (KO) | 90 | 78 | 0.13 | NA | 12 | 9 | 1 | 0 | 6 | 0–27 | |
| Kisangani (KS) | 13 | 11 | 0.15 | NA | 8 | 4 | 3 | 0 | 38 | 9–76 | |
| Lubutu (LU) | 212 | 163 | 0.23 | NA | 18 | 34 | 3 | 27 | 8 | 2–21 | |
| Mawa Gare (MA) | 11 | 7 | 0.36 | NA | 1 | 0 | 0 | 0 | 0 | 0–84 | |
| Mount Mogogoli (MN) | 2 | 1 | 0.50 | NA | 1 | 0 | 0 | 0 | 0 | 0–98 | |
| Mongandjo (MO) | 21 | 15 | 0.29 | NA | 10 | 2 | 1 | 2 | 10 | 0–45 | |
| Mungbere (MU) | 58 | 53 | 0.09 | NA | 11 | 26 | 6 | 3 | 50 | 21–79 | |
| Niangara (NI) | 7 | 4 | 0.43 | NA | 2 | 0 | 0 | 0 | 0 | 0–84 | |
| Onga (ON) | 59 | 40 | 0.32 | NA | 4 | 0 | 0 | 0 | 0 | 0–34 | |
| Opienge (OP) | 85 | 61 | 0.28 | NA | 9 | 26 | 2 | 18 | 14 | 2–43 | |
| Parisi (PA) | 131 | 110 | 0.16 | NA | 16 | 48 | 12 | 5 | 46 | 27–67 | |
| Poko (PO) | 11 | 10 | 0.09 | NA | 6 | 0 | 0 | 0 | 0 | 0–46 | |
| Rungu (RU) | 18 | 13 | 0.28 | NA | 4 | 0 | 0 | 0 | 0 | 0–60 | |
| Ubangi (UB) | 117 | 99 | 0.15 | NA | 23 | 11 | 7 | 3 | 30 | 13–53 | |
| Ubundu (UD) | 5 | 5 | 0.00 | NA | 2 | 0 | 0 | 0 | 0 | 0–84 | |
| Uma (UM) | 100 | 61 | 0.39 | NA | 10 | 0 | 0 | 0 | 0 | 0–23 | |
| Wanie Rukula (WA) | 170 | 151 | 0.11 | NA | 25 | 40 | 8 | 1 | 22 | 10–39 | |
| Wamba (WB) | 91 | 69 | 0.24 | NA | 11 | 8 | 3 | 1 | 19 | 4–46 | |
| Walikale (WK) | 35 | 33 | 0.06 | NA | 10 | 0 | 0 | 0 | 0 | 0–31 | |
| Walengola (WL) | 44 | 39 | 0.11 | NA | 13 | 1 | 1 | 0 | 8 | 0–36 | |
| Budongo (BG) | 20 | 19 | 0.05 | 19 | 9 | 0 | 0 | 0 | 0 | NA | |
| Kyambura Gorge (KY) | 16 | 16 | 0.00 | 13 | 4 | 0 | 0 | 0 | 0 | NA | |
| Gishwati (GI) | 49 | 42 | 0.14 | NA | 6 | 0 | 0 | 0 | 0 | 0–31 | |
| All sites (n = 44) | 2,565 | 2,070 | 0.19 | 567i | 383 | 323 | 76 | 92 | 13.4 | 10.7–16.5 | |
| P. paniscus | Bayandjo (BJ) | 2 | 2 | 0 | NA | 1 | 0 | 0 | 0 | 0 | 0–98 |
| Balanga (BN) | 96 | 85 | 0.12 | NA | 7 | 0 | 0 | 0 | 0 | 0–8 | |
| Ikela (IK) | 56 | 39 | 0.30 | 17 | 7 | 0 | 0 | 0 | 0 | 0–20 | |
| kokolopori (KR) | 78 | 69 | 0.12 | 38 | 12 | 0 | 0 | 0 | 0 | 0–9 | |
| Lui Kotale (LK) | 43 | 38 | 0.12 | 17 | 8 | 0 | 0 | 0 | 0 | 0–20 | |
| Malebo (ML) | 268 | 262 | 0.02 | NA | 6 | 0 | 0 | 0 | 0 | 0–3 | |
| All sites (n = 6) | 543 | 495 | 0.09 | 244i | 41 | 0 | 0 | 0 | 0 | 0–1.5 |
vRNA, virion RNA; NA, not available.
Field site locations are shown in Fig. 1.
Chimpanzees at the BG and KY field sites were individually known. For the AZ, IK, KR, and LK field sites, the number of sampled apes was determined by microsatellite analyses (see Tables S3 and S4 in the supplemental material).
Minimum number of individuals, as indicated by the number of distinct mtDNA haplotypes.
SIVcpz-infected chimpanzees were enumerated for each field site by microsatellite analysis of antibody-positive fecal samples (see Table S3 in the supplemental material).
Prevalence of SIVcpz infection (%) with 95% confidence intervals. Values are based on the proportions of SIVcpz antibody-positive fecal samples, corrected for sample degradation and oversampling.
The single sample from the BU field site was collected from a pet chimpanzee.
Samples from the KS field site include three from pet chimpanzees.
Estimated number of sampled individuals (see Materials and Methods for details).
SIVcpz antibody detection.
All fecal samples were screened for the presence of HIV-1 cross-reactive antibodies as previously described (30, 41, 59, 78). Bonobo samples from the ML field site were tested using the Inno-LIA HIV I/II Score test (Innogenetics, Ghent, Belgium) (78). All other samples were examined by an enhanced chemiluminescence (ECL) Western immunoblot analysis modified for RNAlater-preserved specimens. RNAlater is a high-salt solution (25 mM sodium citrate, 10 mM EDTA, and 70 g ammonium sulfate per 100 ml of solution; pH 5.2) that preserves nucleic acids but precipitates proteins, including immunoglobulin. To prepare extracts suitable for a Western blot analysis, feces-RNAlater mixtures (1.5 ml) were diluted with phosphate-buffered saline (PBS)-Tween 20 (8.5 ml), inactivated for 1 h at 60°C, clarified by centrifugation (3,500 × g for 30 min) to remove solid debris, and then dialyzed against PBS overnight at 4°C to reconstitute fecal immunoglobulin. Reconstituted extracts were subjected to an immunoblot analysis using commercially available HIV-1 antigen-containing strips (Maxim Biomedical, Inc.). Sample integrity was examined using an IgG control.
Amplification of SIVcpz virion RNA.
All Western blot-positive samples were tested for the presence of SIV nucleic acids by reverse transcription-PCR (RT-PCR) amplification as described previously (30, 37, 41, 59, 78). Briefly, fecal RNA was extracted using the RNAqueous-Midi Kit (Life Technologies) and subjected to RT-PCR amplification using SIVcpz-specific gag, pol, vpu, gp41, and gp41/nef consensus primers (see Table S1 in the supplemental material). PCR conditions generally included 60 cycles of denaturation (94°C, 20 s), annealing (50°C, 30 s), and elongation (68°C, 1.5 min) for the first round. Second-round conditions included 50 cycles of denaturation (94°C, 20 s), annealing (52°C, 30 s), and elongation (68°C, 1 min). All amplicons were gel purified and sequenced directly. Samples that failed to yield SIVcpz amplicons were reamplified using pan-SIV-specific primers (see Table S1 in the supplemental material) so as to not miss infection with SIVs other than SIVcpz.
Individual identification.
All fecal samples were subjected to mitochondrial (mt) DNA analysis to confirm their species and subspecies origin. In addition, all antibody-positive P. t. schweinfurthii samples as well as 146 bonobo samples from three different field sites (IK, LK, and KR) were subjected to microsatellite analyses. Fecal DNA was extracted as described and used to amplify a 498-bp fragment of the mitochondrial D-loop region (30, 38, 59). Amplicons were sequenced directly (using one primer which yielded 479 bp of sequence) and classified into distinct mitochondrial haplotypes (see Fig. S1 and Table S2 in the supplemental material). To identify the number of sampled individuals, fecal samples were genotyped at four (P. t. schweinfurthii) or eight (P. paniscus) autosomal microsatellite loci (see Tables S3 and S4 in the supplemental material), with amplification products sized on an automated sequencer using GeneMapper 4.0 (Applied Biosystems). Samples were first grouped by field site and mtDNA haplotype. Within each haplotype, samples were then grouped by microsatellite genotypes and, when possible, also by gender and viral genotype. Due to prolonged storage at ambient temperatures, even mtDNA-positive samples were frequently partially degraded. We thus allowed allelic mismatches at up to four (eastern chimpanzees) or six (bonobos) loci if other markers (mtDNA haplotype, gender) indicated possible identity. This very conservative approach likely resulted in an underestimation of the number of sampled individuals. Samples with evidence of DNA admixture (multiple peaks for the same locus or double peaks in the mtDNA sequence) were excluded.
Gender determination.
For most samples, gender was determined by amplifying a 218-bp fragment of the amelogenin gene that contains a 6-bp insertion in the Y, but not the X, chromosome using primers AMEL-F212 (5′-ACCTCATCCTGGGCACCCTGG-3′) and AMEL-R212 (5′-AGGCTTGAGGCCAACCATCAG-3′) (69). PCR conditions were the same as those for the microsatellite analyses, and fragments were sized on an automated sequencer. For samples that failed this genotyping, a second set of amelogenin primers (AMXY-1F [5′-CTGATGGTTGGCCTCAAGCCTGTG-3′] and AMXY-2R [5′-TAAAGAGATTCATTAACTTGACTG-3′]) were used to amplify a 977-bp fragment from the X chromosome and a 788-bp fragment from the Y chromosome. The resulting PCR products were sized by 1.2% agarose gel electrophoresis (16, 59).
SIVcpz prevalence determination.
The prevalence of SIVcpz infection was estimated for each field site based on the proportion of SIVcpz antibody-positive fecal samples, correcting for degradation and redundant sampling. As shown in Table 1, 19% of eastern chimpanzee and 9% of bonobo fecal samples failed to yield usable mtDNA sequences and were thus excluded from further analysis. A subset of the remainder was then subjected to microsatellite analyses, which provided a quantitative estimate of oversampling for both species. For P. t. schweinfurthii, genotyping of 323 antibody-positive fecal samples from 19 field sites identified 76 infected individuals (Table 1), indicating that each had been sampled, on average, 4.25 times (see Table S3 in the supplemental material). For bonobos, analysis of 146 fecal samples from three field sites identified 72 individuals (Table 1), indicating that each had been sampled, on average, 2.03 times (see Table S4 in the supplemental material). Using these corrections, the proportion of SIVcpz-infected chimpanzees was estimated for each sampling location, taking into account the number of unique mtDNA haplotypes as an indicator of the minimum number of chimpanzees tested. From these determinations, prevalence rates were calculated (95% confidence limits were calculated based on binomial sampling).
Full-length genome amplification.
The full-length genome of one representative SIVcpzPts strain from the DRC (BF1167) was sequenced as described previously (58, 78). Briefly, partially overlapping subgenomic fragments were amplified from fecal RNA, gel purified, and sequenced directly. Chromatograms were examined for positions of base mixtures, and ambiguous sites were resolved as previously reported (7, 55, 72, 73). For positions that did not affect the corresponding amino acid, the predominant nucleotide (highest amplitude in the sequence chromatogram or the most frequent nucleotide in repeat sequencing reactions) was chosen. For positions that affected the corresponding amino acid, the base that encoded the most common amino acid residue in alignments of existing SIVcpz protein sequences was selected. In the absence of an apparent common amino acid (e.g., in hypervariable protein regions), the nucleotide with the highest amplitude in sequence chromatograms was selected. Using these criteria, we were able to infer a unique consensus sequence for BF1167.
Construction of a replication-competent SIVcpzPts molecular clone.
To obtain a full-length infectious molecular clone of BF1167, the consensus sequence was chemically synthesized as three subgenomic fragments (Blue Heron Biotechnology). These included a 3.7-kb 5′ long terminal repeat (LTR)-pol fragment, a 3.9-kb pol-env fragment, and a 2.3-kb env-3′ LTR fragment. To facilitate subsequent cloning, unique MluI and ApaI sites were added to the 5′ and 3′ termini of the provirus, respectively. These, together with internal NcoI and SalI sites at positions 3,728 and 7,596, respectively, were used to assemble the three subgenomic fragments to generate a full-length provirus. Ligation products were used to transform XL2-MRF bacteria (Stratagene). Resulting transformants were screened for appropriately sized inserts, transfected into 293T cells, and tested for infectivity in the TZM-bl assay (73) One functional clone (pBF1167) was identified and grown on a large scale (available from the National Institutes of Health Research and Reference Program, Rockville, MD).
BF1167 infectivity and coreceptor usage.
BF1167 and TAN2 (SIVcpz) and SG3 (HIV-1) reference clones were transfected into 293T cells, and supernatants were equilibrated by particle-associated reverse transcriptase activity as described previously (7, 73). Viral infectivity was assessed in TZM-bl cells, a HeLa-derived line which has been genetically modified to constitutively express human CD4, CCR5, and CXCR4 and to contain integrated luciferase and β-galactosidase reporter genes under the control of an HIV-1 LTR (48, 83). For coreceptor analysis, TZM-bl cells were seeded in 96-well plates at 8,300 cells/well overnight and then treated with the CCR5 antagonist TAK-779 (10 μM), the CXCR4 antagonist AMD3100 (1.2 μM), or a combination of both for 1 h (73). Virus was added in the presence of 40 μg/ml DEAE-dextran and removed 48 h later. Cells were then lysed and analyzed for luciferase activity (Promega) using a Tropix luminometer with WinGlow version 1.24 software.
CD4 T cell cultures.
Blood was obtained from normal human volunteers and healthy (SIV/HIV-1-uninfected) chimpanzees housed at the Yerkes Regional Primate Center as described previously (chimpanzee blood samples were leftover specimens from the annual health check-up) (14). Briefly, peripheral blood mononuclear cells were isolated using Ficoll Hypaque Plus (GE Healthcase). CD4+ T cells were enriched using CD4 microbeads and magnetic cell sorting (Militenyi Biotec), stimulated with staphylococcal enterotoxin B (Sigma-Aldridge) for 12 to 15 h (3 μg/ml), and subsequently cocultivated with autologous monocyte-derived macrophages for optimal activation (14). After 5 to 6 days, CD4+ T cells were removed from the macrophages, placed into Dulbecco's modified Eagle medium (DMEM) with 10% fetal calf serum (FCS), and incubated with 30 U/ml interleukin-2 (IL-2). After 24 h, 5 × 105 CD4+ T cells were incubated with transfection-derived viral stocks at a multiplicity of infection (MOI) of 0.1 (as determined on TZM-bl cells) in 300 μl DMEM containing 10% FCS and 30 U/ml IL-2 for 16 h. CD4+ T cells were washed three times and plated in 24-well plates in DMEM with 10% FCS and 30 U/ml IL-2, and reverse transcriptase activity was measured in culture supernatants every 3 days to monitor viral replication.
Phylogenetic analyses.
Partial pol (232-bp and 892-bp), vpu/env (481- to 514-bp), gp41 (325- to 465-bp), and gp41/nef (699- to 1,259-bp) sequences from the newly characterized viruses were aligned with HIV-1, SIVcpz, and SIVgor reference sequences (GenBank accession numbers K03455 for HIV-1 group M HXB2; AJ006022 for HIV-1 group N YBF30; L20587 for HIV-1 group O ANT70; HQ179987 for HIV-1 group P U14788; DQ373065 for SIVcpzPtt EK505, DQ373063 for SIVcpzPtt MB66, DQ373064 for SIVcpzPtt LB7, DQ373066 for SIVcpzPtt MT145, AF103818 for SIVcpzPtt US, AY169968 for SIVcpzPtt CAM13, X52154 for SIVcpzPtt GAB1, AF115393 for SIVcpzPtt CAM3, AJ271369 for SIVcpzPtt CAM5, FJ424866 for SIVgor CP2139, AF447763 for SIVcpzPts TAN1, DQ374657 for SIVcpzPts TAN2, DQ373065 for SIVcpzPts TAN3, JN091691 for SIVcpzPts TAN5, JQ768416 for SIVcpzPts TAN13, JN091690 for SIVcpzPts UG38, and U42720 for SIVcpzPts ANT) using Clustal W (34). Regions of the sequences that could not be unambiguously aligned were removed from further analyses. For SIVcpzBF1167, deduced Gag, Pol, Vif, and Env sequences were aligned with the corresponding protein sequences of the same HIV-1, SIVcpz, and SIVgor reference strains. Gag/Pol and Pol/Vif protein overlaps were removed from the N and C termini of the deduced Pol protein sequences. In addition, the concatenated Pol and Vif alignment was divided into two regions around a previously reported recombination breakpoint (20, 67). Appropriate evolutionary models for phylogenetic analyses were selected using ModelTest version 3.7 (51) and ProtTest version 2.4 (1). For nucleotide sequence analyses, these were K80+G for the diagnostic 232-bp pol fragment and GTR+I+G for the longer pol, vpu/env, gp41, and gp41/nef fragments. For amino acid sequence analyses, the chosen models were LG+I+G (Gag), RtREV+I+G+F (Pol), LG+I+G+F (Pol/Vif), and WAG+I+G+F (Env). Phylogenetic trees were constructed using maximum likelihood (22) and Bayesian (53) methods, with the latter using a 25% burn-in and using as a convergence criterion an average standard deviation of partition frequencies of <0.01.
For species and subspecies analysis, mtDNA control region (D-loop) sequences were aligned and identical sequences were grouped into haplotypes (see Table S2 in the supplemental material). The evolutionary relationships of the new haplotypes to each other and to reference sequences from the database were then determined by phylogenetic analysis. A neighbor-joining tree is shown in Fig. S1 in the supplemental material.
Nucleotide sequence accession numbers.
New SIVcpzPts sequences have been deposited in GenBank under accession numbers JQ866001, JQ866003 to JQ866011, JQ866013 to JQ866017, JQ866024, JQ866026 to JQ866041, JQ866043 to JQ866045, JQ866047 to JQ866052, JQ866055, JQ866057 to JQ866059, JQ866061 to JQ866066, JQ866068 to JQ866070, and JX178444 to JX178449. The new P. t. schweinfurthii and Pan paniscus mitochondrial D-loop sequences are listed in Table S2 in the supplemental material (deposited in GenBank under accession numbers JQ866072 to JQ866157 and JQ866159 to JQ866296).
RESULTS
SIVcpz infection is endemic and widespread among eastern chimpanzees.
To determine the geographic distribution and prevalence of SIVcpz in the Democratic Republic of the Congo (DRC), we conducted a comprehensive, noninvasive (fecal-based) survey at 41 different collection sites (Fig. 1B). Between January 2001 and May 2011, we obtained a total of 2,480 fecal samples north of the Congo River in an area spanning almost 250,000 km2. Given the vastness of this study area, most samples were collected by local trackers in the vicinity of their villages. While this led to some variation in sample quality, we were able to obtain samples as far south as Kasese (KE), as far east as Gombari (GO) and Walikale (WK), as far north as Bondo (BD) and Niangara (NI), and as far west as Kotakoli (KO) and Bumba (BU) (Fig. 1B). Except for four samples from pet chimpanzees, all other specimens were obtained from wild-living apes within their natural habitat (Table 1). We also obtained samples from habituated P. t. schweinfurthii apes in the Budongo Forest (BG) (52) and the Kyambura Gorge (KY) in Uganda and from a nonhabituated group in the Gishwati Forest (GI) in Rwanda (49). The rationale for these surveys was to examine whether previous studies had missed isolated foci of SIVcpz infection in east Africa. Since the latter field sites were well established, sample collection and storage occurred under more-controlled conditions.
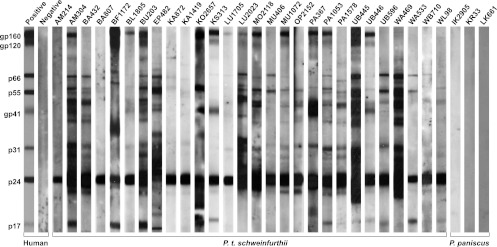
Before testing for SIVcpz antibodies and nucleic acids, all specimens were subjected to mitochondrial (mt) DNA analysis to confirm their species origin and assess their integrity. This analysis identified 495 samples that either were not of chimpanzee origin, represented fecal mixtures from more than one individual, or were too degraded for further analysis (Table 1). The remaining 2,070 samples yielded mtDNA (D-loop) sequences which comprised 252 different haplotypes (see Fig. S1 in the supplemental material). All of these were subjected to enhanced chemiluminescence (ECL) Western blot analysis, a method that detects SIVcpz-specific antibodies in fecal extracts with high sensitivity (0.92) and specificity (1.00) even after prolonged storage at ambient temperatures (30). Consistent with previous findings, none of the samples collected in Uganda and Rwanda were SIVcpz antibody positive (Table 1). Interestingly, however, 323 fecal specimens from 19 different sites in the DRC exhibited clear evidence of SIVcpz infection (Fig. 2 and Table 1). All of these reacted strongly with the HIV-1 p24 core antigen, and 11%, 62%, and 35% also reacted with p17 Gag, reverse transcriptase (p66/p55), and integrase (p31) proteins, respectively. Surprisingly, half of the samples (54%) also exhibited cross-reactivity with the HIV-1 envelope antigens (gp41, gp120, and gp160) which in some cases was as strong as that of the human positive control (Fig. 2).
Fig 2.
Detection of SIVcpz antibodies in chimpanzee fecal samples. Fecal samples from eastern chimpanzees (middle) and bonobos (right) and human controls (left) were tested by an enhanced chemiluminescence Western blot using HIV-1 antigen-containing strips. Samples are numbered, with letters indicating their collection site as shown in Fig. 1B. Molecular weights of HIV-1 proteins are indicated. The banding patterns of plasma from HIV-1-infected (positive) and uninfected (negative) humans are shown for the control.
To determine the number of SIVcpz-infected apes at the 19 sampling locations, we subjected all antibody-positive samples to microsatellite analyses (see Table S3 in the supplemental material). Many of these failed to yield a complete genotype due to partial sample degradation. To guard against allelic drop out, we thus allowed mismatches at up to four microsatellite loci for samples that shared the same mtDNA haplotype. This conservative approach identified a total of 76 SIVcpz-infected apes as a minimum estimate (Table 1). Microsatellite analysis also provided a quantitative measure of redundant sampling which, together with the proportion of degraded specimens, allowed us to calculate the prevalence of SIVcpz infection at each collection site. As shown in Table 1, analysis of an estimated 567 eastern chimpanzees yielded an overall prevalence rate of 13.4% (95% confidence interval, 10.7% to 16.5%). As previously observed in Cameroon and Tanzania, infection rates at individual field sites varied widely (Table 1), with high prevalence rates observed in some communities and low levels or absence of infection observed in others. In general, evidence for infection was observed throughout the study area, indicating that SIVcpz was not restricted to any one geographic region. The only exceptions were field sites in the northern part of the range, near the Uele River, including Buta (BT), Bondo (BD), Ango (AN), Niangara (NI), Dingila (DL), Mawa Gare (MA), Poko (PO), Rungu (RU), and Isiro (IS), which seemed to be free of SIV cpz infection. However, the number of usable samples from these sites was very small (n = 77), representing only a minor fraction (3.7%) of the total survey. Given this and the extent of sample degradation, it is thus possible that infected communities in this northern area were missed. Seven sites exhibited prevalence rates of 30% or higher, including sites in the far western (UB) and eastern (MU) parts of the DRC. These results indicate that SIVcpz is common and widespread among most P. t. schweinfurthii communities in the DRC (Fig. 1B), with prevalence rates similar to, or exceeding, those previously observed in P. t. troglodytes apes in Gabon and Cameroon (30, 35a, 41, 78).
No evidence of SIVcpz infection in wild-living bonobos.
In addition to eastern chimpanzees, the DRC is also home to bonobos (Pan paniscus), whose habitat is restricted to forest areas south of the Congo River (19). Although a limited number of bonobos have previously been tested in captivity (77), wild-living members of this species have never been screened for SIVcpz infection. To examine whether bonobos harbor SIV, we collected 543 fecal samples from six field sites located throughout the species range (Fig. 1B). All of these were subjected to mtDNA analysis, which identified 48 degraded or misidentified samples. The remaining 495 samples yielded mtDNA (D-loop) sequences that grouped into 24 distinct haplotypes (see Fig. S1 in the supplemental material). These were tested by immunoblot analysis, which failed to detect SIV/HIV cross-reactive antibodies. Western blots were completely negative for all samples, lacking even faint reactivity with the Gag p24 antigen, which is usually the most cross-reactive protein (Fig. 2). To estimate the number of sampled individuals, specimens from the Ikela (IK), Kokolopori (KR) and Lui-Kotale (LK) field sites were genotyped at eight microsatellite loci (see Table S4 in the supplemental material). This analysis identified a minimum of 72 individuals and an oversampling factor of 2.03. From this and the proportion of degraded samples, we estimated that we screened approximately 244 individuals, none of whom was SIVcpz infected.
SIVcpzPts strains form a monophyletic clade.
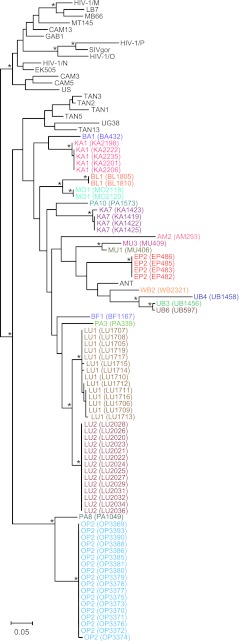
To examine the genetic relationships of the P. t. schweinfurthii viruses from the DRC to each other and to previously characterized SIVcpz strains, all antibody-positive samples were subjected to fecal RNA extraction and reverse transcription-PCR (RT-PCR) amplification. Using a diagnostic (minipol) primer set (see Table S1 in the supplemental material), we amplified viral sequences from 75 antibody-positive specimens. Subsequent screening with gp41 and gp41/nef primers identified SIVcpz virion RNA in 17 additional samples, molecularly confirming infection in 25 of the 76 infected individuals (see Table S3 in the supplemental material). So as to not miss infection by other SIVs, we tested the remaining antibody-positive samples using pan-SIV-specific primers (see Table S1 in the supplemental material). No other sequences were amplified, indicating that the relatively low RT-PCR positivity rate (29%) was the result of partial sample degradation and not infection by other primate lentiviruses. All amplicons were sequenced and phylogenetically analyzed. Although the minipol sequences were relatively short (232 bp), they were of sufficient length to show that all of the new DRC viruses were members of the SIVcpzPts lineage. As shown in Fig. 3, minipol sequences from 20 different individuals grouped with previously characterized P. t. schweinfurthii viruses in a clade supported by significant bootstrap values.
Fig 3.
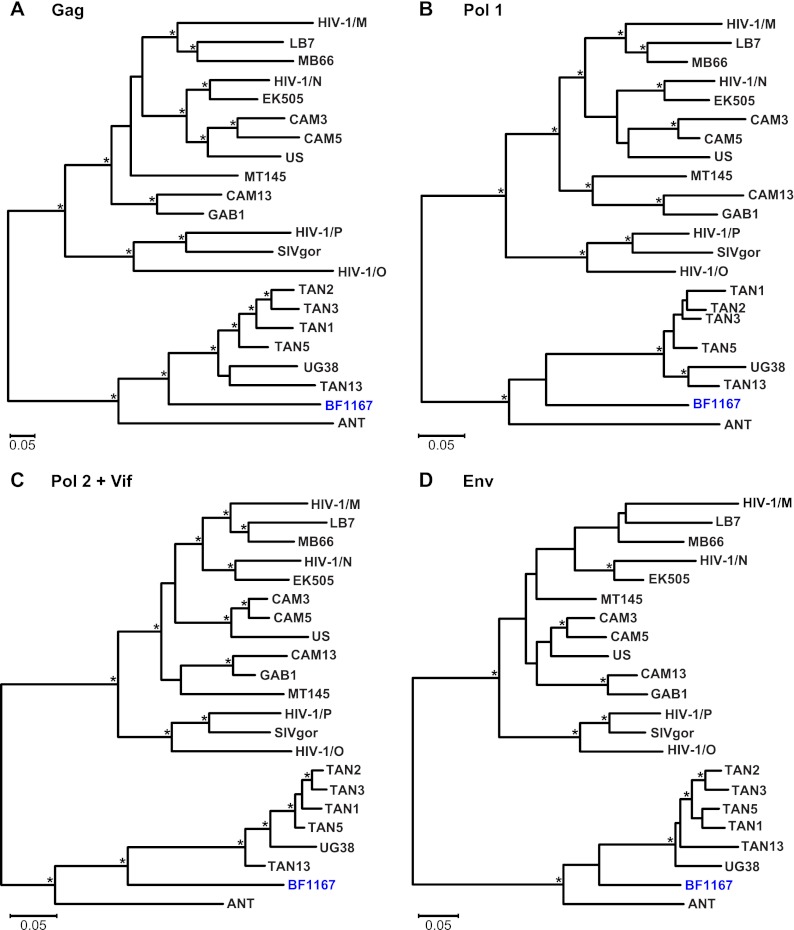
SIVcpz strains from the DRC cluster according to their subspecies of origin. A maximum likelihood tree was constructed from partial (232-bp) pol sequences (spanning HXB2 coordinates 4682 to 4913). Newly characterized SIVcpz strains from the DRC are highlighted, with sequences from the same individual color coded (for individual designations and sample numbers, see Table S3 in the supplemental material). Previously characterized SIVcpz, SIVgor, and HIV-1 strains forming the SIVcpzPtt (top cluster) and SIVcpzPts (bottom cluster) lineages are shown in black. The latter includes reference strains from Gombe (TAN1, TAN2, TAN3, TAN5, and TAN13) and Ugalla (UG38) as well as ANT, which is of unknown origin. Asterisks indicate bootstrap support of ≥70%. The scale bar represents 0.05 substitutions per site.
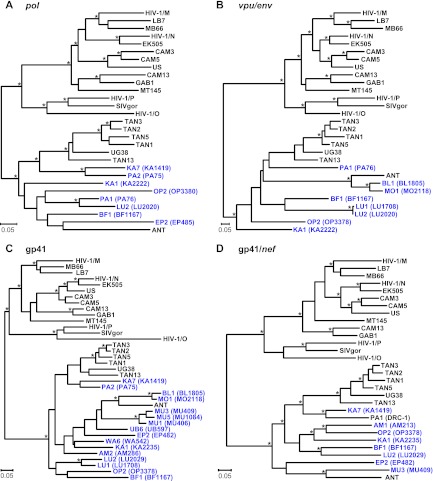
To examine further the phylogenetic relationships of the newly derived SIVcpzPts strains, we targeted regions in pol (892 bp), vpuenv (419 bp), gp41 (405 bp), and gp41/nef (665 bp) for additional amplifications. Although longer sequences could be amplified only from 18 individuals, their phylogeny confirmed the minipol results (Fig. 4). All newly derived sequences clustered according to their subspecies origin, forming a single well-supported viral lineage. In addition, the new SIVcpzPts strains exhibited evolutionary relationships similar to those previously described for SIVcpzPtt strains (30, 41, 78): viruses from distant collection sites generally formed well-separated clades or lineages, while viruses from the same geographic locale were usually closely related. For example, viruses from Lubutu (LU) and Mungbere (MU) each formed discrete clusters, indicating local transmissions. Significant clustering was also observed for viruses from Bongbola (BL) and Mongandjo (MO), suggesting unimpeded virus flow between these neighboring sites. Interestingly, the BL and MO strains were also closely related to SIVcpzANT, suggesting a possible geographic origin for this reference strain. Nonetheless, phylogeographic clustering was not uniform. Ape communities at Kabuka (KA) and Parisi (PA) harbored multiple divergent SIVcpzPts strains, perhaps indicating a greater connectivity of these communities. There was also evidence of recombination, as expected in populations coinfected with divergent viral lineages. One strain from Parisi (PA1) had a mosaic genome, as evidenced by its discordant clustering in the pol and gp41/nef regions (Fig. 4A and D). Importantly, however, there was no evidence of recombination between any of the newly identified DRC viruses and SIVcpzPtt strains. In fact, none of the DRC viruses, including those identified at the westernmost collection sites (UB, BL, MO), were particularly closely related to P. t. troglodytes viruses. Overall, SIVcpzPts strains were quite diverse, with nucleotide sequence distances of up to 25% and 35% in pol and gp41/nef regions, respectively, compared to 22% and 33%, respectively, for members of the SIVcpzPtt group.
Fig 4.
Phylogeny of SIVcpz from the DRC. Maximum likelihood trees were constructed of partial pol (HXB2 coordinates 3887 to 4778) (A), vpu/env (HXB2 coordinates 6062 to 6578) (B), gp41 (HXB2 coordinates 7836 to 8264) (C), and gp41/nef (HXB2 coordinates 8277 to 9047) (D) sequences. Regions of ambiguous alignment were removed from this analysis. New SIVcpzPts strains from the DRC are shown in blue, followed by the sample code in parentheses. Previously characterized SIVcpz, SIVgor, and HIV-1 strains forming the SIVcpzPtt (top cluster) and SIVcpzPts (bottom cluster) lineages are shown in black. Nodes with both bootstrap support of ≥70% and a Bayesian posterior probability of ≥0.95 are indicated by asterisks. The scale bar represents 0.05 substitutions/site.
To obtain at least one full-length SIVcpzPts sequence from the DRC, we selected a sample (BF1167) with a sufficiently high viral load for whole-genome amplification. Using strain-specific primers, we amplified 12 partially overlapping fragments, which together comprised a complete proviral genome (Fig. 5A). Inspection of the BF1167 consensus sequence revealed uninterrupted open reading frames for all structural and regulatory proteins as well as intact regulatory elements. BF1167 also contained all previously identified SIVcpzPts signatures (55, 58, 73), including three amino acid insertions in Gag p24, a conserved PPLP Vif motif, a short Vpr protein of 95 amino acids, a 5-amino-acid deletion at the C terminus of Nef, and an insertion in the ectodomain of gp41. Phylogenetic analysis of full-length Gag, Pol, and Env proteins showed that BF1167 fell within the SIVcpzPts radiation (Fig. 6), confirming the relationships derived from the partial genome sequences.
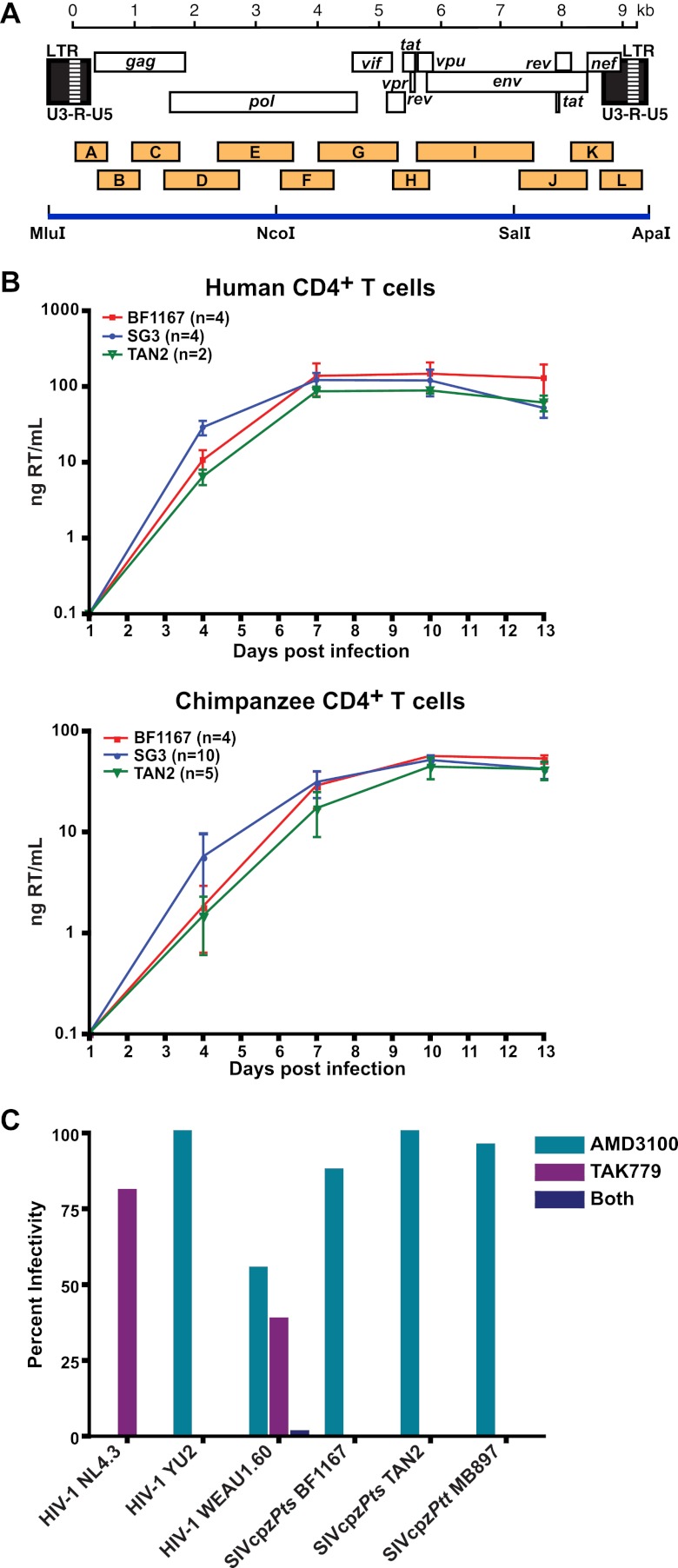
Fig 5.
Generation and biological characterization of a replication-competent SIVcpzPts molecular clone. (A) Individual RT-PCR amplicons (orange boxes) of BF1167 are shown in relation to the SIVcpz genome. Fragments are drawn to scale, with nucleotide sequences numbered starting at the beginning of the R region in the 5′ LTR (see scale bar). Three subgenomic fragments bound by MluI, NcoI, SalI, and ApaI restriction sites were synthesized and then assembled to produce a full-length provirus (blue line). (B) The replication kinetics of BF1167-derived virus in human (top) and chimpanzee (bottom) CD4+ T cells are shown in relation to those of HIV-1 (SG3; blue) and SIVcpzPts (TAN2; green) reference strains (x axis, days postinfection; y axis, nanograms of reverse transcriptase [RT] activity per ml of culture supernatant). Average values (and one standard deviation) from different experiments (indicated in parentheses) are shown. (C) TZM-bl cells were pretreated with AMD3100 (inhibitor of CXCR4), TAK779 (inhibitor of CCR5), or both prior to addition of the virus preparations indicated. Virus infectivity is plotted on the vertical axis as a percentage of the untreated control. Virus derived from the reference clones NL4.3 (X4 tropic), YU2 (R5 tropic), and WEAU1.6 (dual tropic) and SIVcpzPtt (MB897) and SIVcpzPts (TAN2) strains were included as controls. BF1167 is an R5-tropic virus.
Fig 6.
Evolutionary relationships of BF1167 full-length genome sequences. Maximum likelihood trees were inferred from amino acid (aa) sequence alignments of the major proteins, including Gag (420 aa; HXB2 coordinates 790 to 2280) (A), N-terminal Pol (630 aa; HXB2 coordinates 2295 to 4184) (B), C-terminal Pol/Vif (453 aa; HXB2 coordinates 4185 to 5556) (C), and Env (729 aa; HXB2 coordinates 6324 to 8792) (D); the Pol protein was separated into two fragments at a point where a recombination breakpoint was previously identified in HIV-1 group N. The BF1167 sequence is shown in blue. Previously characterized SIVcpz, SIVgor, and HIV-1 strains forming the SIVcpzPtt (top cluster) and SIVcpzPts (bottom cluster) lineages are shown in black. Nodes with both bootstrap support of ≥70% and a Bayesian posterior probability of ≥0.95 are indicated by asterisks. The scale bar represents 0.05 amino acid replacements/site.
Generation of a replication-competent SIVcpzPts clone.
The genomic organization of BF1167 suggested that it may encode a replication-competent provirus. To test this, we synthesized its consensus sequence as three subgenomic fragments and ligated them into a low-copy-number vector (Fig. 5A). The resulting plasmid clone was transfected into 293T cells, and culture supernatant was used to infect CD4+ T cell cultures. As shown in Fig. 5B, BF1167-derived virus replicated efficiently and to high titers in CD4+ T cells from both human (n = 4) and chimpanzee (n = 4) donors, with kinetics similar to those of previously characterized SIVcpzPts (TAN2) and HIV-1 (SG3) strains. Testing its coreceptor usage, we found that the infectivity of BF1167 was completely blocked by the CCR5 antagonist TAK-779 but not by the CXCR4 antagonist AMD3100 (Fig. 5C). This was also true for R5-tropic reference strains of HIV-1 (YU-2), SIVcpzPts (TAN2), and SIVcpzPtt (MB897) but not for X4 (NL4-3) and R5/X4 dual-tropic (WEAU) controls (Fig. 5C). Taken together, these data indicate that the newly derived BF1167 clone encodes an R5-tropic SIVcpzPts strain capable of infecting both primary human and chimpanzee CD4+ T cells.
SIVcpzPts strains require more mutational steps than SIVcpzPtt strains to gain human-specific Gag matrix and Vpu adaptations.
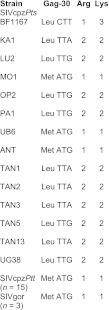
To evaluate the zoonotic potential of the newly derived DRC viruses, we compared their sequences in protein domains known to be under strong host-specific selection pressure. One such site was previously mapped to position 30 (Gag-30) of the viral matrix protein (82). Inspection of the proteome of all available SIVcpz and SIVgor strains identified a Met or Leu at this position. However, when these viruses infected humans, this residue was changed to an Arg in the inferred ancestors of HIV-1 groups M, N, and O and subsequently to a Lys in some M, N, and O strains (82). To determine the nature of Gag-30 in the DRC viruses, we amplified and sequenced the corresponding gag fragment from seven strains (see Table S3 in the supplemental material). Interestingly, we found that two of the new viruses (MO1 and UB6) encoded a Met at Gag-30, similar to SIVcpzANT and all known SIVcpzPtt and SIVgor strains (Fig. 7). However, the other five DRC strains encoded a Leu at Gag-30, similar to SIVcpzPts strains from Gombe (TAN) and Ugalla (UG). We then counted how many nucleotide substitutions would be required to change these Gag-30 codons to the HIV-1-specific residues. Arg is encoded by either CGN (where N is A, C, T, or G) or AGR (where R is A or G), and Lys is encoded by AAR. Thus, changing a Met (ATG) to Arg or Lys requires only a single nucleotide substitution. In contrast, Leu is encoded by CTN or TTR, and changing a Leu to Arg or Lys can therefore require one to three nucleotide substitutions (Fig. 7). Examining the Gag-30 codon in all known SIVcpzPts strains, we found that 10 of 14 viruses, including four of seven DRC strains, encoded Leu using TTA or TTG codons and thus required at least two mutational steps to acquire the basic Arg or Lys residue at Gag-30 (Fig. 7).
Fig 7.
Adaptive requirements of SIVcpz at Gag-30. The codon at position 30 of the Gag matrix protein is shown for 14 SIVcpzPts strains, including seven new viruses from the DRC. Ten of these 14 viruses encoded a Leu using TTA or TTG codons at Gag-30, codons which require at least two nucleotide changes to become Arg (CGN or AGR) or Lys (AAR) codons. In contrast, all sequenced SIVcpzPtt (n = 15) and SIVgor (n = 3) strains contain a Met ATG codon, which requires only a single substitution to change to either human-specific signature.
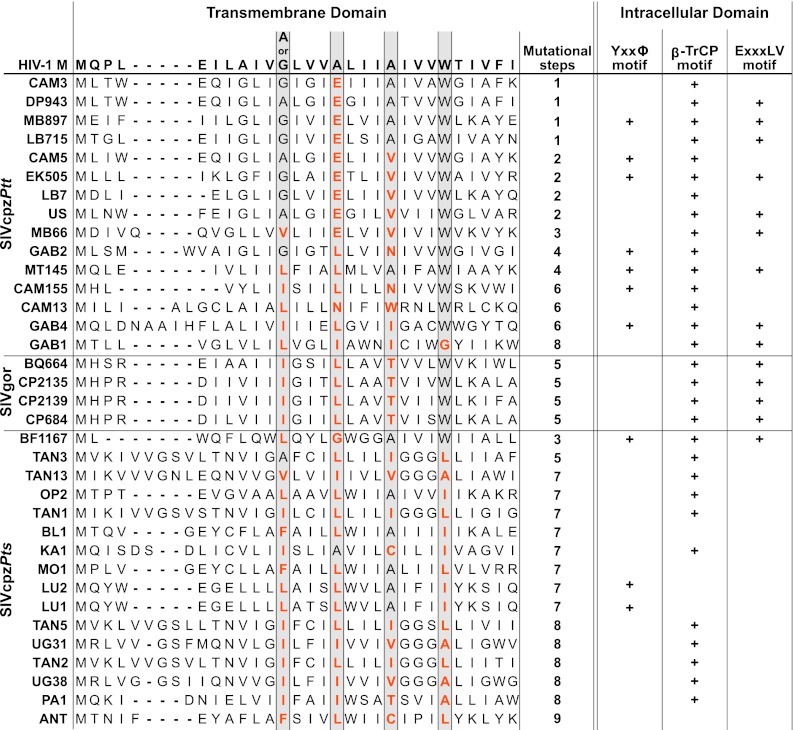
Human-specific adaptation has also shaped the function of the HIV-1 Vpu protein (18, 36, 61). Vpu modulates the cell surface expression of a number of immunoregulatory proteins, including the CD4 receptor, the natural killer (NK) cell ligand NTB-A, and the lipid antigen-presenting protein CD1d (13, 40, 63). In HIV-1, Vpu also counteracts tetherin, an innate restriction factor that inhibits the release of nascent virus particles from infected cells by “tethering” them to the cell surface (42, 46, 75). In SIVcpz and SIVgor, Vpu lacks this function and these viruses antagonize tetherin via their Nef proteins (61). However, these same Nef proteins are inactive against human tetherin due to a five-amino-acid deletion that confers resistance (61). To gauge how difficult or easy it would be for the newly characterized DRC strains to acquire antihuman tetherin activity, we aligned their Vpu sequences, as well as those of available SIVcpzPtt and SIVgor strains, to the HIV-1 group M Vpu consensus sequence (Fig. 8). Focusing in particular on the N-terminal transmembrane domain (TMD), which has been shown to interact directly with tetherin via four amino acid residues on the same face of its membrane-spanning helix (68), we counted the number of nucleotide substitutions that would be required to gain a functional A/GxxxAxxxAxxxW motif (15, 31, 68, 80, 81). The results ranged from a single substitution for a subset of SIVcpzPtt strains to nine changes for the SIVcpzPts strain ANT, with the closest chimpanzee relatives of HIV-1 groups M (MB879 and LB715) and N (EK505) requiring only one and two substitutions, respectively (Fig. 8). Although the adaptive distance depended on the particular virus, SIVcpzPtt and SIVgor strains typically required fewer changes (median, 4; range, 1 to 8) than SIVcpzPts strains (median, 7; range, 3 to 9) to gain the helix-helix interaction motif, suggesting that they might be more prone to human adaptation. We also examined the cytoplasmic domains of the various Vpu proteins for the presence or absence of functional motifs, including a YxxΦ motif (56), a DSGxxS β-TrCP binding site (39), and a putative ExxxLV trafficking signal (33), that have previously been shown to play a role in tetherin trafficking and/or degradation. Again, all of these domains were more commonly found in the Vpu proteins of SIVcpzPtt and SIVgor strains than in the Vpu proteins of SIVcpzPts strains (Fig. 8), with only one of 16 P. t. schweinfurthii viruses containing the ExxxLV motif that was recently shown to be required for efficient cell-free virion release from CD4 T cells (33). Thus, as a group, SIVcpzPts strains seem to require a much greater number of mutational steps to gain human-specific adaptations than do SIVcpzPtt and SIVgor strains.
Fig 8.
Adaptive requirements of SIVcpz in transmembrane and cytoplasmic domains of Vpu. Vpu amino acid sequences of SIVcpzPtt, SIVgor, and SIVcpzPts strains are aligned in their transmembrane domain to the corresponding region of the HIV-1 group M consensus as previously described (36). Dashes indicate gaps introduced to optimize the alignment. Gray boxes highlight residues of a conserved helix-helix interaction motif (G/AxxxAxxxAxxxW, where “x” may be any amino acid) that is required to counteract human tetherin (68, 80). The minimum number of mutational steps needed to change the corresponding amino acid to that of the human residue is indicated on the right. Columns on the far right indicate the presence (+) or absence (blank) of previously described transport and/or degradation motifs in the intracellular domain of Vpu. These include a YxxΦ motif scored as Yxx(L/M/V/I/F/W) (8, 21, 56), a β-TrCP ubiquitin-dependent degradation signal scored as D(S/D/E)Gxx(S/D/E) (28, 39), and a putative trafficking signal scored as (D/E)xxxL(L/V/I/M) (33).
DISCUSSION
Although long known to harbor SIVcpz in the wild (59, 60), wild-living eastern chimpanzees have not been thought to represent a virus reservoir. This is because previous field studies in Uganda, Rwanda, and Tanzania failed to uncover infected apes at most locations, except for communities in Gombe National Park and the Masito-Ugalla region of western Tanzania (29, 54, 55, 59, 60, 66). However, since these areas of east Africa comprise only a small part of the P. t. schweinfurthii range (Fig. 1), we reasoned that the observed paucity of infections might not be representative of the entire subspecies. To test this hypothesis, we targeted wild-living ape populations in the DRC, a country believed to be home to as many as 200,000 P. t. schweinfurthii apes and 50,000 bonobos (10, 19). To cover this vast area, we recruited local teams of trackers to collect ape fecal samples in the vicinity of their villages. Although this resulted at times in prolonged sample storage at ambient temperatures and, thus, partial specimen degradation, we were able to procure an unprecedented number of specimens from a wide variety of different locales (Table 1). This allowed us to assess the prevalence and geographic distribution of SIVcpz throughout the Congo Basin and to determine whether wild-living bonobos, which have not previously been tested for SIV infection, are natural carriers of this virus. We also screened additional communities in Uganda and Rwanda to determine whether isolated foci of SIVcpz infection had previously been missed.
Testing 2,070 fecal samples from an estimated 567 P. t. schweinfurthii apes, we identified 323 specimens from 76 individuals that harbored SIVcpz-specific antibodies, yielding an overall prevalence of 13.4% (95% confidence interval, 10.7% to 16.5%). Since chimpanzees in the northern DRC differ from their east African counterparts in a number of key behavioral traits (such as ground nesting and lack of termite fishing), which suggests a longstanding cultural separation (25), we also analyzed the DRC populations separately. Excluding samples from Rwanda and Uganda, we found that the remaining sites exhibited a prevalence of 14.9%. Remarkably, this estimate is 2.5 times higher than the prevalence previously determined for P. t. troglodytes apes in Cameroon (41). In the latter study, analysis of 1,217 fecal samples from 25 different field sites south of the Sanaga River yielded an overall prevalence of 5.9% (95% confidence interval, 4.3% to 7.9%). Although infected apes were identified at 10 of the 25 field sites, local infection levels were relatively low (41). Only one Cameroonian site exhibited a prevalence of greater than 30% (41), compared to seven such sites in the DRC (Table 1). Since the methods of fecal collection and diagnosing SIVcpz were very similar, the observed differences cannot be explained by ascertainment biases or technical differences. Moreover, infection rates in the DRC represent minimum estimates, given that many samples were partially degraded. Although SIVcpz is generally unevenly distributed among wild-living chimpanzees (29, 35a, 55, 59), our data indicate that P. t. schweinfurthii apes are at least as widely and commonly infected as P. t. troglodytes apes. A recent study of wild-living P. t. troglodytes apes in Gabon confirmed this, identifying SIVcpz infection at only three of 10 locations, with high-level infection detected at only one of these sites (35a). In contrast, the newly screened communities in the Budongo Reserve (BG), the Kyambura Gorge (KY), and the Gishwati Forest (GI) were all virus negative, further supporting the notion that SIVcpz is absent from the extreme eastern edge of the P. t. schweinfurthii range except for isolated communities in western Tanzania. It remains unknown whether SIVcpz was once present there and has subsequently gone extinct or whether its eastward spread was obstructed by habitat loss and/or other barriers such that certain communities were never exposed.
In contrast to eastern chimpanzees, none of the 495 bonobo samples from an estimated 244 individuals were SIV antibody positive (Table 1). Although bonobos were sampled at fewer locations than P. t. schweinfurthii apes, the six field sites were widely distributed throughout the bonobo range (Fig. 1B). Moreover, for all but one field site (BJ), significant numbers of samples were tested (Table 1), arguing against the possibility that low-level infections were missed. There were also no differences in the ways bonobo and eastern chimpanzee samples were collected, stored, and transported. Thus, sample degradation cannot be invoked as an explanation for the negative Western blot results. Based on these data, it seems likely that bonobos are free of SIVcpz infection. This is consistent with a previous study that failed to detect SIVcpz antibodies in the blood of 26 captive bonobos (77). It is also consistent with the fact that bonobos and eastern chimpanzees have nonoverlapping ranges (Fig. 1). However, given that western gorillas are only rarely infected with SIVgor (41), it will be important to exclude isolated infections of bonobos by testing additional individuals from a wider range of locations. It should also be noted that bonobos, like chimpanzees, are exposed to a variety of SIVs because they hunt and eat smaller primates, including different species of guenons (70, 71), which carry their own types of SIV (2). These monkey SIVs are genetically quite divergent from HIV-1/SIVcpz and may elicit antibodies that do not cross-react with HIV-1 proteins (2, 43). It will thus be important to include additional SIV antigens in future noninvasive screening approaches to examine whether bonobos harbor such viruses.
Previous evolutionary studies have shown that SIVcpz sequences form two highly divergent lineages, SIVcpzPtt and SIVcpzPts, according to their subspecies of origin (64–66). These studies also revealed that all groups of HIV-1 and SIVgor cluster within the SIVcpzPtt lineage, thus identifying P. t. troglodytes apes as the original source of both human and gorilla viruses (24, 47, 72, 79). However, except for ANT (76) and the only partially characterized DRC1 virus (84), all other SIVcpzPts strains were derived from infected apes in Gombe and Ugalla (29, 54, 55), thus leaving the evolutionary history of viruses from the remaining parts of the P. t. schweinfurthii range open to question (45). In this study, we tested all antibody-positive fecal samples from the DRC for the presence of SIVcpz sequences. Using gag, pol, vpu, and env primers, we were able to amplify virion RNA from 25 of the 76 infected individuals (see Table S3 in the supplemental material). Although this recovery rate was lower than in previous field studies, we obtained SIVcpz sequences from 14 of the 19 positive locations (Fig. 1B). Importantly, we found that all of the new viruses clustered with previously characterized SIVcpzPts strains in all genomic regions analyzed (Fig. 3, 4, and 6). While some grouped according to their field site of origin, phylogeographic clustering seemed less pronounced in the DRC than previously observed in Cameroon (30, 78). This may be because the largely contiguous forests in the DRC have provided for greater connectivity and, thus, exchange of divergent viruses over longer distances. Nonetheless, all DRC viruses fell within the SIVcpzPts radiation, forming a single well-supported phylogenetic lineage that also included viruses from Gombe and Ugalla (Fig. 3, 4, and 6). This strict subspecies-specific clustering indicates that P. t. schweinfurthii apes have been effectively isolated from P. t. troglodytes apes for a considerable period of time. It also confirms that chimpanzees in the DRC were not the source of any known strain of HIV-1 (84).
Given that SIVcpz infection rates in wild-living chimpanzees in the DRC are at least as high, if not higher, than in Cameroon and Gabon, it seems striking that SIVcpzPts strains have never been found in humans. There are at least three potential explanations which are not mutually exclusive. One possibility is that humans in the DRC are less frequently exposed to SIVcpz. Although it remains unknown exactly how humans acquired the ape precursors of HIV-1 groups M, N, O, and P, cross-species transmission must have occurred through cutaneous or mucous membrane exposure to infected ape blood and/or body fluids which occurs most frequently in the context of bushmeat hunting (24). While firm data concerning the frequencies and types of human-ape interactions in the DRC are lacking, it is believed that hunting varies regionally due to differences in local traditions and preferences, with some tribes having taboos against the consumption of apes (45). Thus, human exposure to SIVcpz may have historically been lower in the DRC than in west central Africa, and this may explain the lack of SIVcpzPts zoonoses. However, even if this were the case, this barrier is clearly no longer in place. Bushmeat hunting in the DRC, including the poaching of chimpanzees, has been on the rise in the past decade due to political turmoil and economic changes (5). A recent study of apes north and south of the Uele River documented a major increase in chimpanzee killing due to an influx of artisanal diamond and gold miners (26). Thus, increased surveillance of humans in these areas for SIVcpzPts and other ape-derived infections may be warranted (6, 11, 38).
A second explanation is that SIVcpzPts infections of humans may, in fact, have occurred but gone unrecognized because of limited human sampling and a lack of specific tests. The great majority of HIV-1 infections in the DRC and elsewhere are diagnosed serologically, using enzyme-linked immunosorbent assays (ELISAs) or rapid-test immunoblot approaches. Despite their genetic diversity, many of the ape infections characterized here exhibited a Western blot profile indistinguishable from that of the positive HIV-1 control (Fig. 2). It is thus possible that a human infected with such a virus may be misdiagnosed as being infected by HIV-1 group M, especially since the vast majority of HIV-1 infections in the DRC are not molecularly confirmed. However, such SIVcpzPts zoonoses—if they have indeed occurred—are unlikely to have infected large numbers of people, because more-substantive outbreaks would likely have been detected by existing surveillance programs, such as those that discovered the very rare N and P groups of HIV-1 (47, 67). The apparent lack of SIVcpzPts zoonoses may also reflect regional differences in human transmission networks, since sporadic introductions into less-dense, less-urban, and/or less-well-connected populations would be more likely to result in dead-end infections.
A third possibility is that SIVcpzPts strains face greater adaptive hurdles before they can replicate and spread efficiently in the human host. Examining SIVcpz proteomes for amino acids that were highly conserved in the ape precursors of HIV-1 but that changed each time these viruses crossed the species barrier to humans, we previously found a Met or Leu in all SIVcpz/SIVgor strains but an Arg in the inferred ancestors of HIV-1 groups M, N, and O and a Lys in some HIV-1 strains (82). We also showed that changing Met/Leu at Gag-30 to Arg/Lys greatly enhanced the replication fitness of SIVcpz strains in human tonsil cultures, while the opposite was true for HIV-1 strains that contained the ape-specific Gag-30 residues (7). These studies provided compelling evidence that host-specific adaptation at Gag-30 is required for efficient replication of SIVcpz in human lymphatic tissue. When we determined the nature of the Gag-30 residue in all available SIVcpzPts strains, including the new viruses from the DRC, we found that most require twice as many mutational steps as SIVcpzPtt and SIVgor strains to adapt at Gag-30 (Fig. 7). Substantial adaptive hurdles were also identified for the SIVcpz Vpu protein (18, 36, 61). It has been shown that upon cross-species transmission, the ape precursors of HIV-1 had to switch from Nef- to Vpu-mediated tetherin antagonism (36, 61). However, only the pandemic M group viruses acquired efficient antitetherin activity, while the much-less-prevalent group N, O, and P viruses either failed to gain this activity or lost other Vpu functions (36, 61, 62, 81, 85, 86). These findings have been taken to indicate that successful SIV zoonoses require effective tetherin antagonism (23). When we compared ape virus Vpu sequences to that of the HIV-1 group M consensus, we found that some SIVcpzPtt Vpu proteins required only very few changes to gain key functional motifs (Fig. 8). In contrast, most SIVcpzPts Vpu proteins required a substantially larger number of substitutions to acquire these same human-specific signatures (Fig. 8). Although counting numbers of mutational steps represents a gross oversimplification of the adaptation process, our analyses suggest that certain SIVcpzPtt strains are better equipped to become new human pathogens than most SIVcpzPts strains. The fact that the reconstructed BF1167 genome (as well as those of other SIVcpzPts strains) replicates well in human CD4+ T cells (Fig. 5) does not argue against this, since such maximally stimulated cultures do not accurately recapitulate the conditions of viral replication and transmission in vivo (7).
In summary, we report here that wild-living P. t. schweinfurthii populations are much more widely and commonly infected with SIVcpz than previously appreciated. This is particularly true for communities in the northern DRC which represent a large continuous population that seems to provide opportunity for virus flow across vast areas. Whether these viruses have a reduced potential to infect humans and cause epidemic outbreaks is not known but should be investigated. In particular, studies of host restriction mechanisms that may have prevented the spread of these viruses in the human population would be informative. This is now possible, since well-characterized reagents, including a large panel of SIVcpzPtt and SIVcpzPts infectious molecular clones, are available for study. The high prevalence of SIVcpzPts infection also has implications for conservation efforts. SIVcpz is quite pathogenic and has been shown to negatively impact chimpanzee population growth (17, 29, 54, 74). Given that the DRC is home to half of the world's remaining chimpanzees, it will be critical to determine in much greater detail to what extent SIVcpz has penetrated these populations so that the impact of this infection on the long-term survival of this species can be determined. Finer-grained prevalence and natural history data will also be critical for attempts to limit the spread of SIVcpz in wild ape populations, for example, through the use of adeno-associated virus (AAV)-mediated gene transfer of antibodies that neutralize SIVcpz as a prophylactic or therapeutic vaccine (4, 27) which can potentially be administered in wild settings (57).
Supplementary Material
ACKNOWLEDGMENTS
We thank the following individuals and groups: Claude Kitoko and Bola Iyoka for collection of chimpanzee and bonobo fecal samples in the Parisi Forest and the Ikela region (DRC), respectively; the staff of the World Wildlife Fund (WWF) for collecting bonobo fecal samples in the Lac Tumba area (DRC); James Kakura, Geresomu Muhumuza, Monday Gideon, and Raymond Ogen for field work in the Budongo Forest (Uganda); Sylvain Nyandwi, Thomas Safari, Samuel Uwimana, Patience Mwiseneza, Alex Ndayambaje, Isaac Ngayincyuro, Olivier Ngabonziza, and Eric Munyeshuli for field work in the Gishwati Reserve (Rwanda); Cleve Hicks for unpublished behavioral data from eastern chimpanzees in the northern DRC; Frank Kirchhoff and Daniel Sauter for helpful discussions; Patricia Crystal for artwork and preparation of the manuscript; the Ministry of Scientific Research and Technology, the Department of Ecology and Management of Plant and Animal Resources of the University of Kisangani, the Ministries of Health and Environment, and the National Ethics committee for permission to collect samples in the DRC; the Uganda Wildlife Authority and the Uganda National Council for Science and Technology for permission to conduct research in the Budongo Forest and the Kyambura Gorge of Queen Elizabeth National Park; and the Rwandan Office of Tourism and National Parks for permission to collect samples in the Gishwati Forest Reserve.
This work was supported in part by grants from the National Institutes of Health (R01 AI50529, R01 AI58715, and P30 AI 27767), the Agence Nationale de Recherches sur le SIDA (ANRS 12255), and the Great Ape Trust. R.S.R. was funded by a Howard Hughes Medical Institute Med-into-Grad Fellowship, and S.A.-M. was funded by a grant from Infectiopole Sud, France.
Footnotes
Published ahead of print 25 July 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Abascal F, Zardoya R, Posada D. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104–2105 [DOI] [PubMed] [Google Scholar]
- 2. Ahuka-Mundeke S, et al. 2011. Novel multiplexed HIV/simian immunodeficiency virus antibody detection assay. Emerg. Infect. Dis. 17:2277–2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bailes E, et al. 2003. Hybrid origin of SIV in chimpanzees. Science 300:1713. [DOI] [PubMed] [Google Scholar]
- 4. Balazs AB, et al. 2012. Antibody-based protection against HIV infection by vectored immunoprophylaxis. Nature 481:81–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bennett EL, et al. 2007. Hunting for consensus: reconciling bushmeat harvest, conservation, and development policy in West and Central Africa. Conserv. Biol. 21:884–887 [DOI] [PubMed] [Google Scholar]
- 6. Betsem E, Rua R, Tortevoye P, Froment A, Gessain A. 2011. Frequent and recent human acquisition of simian foamy viruses through apes' bites in central Africa. PLoS Pathog. 7:e1002306 doi:10.1371/journal.ppat.1002306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bibollet-Ruche F, et al. 2012. Efficient SIVcpz replication in human lymphoid tissue requires viral matrix protein adaptation. J. Clin. Invest. 122:1644–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bonifacino JS, Traub LM. 2003. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72:395–447 [DOI] [PubMed] [Google Scholar]
- 9. Bowden R, et al. 2012. Genomic tools for evolution and conservation in the chimpanzee: Pan troglodytes ellioti is a genetically distinct population. PLoS Genet. 8:e1002504 doi:10.1371/journal.pgen.1002504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Butynski TM. 2001. Africa's great apes, p 3–56 In Beck B, et al. (ed), Great apes and humans—the ethics of coexistence. Smithsonian Institution Press, Washington, DC [Google Scholar]
- 11. Calvignac-Spencer S, Leendertz SAJ, Gillespie TR, Leendertz FH. 2012. Wild great apes as sentinels and sources of infectious disease. Clin. Microbiol. Infect. 18:521–527 [DOI] [PubMed] [Google Scholar]
- 12. Chancellor RL, Langergraber K, Ramirez S, Rundus AS, Vigilant L. 2012. Genetic sampling of unhabituated chimpanzees (Pan troglodytes schweinfurthii) in Gishwati forest reserve, an isolated forest fragment in western Rwanda. Int. J. Primatol. 33:479–488 [Google Scholar]
- 13. Chen BK, Gandhi RT, Baltimore D. 1996. CD4 down-modulation during infection of human T cells with human immunodeficiency virus type 1 involves independent activities of vpu, env, and nef. J. Virol. 70:6044–6053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Decker JM, et al. 2009. Effective activation alleviates the replication block of CCR5-tropic HIV-1 in chimpanzee CD4+ lymphocytes. Virology 394:109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dube M, et al. 2010. Antagonism of tetherin restriction of HIV-1 release by Vpu involves binding and sequestration of the restriction factor in a perinuclear compartment. PLoS Pathog. 6:e1000856 doi:10.1371/journal.ppat.1000856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Eng B, Ainsworth P, Waye JS. 1994. Anomalous migration of PCR products using nondenaturing polyacrylamide gel electrophoresis: the amelogenin sex-typing system. J. Forensic Sci. 39:1356–1359 [PubMed] [Google Scholar]
- 17. Etienne L, et al. 2011. Characterization of a new simian immunodeficiency virus strain in a naturally infected Pan troglodytes troglodytes chimpanzee with AIDS related symptoms. Retrovirology 8:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Evans DT, Serra-Moreno R, Singh RK, Guatelli JC. 2010. BST-2/tetherin: a new component of the innate immune response to enveloped viruses. Trends Microbiol. 18:388–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fruth B, et al. 2008. Pan paniscus. The IUCN Red List of Threatened Species, IUCN, Cambridge, United Kingdom: http://www.iucnredlist.org/details/15932/0 [Google Scholar]
- 20. Gao F, et al. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436–441 [DOI] [PubMed] [Google Scholar]
- 21. Gough NR, et al. 1999. Utilization of the indirect lysosome targeting pathway by lysosome-associated membrane proteins (LAMPs) is influenced largely by the C-terminal residue of their GYXXΦ targeting signals. J. Cell Sci. 112(Part 23):4257–4269 [DOI] [PubMed] [Google Scholar]
- 22. Guindon S, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321 [DOI] [PubMed] [Google Scholar]
- 23. Gupta RK, Towers GJ. 2009. A tail of tetherin: how pandemic HIV-1 conquered the world. Cell Host Microbe 6:393–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hahn BH, Shaw GM, De Cock KM, Sharp PM. 2000. AIDS as a zoonosis: scientific and public health implications. Science 287:607–614 [DOI] [PubMed] [Google Scholar]
- 25. Hicks TC. 2010. A chimpanzee mega-culture? Exploring behavioral continuity in Pan troglodytes schweinfurthii across northern DR Congo. Ph.D. thesis University of Amsterdam, Amsterdam, The Netherlands [Google Scholar]
- 26. Hicks TC, et al. 2010. Trade in orphans and bushmeat threatens one of the Democratic Republic of the Congo's most important populations of eastern chimpanzees (Pan troglodytes schweinfurthii). Afr. Primates 7:1–18 [Google Scholar]
- 27. Johnson PR, et al. 2009. Vector-mediated gene transfer engenders long-lived neutralizing activity and protection against SIV infection in monkeys. Nat. Med. 15:901–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kanemori Y, Uto K, Sagata N. 2005. Beta-TrCP recognizes a previously undescribed nonphosphorylated destruction motif in Cdc25A and Cdc25B phosphatases. Proc. Natl. Acad. Sci. U. S. A. 102:6279–6284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Keele BF, et al. 2009. Increased mortality and AIDS-like immunopathology in wild chimpanzees infected with SIVcpz. Nature 460:515–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Keele BF, et al. 2006. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science 313:523–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kobayashi T, et al. 2011. Identification of amino acids in the human tetherin transmembrane domain responsible for HIV-1 Vpu interaction and susceptibility. J. Virol. 85:932–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Krüger O, Affeldt E, Brackmann M, Milhahn K. 1998. Group size and composition of Colobus guereza in Kyambura Gorge, southwest Uganda, in relation to chimpanzee activity. Int. J. Primatol. 19:287–297 [Google Scholar]
- 33. Kueck T, Neil SJ. 2012. A cytoplasmic tail determinant in HIV-1 Vpu mediates targeting of tetherin for endosomal degradation and counteracts interferon-induced restriction. PLoS Pathog. 8:e1002609 doi:10.1371/journal.ppat.1002609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Larkin MA, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 35. Leendertz SA, et al. 2011. No evidence for transmission of SIVwrc from western red colobus monkeys (Piliocolobus badius badius) to wild West African chimpanzees (Pan troglodytes verus) despite high exposure through hunting. BMC Microbiol. 11:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35a. Liegeois F, et al. 2012. Identification of new divergent SIVcpzPtt strains in wild living chimpanzees in Gabon, poster 528. The Conference on Retroviruses and Opportunistic Infections, Seattle, WA http://www.retroconference.org/2012b/PDFs/528.pdf [Google Scholar]
- 36. Lim ES, Malik HS, Emerman M. 2010. Ancient adaptive evolution of tetherin shaped the functions of Vpu and Nef in human immunodeficiency virus and primate lentiviruses. J. Virol. 84:7124–7134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ling B, et al. 2003. Noninvasive detection of new simian immunodeficiency virus lineages in captive sooty mangabeys: ability to amplify virion RNA from fecal samples correlates with viral load in plasma. J. Virol. 77:2214–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu W, et al. 2008. Molecular ecology and natural history of simian foamy virus infection in wild-living chimpanzees. PLoS Pathog. 4:e1000097 doi:10.1371/journal.ppat.1000097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Margottin F, et al. 1998. A novel human WD protein, h-βTrCp, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol. Cell 1:565–574 [DOI] [PubMed] [Google Scholar]
- 40. Moll M, Andersson SK, Smed-Sorensen A, Sandberg JK. 2010. Inhibition of lipid antigen presentation in dendritic cells by HIV-1 Vpu interference with CD1d recycling from endosomal compartments. Blood 116:1876–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Neel C, et al. 2010. Molecular epidemiology of simian immunodeficiency virus infection in wild-living gorillas. J. Virol. 84:1464–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Neil SJ, Zang T, Bieniasz PD. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425–430 [DOI] [PubMed] [Google Scholar]
- 43. Peeters M, et al. 2002. Risk to human health from a plethora of simian immunodeficiency viruses in primate bushmeat. Emerg. Infect. Dis. 8:451–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Peeters M, et al. 1992. Isolation and characterization of a new chimpanzee lentivirus (simian immunodeficiency virus isolate cpz-ant) from a wild-captured chimpanzee. AIDS 6:447–451 [DOI] [PubMed] [Google Scholar]
- 45. Pepin J. 2011. The origins of AIDS. Cambridge University Press, New York, NY [Google Scholar]
- 46. Perez-Caballero D, et al. 2009. Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell 139:499–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Plantier JC, et al. 2009. A new human immunodeficiency virus derived from gorillas. Nat. Med. 15:871–872 [DOI] [PubMed] [Google Scholar]
- 48. Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 72:2855–2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Plumptre AJ, Masozera M, Vedder A. 2001. The impact of civil war on the conservation of protected areas in Rwanda. Biodiversity Support Program, Washington, DC: http://www.worldwildlife.org/bsp/publications/africa/145/pdf/Rwanda.pdf [Google Scholar]
- 50. Plumptre AJ, et al. 2010. Eastern chimpanzee (Pan troglodytes schweinfurthii): status survey and conservation action plan 2010–2020. IUCN/SSC Primate Specialist Group, Arlington, VA: http://data.iucn.org/dbtw-wpd/edocs/2010-023.pdf [Google Scholar]
- 51. Posada D, Crandall KA. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818 [DOI] [PubMed] [Google Scholar]
- 52. Reynolds V. 2005. The chimpanzees of the Budongo forest: ecology, behaviour, and conservation. Oxford University Press, New York, NY [Google Scholar]
- 53. Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574 [DOI] [PubMed] [Google Scholar]
- 54. Rudicell RS, et al. 2010. Impact of simian immunodeficiency virus infection on chimpanzee population dynamics. PLoS Pathog. 6:e1001116 doi:10.1371/journal.ppat.1001116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rudicell RS, et al. 2011. High prevalence of simian immunodeficiency virus infection in a community of savanna chimpanzees. J. Virol. 85:9918–9928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ruiz A, Hill MS, Schmitt K, Guatelli J, Stephens EB. 2008. Requirements of the membrane proximal tyrosine and dileucine-based sorting signals for efficient transport of the subtype C Vpu protein to the plasma membrane and in virus release. Virology 378:58–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ryan SJ, Walsh PD. 2011. Consequences of non-intervention for infectious disease in African great apes. PLoS One 6:e29030 doi:10.1371/journal.pone.0029030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Santiago ML, et al. 2003. Amplification of a complete simian immunodeficiency virus genome from fecal RNA of a wild chimpanzee. J. Virol. 77:2233–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Santiago ML, et al. 2003. Foci of endemic simian immunodeficiency virus infection in wild-living eastern chimpanzees (Pan troglodytes schweinfurthii). J. Virol. 77:7545–7562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Santiago ML, et al. 2002. SIVcpz in wild chimpanzees. Science 295:465. [DOI] [PubMed] [Google Scholar]
- 61. Sauter D, et al. 2009. Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe 6:409–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sauter D, Specht A, Kirchhoff F. 2010. Tetherin: holding on and letting go. Cell 141:392–398 [DOI] [PubMed] [Google Scholar]
- 63. Shah AH, et al. 2010. Degranulation of natural killer cells following interaction with HIV-1-infected cells is hindered by downmodulation of NTB-A by Vpu. Cell Host Microbe 8:397–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sharp PM, Hahn BH. 2010. The evolution of HIV-1 and the origin of AIDS. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365:2487–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sharp PM, Hahn BH. 2011. Origins of HIV and the AIDS pandemic. Cold Spring Harb. Perspect. Med. 1:a006841 doi:10.1101/cshperspect/a006841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sharp PM, Shaw GM, Hahn BH. 2005. Simian immunodeficiency virus infection of chimpanzees. J. Virol. 79:3891–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Simon F, et al. 1998. Identification of a new human immunodeficiency virus type 1 distinct from group M and group O. Nat. Med. 4:1032–1037 [DOI] [PubMed] [Google Scholar]
- 68. Skasko M, et al. 2012. HIV-1 Vpu protein antagonizes innate restriction factor BST-2 via lipid-embedded helix-helix interactions. J. Biol. Chem. 287:58–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sullivan KM, Mannucci A, Kimpton CP, Gill P. 1993. A rapid and quantitative DNA sex test: fluorescence-based PCR analysis of X-Y homologous gene amelogenin. Biotechniques 15:636–638, 640–641 [PubMed] [Google Scholar]
- 70. Surbeck M, Fowler A, Deimel C, Hohmann G. 2009. Evidence for the consumption of arboreal, diurnal primates by bonobos (Pan paniscus). Am. J. Primatol. 71:171–174 [DOI] [PubMed] [Google Scholar]
- 71. Surbeck M, Hohmann G. 2008. Primate hunting by bonobos at LuiKotale, Salonga National Park. Curr. Biol. 18:R906–R907 [DOI] [PubMed] [Google Scholar]
- 72. Takehisa J, et al. 2009. Origin and biology of simian immunodeficiency virus in wild-living western gorillas. J. Virol. 83:1635–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Takehisa J, et al. 2007. Generation of infectious molecular clones of simian immunodeficiency virus from fecal consensus sequences of wild chimpanzees. J. Virol. 81:7463–7475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Terio KA, et al. 2011. Pathologic lesions in chimpanzees (Pan trogylodytes schweinfurthii) from Gombe National Park, Tanzania, 2004-2010. J. Zoo Wildl. Med. 42:597–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Van Damme N, et al. 2008. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3:245–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Vanden Haesevelde MM, et al. 1996. Sequence analysis of a highly divergent HIV-1-related lentivirus isolated from a wild captured chimpanzee. Virology 221:346–350 [DOI] [PubMed] [Google Scholar]
- 77. Van Dooren S, et al. 2002. Lack of evidence for infection with simian immunodeficiency virus in bonobos. AIDS Res. Hum. Retroviruses 18:213–216 [DOI] [PubMed] [Google Scholar]
- 78. Van Heuverswyn F, et al. 2007. Genetic diversity and phylogeographic clustering of SIVcpzPtt in wild chimpanzees in Cameroon. Virology 368:155–171 [DOI] [PubMed] [Google Scholar]
- 79. Van Heuverswyn F, et al. 2006. Human immunodeficiency viruses: SIV infection in wild gorillas. Nature 444:164. [DOI] [PubMed] [Google Scholar]
- 80. Vigan R, Neil SJ. 2010. Determinants of tetherin antagonism in the transmembrane domain of the human immunodeficiency virus type 1 Vpu protein. J. Virol. 84:12958–12970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Vigan R, Neil SJ. 2011. Separable determinants of subcellular localization and interaction account for the inability of group O HIV-1 Vpu to counteract tetherin. J. Virol. 85:9737–9748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wain LV, et al. 2007. Adaptation of HIV-1 to its human host. Mol. Biol. Evol. 24:1853–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wei X, et al. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312 [DOI] [PubMed] [Google Scholar]
- 84. Worobey M, et al. 2004. Origin of AIDS: contaminated polio vaccine theory refuted. Nature 428:820. [DOI] [PubMed] [Google Scholar]
- 85. Yang SJ, Lopez LA, Exline CM, Haworth KG, Cannon PM. 2011. Lack of adaptation to human tetherin in HIV-1 group O and P. Retrovirology 8:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Yang SJ, et al. 2010. Anti-tetherin activities in Vpu-expressing primate lentiviruses. Retrovirology 7:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.