Abstract
We have exploited the ability of transmembrane domains to engage in highly specific protein-protein interactions to construct a new class of small proteins that inhibit HIV infection. By screening a library encoding hundreds of thousands of artificial transmembrane proteins with randomized transmembrane domains (termed “traptamers,” for transmembrane aptamers), we isolated six 44- or 45-amino-acid proteins with completely different transmembrane sequences that inhibited cell surface and total expression of the HIV coreceptor CCR5. The traptamers inhibited transduction of human T cells by HIV reporter viruses pseudotyped with R5-tropic gp120 envelope proteins but had minimal effects on reporter viruses with X4-tropic gp120. Optimization of two traptamers significantly increased their activity and resulted in greater than 95% inhibition of R5-tropic reporter virus transduction without inhibiting expression of CD4, the primary HIV receptor, or CXCR4, another HIV coreceptor. In addition, traptamers inhibited transduction mediated by a mutant R5-tropic gp120 protein resistant to maraviroc, a small-molecule CCR5 inhibitor, and they dramatically inhibited replication of an R5-tropic laboratory strain of HIV in a multicycle infection assay. Genetic experiments suggested that the active traptamers specifically interacted with the transmembrane domains of CCR5 and that some of the traptamers interacted with different portions of CCR5. Thus, we have constructed multiple proteins not found in nature that interfere with CCR5 expression and inhibit HIV infection. These proteins may be valuable tools to probe the organization of the transmembrane domains of CCR5 and their relationship to its biological activities, and they may serve as starting points to develop new strategies to inhibit HIV infection.
INTRODUCTION
Despite the recognized importance of G protein-coupled receptors (GPCRs) in many biological processes and as therapeutic targets, our understanding of their structure and function remains incomplete. The hydrophobic core of these multipass transmembrane (TM) proteins is flexible, suggesting that essential interactions between the TM domains could be disrupted with specific hydrophobic proteins (23). Other laboratories have modulated GPCR activity using TM peptides derived from native receptor sequences (16, 19, 41). As an alternative approach, we have developed genetic selections to identify proteins with the desired activity from a large collection of small, randomized TM proteins, also called traptamers (for transmembrane aptamers), modeled on the 44-amino-acid bovine papillomavirus (BPV) E5 protein, which targets the platelet-derived growth factor β receptor (PDGFβR) (40). These proteins might be preferable to those derived from naturally occurring TM domains because artificial proteins are not subject to evolutionary constraints that might limit activity or affect specificity. Until now, this approach has been restricted to isolating traptamers that stimulate the activity of single-pass TM proteins (7, 14).
Here, we constructed traptamers that inhibited expression of the human immunodeficiency virus (HIV) coreceptor, CCR5, a chemokine receptor with seven membrane-spanning domains. HIV infects human immune cells through an initial interaction between the viral envelope glycoprotein gp120 and the host cell surface protein CD4. This is followed by binding of gp120 to an additional cellular receptor, typically CCR5 or CXCR4, and subsequent fusion of viral and cellular membranes (4, 11, 37). CCR5 is the main coreceptor used by HIV during transmission, and individuals homozygous for a nonfunctional CCR5 deletion mutant (CCR5Δ32) are largely resistant to infection (21, 29, 34, 36, 43). As a result, major research efforts have been devoted to identifying inhibitors of CCR5 (25). Such inhibitors are attractive because they act at an early step in the HIV life cycle and are aimed at a cellular target that is not essential for organismal viability nor subject to the high mutation rate that affects HIV genes (30). The only CCR5 inhibitor approved for clinical use is maraviroc, a small-molecule allosteric inhibitor (15, 17, 24). Because HIV gp120 mutants can arise that recognize the drug-induced altered conformation of CCR5 at the cell surface, thereby causing drug resistance, strategies that reduce CCR5 expression may be preferable to those that primarily affect CCR5 conformation or activity (32). Indeed, genetic approaches are being developed to block CCR5 expression using a variety of techniques, including RNA interference and zinc finger nucleases (1–3, 20, 28, 38, 39). Since the effectiveness of these treatments in humans is still being evaluated, the development of additional methods to target CCR5 remains important.
We screened a library expressing many different artificial TM proteins and identified several unique proteins that specifically downregulated CCR5 expression and inhibited infection by R5-tropic HIV. Further study of these traptamers may provide insight into how the TM domains of CCR5 interact to specify its structure and function and may contribute to the design of novel therapeutic strategies against HIV.
MATERIALS AND METHODS
Cells and viruses.
Human 293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% fetal bovine serum (FBS), 5% bovine calf serum, 2 mM l-glutamine, 20 mM HEPES (pH 7.3), 1× penicillin-streptomycin (P-S), and 0.5 μg/ml amphotericin B (DMEM-10). Recombinant retroviruses were prepared by using calcium phosphate precipitation to cotransfect 293T cells with pVSV-G (where VSV-G is vesicular stomatitis virus G protein) (Clontech) and pCL-Eco (Imgenex) packaging plasmids and a retroviral plasmid or a pooled plasmid library (6, 33, 44). After transfection, the cells were cultured in Opti-MEM (Gibco) or DMEM-10 for 48 h at 37°C. The viral supernatant was then filtered through a 0.45-μm-pore-size filter (Millipore) and either used immediately or concentrated approximately 20× using Amicon Ultra-15 columns or Retro-Concentin Virus Precipitation Solution (System Biosciences). Pseudotyped HIV enhanced yellow fluorescent protein (eYFP) reporter viruses were generated similarly by cotransfection of 293T cells with various HIV env expression vectors and an env-deficient reporter vector based on a primary HIV isolate (pNL4-3) with deletions in the env, vif, and vpr genes and with an internal ribosome entry site (IRES)-eYFP cassette replacing the nef gene (10) (for the source of env genes and other details, see the paragraph “Reporter virus assays” below). pNL-BaL-HSA-R- virus, designated here pNL-BaL, was obtained from Ned Landau (New York University), and pNL4-3 virus was obtained from the NIH AIDS Research and Reference Reagent Program (NARRRP; catalog number 114, deposited by Malcolm Martin).
Murine BaF3 cells were maintained in RPMI 1640 medium supplemented with 10% heat-inactivated FBS, 5% WEHI-3B cell-conditioned medium (as a source of interleukin-3 [IL-3]), 2 mM l-glutamine, 0.05 mM β-mercaptoethanol, 1× P–S, and 0.5 μg/ml amphotericin B (RPMI-IL-3 medium). Human PM1 and CEM.NKR-CCR5 cells were maintained in RPMI 1640 medium supplemented with 10% FBS and 1× P-S (RPMI-10 medium), containing 2 mM l-glutamine for CEM.NRK-CCR5 cells. TZM-bl cells were maintained in DMEM supplemented with 10% FBS and 1× P-S (DMEM-10T). The last three cell lines were obtained from the NARRRP: PM1, catalog number 3038, deposited by Paulo Lusso and Robert Gallo; CEM.NKR.CCR5, catalog number 4376, deposited by Alexandra Trkola; and TZM-bl, catalog number 8129, deposited by John C. Kappes, and Xiaoyun Wu (Tranzyme, Inc.).
Retroviral library construction.
The YX4 library was constructed using a degenerate oligonucleotide in which codons 19 to 42, 45, and 46 were randomized. The nonrandomized segments were derived from the BPV E5 protein. Sequences of all oligonucleotides used in this study are shown in Table S1 in the supplemental material. To encode primarily hydrophobic amino acids at randomized residues 19 to 42, the composition ratio of A to C to G to T was 1:1:1:0.5 at the first position of each codon; 0.1:0.25:0.1:1 at the second position, and 0:1:0.1:0 at the third position. In addition, codon 45 was randomized using an equimolar mixture of A, C, G, and T at the first and third positions and an equimolar mixture of A and G at the second position, and codon 46 was randomized using an equimolar mixture of C and T at the first position, only A at the second position, and an equimolar mixture of A, C, G, and T at the third position. The randomization of positions 45 and 46 inserted stop codons in approximately 35% of the clones so that proteins ending at residues 44 or 45 could be expressed from the library. This degenerate oligonucleotide was annealed to a nondegenerate oligonucleotide, which was complementary to the 3′ fixed sequence of the degenerate oligonucleotide and encoded a stop codon and downstream restriction sites. These oligonucleotides were extended and amplified by PCR using short primers that annealed to the fixed regions at each end, and the resulting product was digested using BstXI and EcoRI and ligated into a pMSCV-puro (where MSCV is murine stem cell virus and puro is puromycin) vector (Clontech) modified to encode a hemagglutinin (HA) tag at the N termini of the library proteins (HA-pMSCV-puro). The ligation reaction was used to transform Escherichia coli strain DH10-β (Invitrogen). DNA from randomly picked ampicillin-resistant colonies was sequenced to confirm the desired amino acid composition and structure of clones in the library. Approximately 6.6 × 105 colonies of transformed bacteria were pooled, and plasmid DNA isolated from the pooled bacteria was used to generate the YX4 library.
The BY1 and BY6 limited random mutagenesis (LRM) libraries were constructed using degenerate oligonucleotides with fixed upstream sequences encoding amino acids 12 to 18 of the two traptamers, followed by a mutagenized sequence encoding amino acids 19 to 42 of each protein and a downstream sequence encoding Tyr43 and Trp44, a stop codon, and restriction sites for cloning. Each mutagenized position was synthesized with a nucleotide mixture consisting of 94% of the wild-type base from BY1 or BY6 and 2% each of the three remaining bases. This randomization scheme is predicted to yield an average of three substitutions per TM domain sequence. Each degenerate oligonucleotide was annealed to another oligonucleotide that was complementary to its fixed 3′ sequence and encoded additional downstream restriction sites. The resulting annealed products were extended and amplified by PCR using short primers complementary to the fixed regions at each end, digested using BstXI and EcoRI, and ligated into the HA-pMSCV-puro vector. The ligation reaction was used to transform E. coli strain DH10-β. DNA from randomly picked ampicillin-resistant colonies was sequenced to confirm the desired amino acid composition and structure of clones in each library. We pooled approximately 4 × 105 and 1 × 106 bacterial colonies for the BY1 and BY6 LRM transformations, respectively, and the plasmid DNA isolated from the pooled bacteria was used to generate the BY1 and BY6 LRM libraries.
Generation of BaF3 cell lines expressing individual chemokine receptors.
To generate BaF3 cells expressing human CCR5, cells were infected with a pMSCV-CCR5-neo retroviral expression vector, constructed by cloning the CCR5 gene from the pBABE-CCR5-puro plasmid (obtained through NARRRP; pBABE.CCR-5, catalog number 3331, deposited by Nathaniel Landau) into the pMSCV-neo retroviral vector (Clontech). Retroviral stocks were prepared as described above. Parental BaF3 cells were plated in a 12-well plate (5 × 105 cells per well in 500 μl of RPMI-IL-3 medium) and infected with 500 μl of concentrated stock of the MSCV-CCR5-neo virus or empty MSCV-neo vector, and one well was mock infected with medium only. Polybrene was added to each well at a final concentration of 4 μg/ml. After 4 h at 37°C, cells were transferred to individual 25-cm2 flasks containing 9 ml of RPMI-IL-3 medium with Polybrene. Two days later, 2 × 106 cells from each sample were collected and resuspended in RPMI-IL-3 medium containing 1 mg/ml G418 (selection medium). Six days later, when the mock-infected cells were dead, cells from the remaining samples were resuspended in medium without drug.
Fluorescence-activated cell sorting (FACS) was used to isolate a cell population expressing high levels of cell surface CCR5. Briefly, 2.5 × 106 G418-resistant cells were washed twice with phosphate-buffered saline (PBS) and incubated with blocking buffer (0.5% bovine serum albumin [BSA] in PBS, with azide) for 10 min. The cells were pelleted and incubated without permeabilization for 1 h at 4°C in blocking buffer with the mouse monoclonal anti-CCR5 antibody 45523 (antibodies used for flow cytometry are listed in Table S2 in the supplemental material), which recognizes a conformation-specific, extracellular epitope of CCR5 (27). After two washes with blocking buffer, the cells were resuspended in blocking buffer with an Alexa Fluor 488-conjugated donkey anti-mouse IgG secondary antibody (10 μg/ml final concentration) and incubated for 1 h at 4°C. The cells were then washed twice with blocking buffer and once with PBS and resuspended in PBS prior to sorting with a FACS Aria (BD Biosciences). The 5% of cells with the highest levels of cell surface CCR5 expression were isolated. This pooled cell line was used for the YX4 library screen. A clonal cell line with confirmed CCR5 expression was subsequently established from this sorted pool of cells and used for all experiments with individual clones isolated from the library.
A similar procedure was used to generate the BaF3-CCR2b cell line. The human CCR2b gene was cloned from a pcDNA3-CCR2b plasmid (12) (obtained from Robert Doms, University of Pennsylvania) into pMSCV-neo. Unconcentrated retrovirus was prepared as described above and used to infect BaF3 cells. Following selection in medium containing G418, cell surface CCR2b expression was confirmed by flow cytometry using the CCR2b-specific antibody 48607. A clonal cell line with high receptor expression was established from these transduced cells and used for all experiments.
To generate a chimeric receptor with CCR5 TM domain 5 (TM5) replaced with the corresponding CCR2b sequence (designated 5555255), site-directed mutagenesis was used to introduce four amino acid substitutions (K197M/I198R/V199N/V209I) into pMSCV-CCR5-neo. A gene encoding a chimera in which TM domains 4, 5, and 6 (along with the second and third intracellular loops and the second extracellular loop) of CCR2b were replaced with the corresponding CCR5 sequences (2225552) (35) (obtained from Robert Doms) was also cloned into pMSCV-neo. Unconcentrated retroviral stocks of each chimera were used to infect BaF3 cells, and clonal lines were generated with high cell surface expression of each chimera, as assessed by flow cytometry with either CTC5 or 48607 antibody as appropriate.
YX4 library infection and selection.
BaF3-CCR5 cells were plated in a 12-well plate (5 × 105 cells per well in 500 μl medium) and infected with 500 μl concentrated virus stocks of the YX4 retroviral plasmid library (eight wells) or the HA-pMSCV-puro vector (one well) as described above. Two days after infection, 2 × 106 cells from each sample were resuspended in selection medium containing 1 μg/ml puromycin. Three days later, when mock-infected cells were dead, cells from each remaining sample were resuspended in medium without drug. The following day, an aliquot of 2 × 106 cells from each original infected well was incubated with the conformation-specific 2D7 or 45523 antibody as described above for antibody 45523 (four pools of cells per antibody). FACS was used to recover the 10% of cells from each infected pool that displayed the lowest binding to the antibodies. The sorted cells were expanded, and genomic DNA was isolated (DNeasy Blood and Tissue Kit; Qiagen) from each of the eight pools (approximately 2 × 106 cells per pool) and combined. Library insert sequences were recovered from the genomic DNA by PCR amplification (Expand Long Template PCR kit; Roche) using primers specific for the invariant portions of the E5 gene and the vector. The resulting PCR products were purified, digested using XhoI and EcoRI, and ligated into the HA-pMSCV-puro vector. The ligation product was used to transform E. coli strain DH10-β (Invitrogen), and the plasmid DNA isolated from pooled lawns of ampicillin-resistant bacteria was used to generate the secondary library.
The secondary library was packaged, concentrated, and used to infect four wells of naïve BaF3-CCR5 cells as described above. Following puromycin selection, the cells were incubated with the 2D7 or 45523 antibody (two pools per antibody), and the 2.5% of cells from each pool that displayed the lowest antibody binding were recovered by FACS. The genomic DNA isolated from expanded, sorted cells was kept in four separate pools for recovery and amplification of the library sequences, and four tertiary libraries were prepared as described above. The tertiary libraries were packaged separately, concentrated, and used to infect one well each of naïve BaF3-CCR5 cells. Following drug selection and recovery, the cells were sorted as before with the antibody used in the previous round for a given pool. The population of cells displaying minimal antibody binding was recovered (approximately 1 × 105 to 2 × 105 cells per pool), and the genomic DNA from each pool was used to generate separate quaternary libraries. The quaternary libraries were packaged, used to infect separate pools of naïve BaF3-CCR5 cells, and sorted as described above. The cells with minimal antibody binding were recovered (approximately 1.5 × 105 to 4 × 105 per pool), genomic DNA from each pool was isolated, and library inserts were amplified and ligated into the HA-pMSCV-puro vector and used for bacterial transformation. Individual colonies were sequenced after the third and fourth rounds of selection.
Testing clones for inhibition of CCR5 expression.
Sequences that were repeated multiple times in pools from the tertiary and/or quaternary libraries, as well as a clone from the unselected primary library (US7), were packaged individually into retrovirus and used to infect BaF3-CCR5, PM1, or CEM.NKR-CCR5 cells as described above for BaF3-CCR5 cells. For these infections, a single well of cells was infected with 1 ml of unconcentrated virus for each clone. Following drug selection (1 μg/ml puromycin for BaF3-CCR5 and 0.5 μg/ml for PM1 and CEM.NKR-CCR5 cells), cells were incubated with a conformation-specific (2D7, 45523, or 45531) or non-conformation-specific (CTC5) CCR5 antibody and analyzed by flow cytometry using a FACSCalibur (BD Biosciences). The above procedure was also used to express the traptamers in BaF3 cells expressing CCR2b or the chimeric receptors, and drug-selected cells were analyzed by flow cytometry using anti-CCR5 (CTC5) and anti-CCR2b (48607) antibodies as described. The CXCR4-specific monoclonal antibodies 12G5, 44708, and 44717 and the CD4-specific monoclonal antibody B4 were used to analyze CXCR4 and CD4 levels, respectively, by flow cytometry.
BY1 and BY6 limited random mutagenesis library selections.
For infection with the BY1 LRM library, PM1 cells were infected as described above with 500 μl of concentrated virus stocks of the BY1 LRM library (eight wells) or the HA-pMSCV-puro vector (one well) and selected with 0.5 μg/ml puromycin. Four days later, cells were resuspended in medium without drug. The following day, 2 × 106 cells per pool were incubated with the conformation-specific 2D7 antibody as described above. FACS was used to recover the 10% of cells from each infected pool that displayed the lowest binding to the antibody. The sorted cells were expanded, and genomic DNA was isolated from each of the eight pools and combined. Library insert sequences were recovered from the genomic DNA and cloned into HA-pMSCV-puro. The ligation product was used to transform E. coli strain DH10-β, and the plasmid DNA isolated from lawns of ampicillin-resistant bacteria was used to generate the secondary library.
The secondary library was packaged into retrovirus, concentrated, and used to infect four pools of naïve PM1 cells. Following puromycin selection, the cells were incubated with the 2D7 antibody, and the 5% of cells from each pool that displayed the lowest antibody binding were recovered by FACS. The genomic DNA isolated from the expanded, sorted cells was kept in four separate pools for recovery of the library sequences, and four tertiary libraries were prepared as described above. Each was packaged separately, concentrated, and used to infect one pool of naïve PM1 cells. Following puromycin selection, the cells were sorted as before to recover the 5% of cells from each pool that displayed the lowest antibody binding. The genomic DNA from each sorted pool was used to generate four quaternary libraries, which were packaged into retrovirus and used to infect four separate pools of CEM.NKR-CCR5 cells. These cells were drug selected and sorted for reduced antibody binding to CCR5 as in previous rounds, and those with lowest antibody binding were recovered (2 × 105 to 5 × 105 per pool). The genomic DNA from each pool was isolated separately, and the library inserts were cloned into the HA-pMSCV-puro vector and used for bacterial transformation. Individual colonies were selected for DNA sequencing. Sequences that were repeated multiple times were packaged individually into retrovirus and used to infect PM1 or CEM.NKR-CCR5 cells as described above. Following drug selection and recovery, these cells were incubated with the 2D7 or CTC5 anti-CCR5 antibody for flow cytometry.
The BY6 LRM library was screened in CEM.NKR-CCR5 cells as described above for the BY1 LRM library screen. In the first round of the screen, the 10% of cells from each of eight infected pools with the lowest binding to the 2D7 antibody were isolated by FACS. The secondary library was prepared and used to infect naïve CEM.NKR-CCR5 cells. In this round, the 5% of cells from each of four infected pools with the lowest antibody binding were isolated. Four tertiary libraries were prepared and used to infect naïve CEM.NKR-CCR5 cells. The 10% of the cells in each pool with minimal antibody binding were isolated by FACS. Inserts were cloned from genomic DNA, and sequences that were repeated multiple times were packaged individually into retrovirus for testing.
qRT-PCR and immunoblotting.
Total RNA was isolated from approximately 1.5 × 106 cells with TRIzol (Ambion) and purified using a phenol-chloroform protocol and an Ambion Turbo DNA-Free kit. cDNA synthesis was performed using a Bio-Rad iScript cDNA synthesis kit, and quantitative real-time PCR (qRT-PCR) was carried out using a Bio-Rad MyiQ Thermal Cycler, Bio-Rad iQ SYBR green Supermix, and primers specific for CCR5 or glyceraldehyde-3-phosphate dehydrogenase (GAPDH; used as a control) (see Table S1 in the supplemental material).
For immunoblotting, detergent extracts were made using radioimmunoprecipitation assay-morpholinepropanesulfonic acid (RIPA-MOPS) lysis buffer (20 mM MOPS, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS, 1 mM phenylmethylsulfonyl fluoride [PMSF], activated sodium metavanadate, leupeptin, and aprotinin). Lysates were incubated on ice for 20 min and then centrifuged at >14,000 × g for 30 min at 4°C. The concentration of protein in the supernatant was quantified using a Pierce bicinchoninic acid (BCA) protein assay kit. For CCR5 immunoblotting, 25 μg of extracted protein was transferred to a new tube for each sample, 2× protein sample buffer (with β-mercaptoethanol and dithiothreitol [DTT]) (2× PSB) was added, and samples were heated at 72°C for 5 min. Proteins were separated by electrophoresis on a 10% SDS-polyacrylamide gel and transferred to a 0.2-μm-pore-size polyvinylidene difluoride (PVDF) membrane in transfer buffer without SDS (25 mM Tris-base, 192 mM glycine, 20% methanol) for 1 h at 100 V. Membranes were blocked in 5% milk and Tris-buffered saline (TBS) containing 0.1% Tween 20 (TBST) with azide for 1 h at room temperature and then incubated overnight with primary antibody diluted in 5% milk-TBST with azide. To detect CCR5 by immunoblotting, a rat monoclonal antibody against CCR5, CKR-5 (HEK/1/85a; Santa Cruz Biotechnology), was used at a 1:1,000 dilution in 5% milk-TBST as a primary antibody, followed by a goat anti-rat horseradish peroxidase (HRP)-conjugated secondary antibody (Jackson ImmunoResearch). To detect actin as a loading control, membranes were stripped in Restore Western Stripping Buffer (Thermo Scientific) for 30 min at 37°C with vigorous shaking, washed five times in TBST, blocked in 5% milk-TBST with azide for 1 h at room temperature, and incubated overnight with a polyclonal goat antibody against actin (C11; Santa Cruz Biotechnology) diluted 1:1,000 in 5% milk-TBST with azide, followed by a donkey anti-goat-HRP secondary antibody (Jackson ImmunoResearch). To detect the traptamers, 500 μg of total protein was incubated with 30 to 50 μl of Roche anti-HA affinity matrix (immobilized rat monoclonal, clone 3F10) and rotated overnight at 4°C. Beads were then pelleted at >14,000 × g for 1 min and washed three times in 1 ml of NET-N (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, 0.5% NP-40) with PMSF. After the final wash, 20 μl of 2× PSB was added to each pellet. Samples were separated by electrophoresis on a 20% SDS-polyacrylamide gel or a 4 to 20% Mini-Protean TGX gel (Bio-Rad). After electrophoretic separation, proteins were transferred to PVDF membranes as described above for CCR5. Blocked membranes were incubated overnight with a 1:1,000 dilution of a mouse anti-HA antibody (clone 12CA5), followed by a 1-h incubation with a donkey anti-mouse-HRP secondary antibody. All blots were visualized using either a SuperSignal West Pico Chemiluminescent Substrate or a SuperSignal West Femto Maximum Sensitivity Substrate detection kit (Thermo Scientific).
Cell surface biotinylation assay.
To biotinylate cell surface proteins, 2.5 × 106 BaF3-CCR5 cells expressing various traptamers were washed once in PBS and incubated in 5 ml polystyrene round-bottom tubes (BD Falcon) with 750 μg of EZ-Link sulfo-NHS-SS-biotin (sulfosuccinimidyl 2-[biotinamido]ethyl-1,3-dithiopropionate) reagent (diluted in dimethyl sulfoxide [DMSO]; Thermo Scientific) in 1 ml of biotinylation buffer (10 mM triethanolamine, 2 mM CaCl2, 125 mM NaCl, pH 8.9) and rocked for 25 min on ice at 4°C. Cells were then pelleted by centrifugation at 250 × g at 4°C for 5 min, and the supernatant was aspirated. Biotinylation was repeated once more as above with fresh biotin and biotinylation buffer. Unreacted biotin was quenched by the addition of 1 ml of 100 mM glycine in phosphate-buffered saline with 0.1 mM CaCl2 and 1 mM MgCl2 (PBS2+) and incubation with rocking for 20 min on ice at 4°C. Cells were then washed once in cold PBS2+ and lysed in 1 ml of RIPA-MOPS lysis buffer, transferred to an Eppendorf tube, and incubated on ice for 20 min. To precipitate biotinylated proteins, 60 μl of Pierce streptavidin-agarose resin beads (Thermo Scientific), washed three times in RIPA-MOPS lysis buffer, was added to each lysate and rotated overnight at 4°C. The following day, the streptavidin beads were pelleted by centrifugation at >14,000 × g for 1 min, and the supernatant was transferred to a new Eppendorf tube and saved. The streptavidin beads were then washed three times in NET-N containing PMSF and resuspended in 20 μl of 2× PSB. To determine the amount of receptor that was not biotinylated (intracellular pool), CCR5 was immunoprecipitated from the recovered supernatant with the rat monoclonal antibody CKR-5 and protein A/G Plus-agarose beads (Santa Cruz Biotechnology) and washed as above. CCR5 was detected by immunoblotting as described above with CKR-5.
Reporter virus assays.
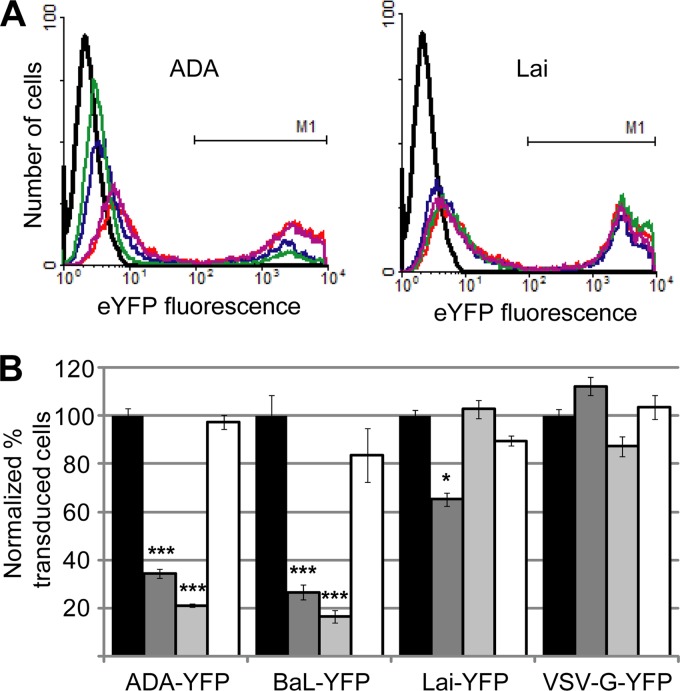
Calcium phosphate precipitation was used to cotransfect 293T cells with the HIV-eYFP reporter plasmid and a plasmid expressing R5-tropic (ADA, BaL, JRFL, SF162, B1206, BK184, and MG505), R5X4-tropic (89.6), X4-tropic (Lai and HXB2), or control (VSV-G) Env proteins (10). The BaL, 89.6, and Lai envelope plasmids were provided by Robert Doms, and the ADA, JRFL, SF162, and HXB2 plasmids were provided by Dan Littman. B1206 (catalog number 11519), BK184 (catalog number 11522), and MG505 (catalog number 11528) plasmids were obtained from the NARRRP (deposited by Julie Overbaugh). All of the envelopes used were cloned from subtype B viruses with the exception of BK184 (subtype C/D) and B1206 and MG505 (both subtype A). Pseudoviruses were harvested from transfected cells as described above for other retroviruses. PM1 cells stably expressing individual traptamers or a vector control were plated in six-well plates (1 × 105 cells per well in 500 μl of RPMI-10 medium) and infected with 2 ml of unconcentrated reporter virus for pseudotypes with HIV env or 500 μl of unconcentrated virus plus 1.5 ml of RPMI-10 medium for the VSV-G-pseudotyped virus in the absence of Polybrene. After 72 h, cells were analyzed by flow cytometry to measure eYFP fluorescence. The percentage of infected cells for each sample was calculated as the fraction of the total number of cells analyzed that were eYFP positive. In these experiments, in addition to the appearance of a peak of fluorescent cells expressing eYFP (M1 gate shown in Fig. 5A), a second peak of weakly eYFP-positive cells was observed whose fluorescence partially overlapped with that of uninfected cells (shown at the left of each plot in Fig. 5A). The latter shift appears to be attributable to pseudotransduction (i.e., delivery of eYFP packaged in virus particles) (18) and was not observed in experiments where the reporter virus was removed and replaced with fresh medium 4 h after the initial infection. Therefore, we quantified only the peak shown at the right of each plot in Fig. 5A by using a gate which excluded 99% of mock-infected cells.
Fig 5.
Traptamers inhibit transduction by pseudotyped HIV reporter viruses. (A) PM1 cells expressing empty vector (red), clone BY1 (blue), clone BY6 (green), or unselected clone US7 (purple) were infected with HIV-based eYFP reporter viruses pseudotyped with a CCR5-specific envelope protein (ADA, left, and BaL [data not shown]), a CXCR4-specific envelope protein (Lai; right), or the VSV-G protein (data not shown). Mock-infected cells are represented in black. HIV reporter virus transduction was assessed by flow cytometry 72 h after infection by measuring eYFP fluorescence as indicated by the M1 gates shown on the right of each panel. The ranges of PM1 cells stably transduced in a typical experiment are as follows: ADA, 40 to 50%; BaL, 15 to 25%; Lai, 35 to 45%; VSV-G, 40 to 50%. The peaks at the left in each plot represent uninfected cells or cells undergoing pseudotransduction (18) and were not quantified here. However, we note that pseudotransduction by the ADA particles was also reduced by the active traptamers. (B) PM1 cells expressing empty vector (black bars), clone BY1 (dark gray bars), clone BY6 (light gray bars), or unselected clone US7 (white bars) were infected with HIV-based eYFP reporter viruses and analyzed as described above. Reporter virus transduction was quantified as the percentage of cells that expressed eYFP, normalized to transduction of cells expressing empty vector. Results shown are the average of at least three independent trials, with error bars indicating standard error. Significance relative to cells expressing vector only was determined by a two-tailed t test: *, P < 0.05; ***, P < 0.001.
To measure transduction of TZM-bl cells, we generated reporter viruses pseudotyped with the R5-tropic Env protein ADA or R3, a maraviroc-resistant R5-tropic Env protein (provided by Robert Doms) (42). TZM-bl cells stably expressing empty vector or individual traptamers were plated in six-well plates (1 × 105 cells per well) 1 day prior to infection. One hour prior to infection, the medium in each well was replaced with 500 μl of fresh DMEM-10T medium. For the maraviroc-treated control samples, maraviroc (obtained from NARRRP; catalog number 11580, deposited by the Division of AIDS, NIAID) was added to a final concentration of 100 nM. After 1 h at 37°C, the cells were infected with 2 ml of unconcentrated reporter virus per well and incubated at 37°C in the absence of Polybrene. After 4 h, the virus and medium on each well were replaced with 2 ml of fresh DMEM-10T medium. After 72 h, cells were analyzed by flow cytometry for eYFP expression as described above.
HIV replication assay.
PM1 cells stably expressing BY1PC2 or the unselected clone US7 (5 × 105 cells per well) were infected with 1 μl of pNL-BaL or pNL4-3 virus (p24 capsid concentrations, 0.44 μg/ml and 1.58 μg/ml, respectively) in a 12-well plate. On alternating days postinfection (from 2 days to 24 days), supernatant was removed from each well of infected PM1 cells, centrifuged at 15,000 rpm for 4 min at room temperature, and used to infect TZM-bl cells (500 μl of supernatant to each well of cells plated 1 day prior as described for PM1 cells). The remaining PM1 cells in each well were split at a ratio of 1:4. TZM-bl cells were harvested 3 days after infection by washing with 500 μl of PBS and lysis in 250 μl of luciferase lysis buffer. Lysates were stored at −20°C and analyzed simultaneously at the end of the experiment time course for luciferase activity generated by Tat-mediated induction of the integrated HIV promoter in the TZM-bl cells. Each experiment was performed in triplicate.
RESULTS
Library design and cells for screening.
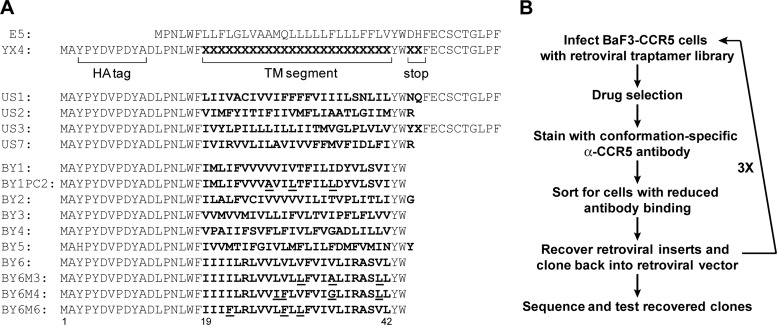
To isolate TM protein inhibitors of CCR5, we constructed a retroviral library, YX4, encoding a large number of small, artificial hydrophobic proteins with randomized TM domains (Fig. 1A). As described in Materials and Methods, the TM domain of the traptamers (amino acids 19 to 42) was randomized by synthesizing a degenerate oligonucleotide using nucleotide mixtures that encoded a composition of amino acids similar to that of naturally occurring TM domains (i.e., ∼80% hydrophobic and ∼20% hydrophilic) (13). In addition, positions 45 and 46 were randomized to insert stop codons in approximately 35% of the clones since disulfide-linked dimers, mediated by the cysteines in the C terminus of BPV E5, might not be ideal for targeting a multipass TM protein. The proteins expressed by the library also contained an N-terminal influenza virus hemagglutinin (HA) tag. Sequencing of individual library members confirmed the design of the library, with approximately 40% of the clones encoding truncated proteins (Fig. 1A). Based on deep sequencing of a similar library, we estimate that YX4 expresses several hundred thousand unique, small TM proteins (31). To generate cells for the inhibitor screen, we stably expressed human CCR5 in murine BaF3 cells and used fluorescence-activated cell sorting (FACS) to isolate BaF3-CCR5 cells expressing a high and homogeneous level of cell surface CCR5.
Fig 1.
Sequence of traptamers and selection scheme. (A) The top line shows the sequence of the wild-type BPV E5 protein. The second line shows the design of the YX4 library, with bold X representing randomized positions. US1 to US3 and US7 are examples of full-length and truncated sequences present in the unselected library, with the randomized sequences shown in bold. The remaining lines are the sequences of traptamers that downregulate CCR5 expression. Mutations in clones BY1PC2, BY6M3, BY6M4, and BY6M6 are underlined. (B) Schematic diagram of the genetic screen used to isolate traptamers that downregulate CCR5 expression. α, anti.
Isolation of CCR5 inhibitors using a FACS-based screen.
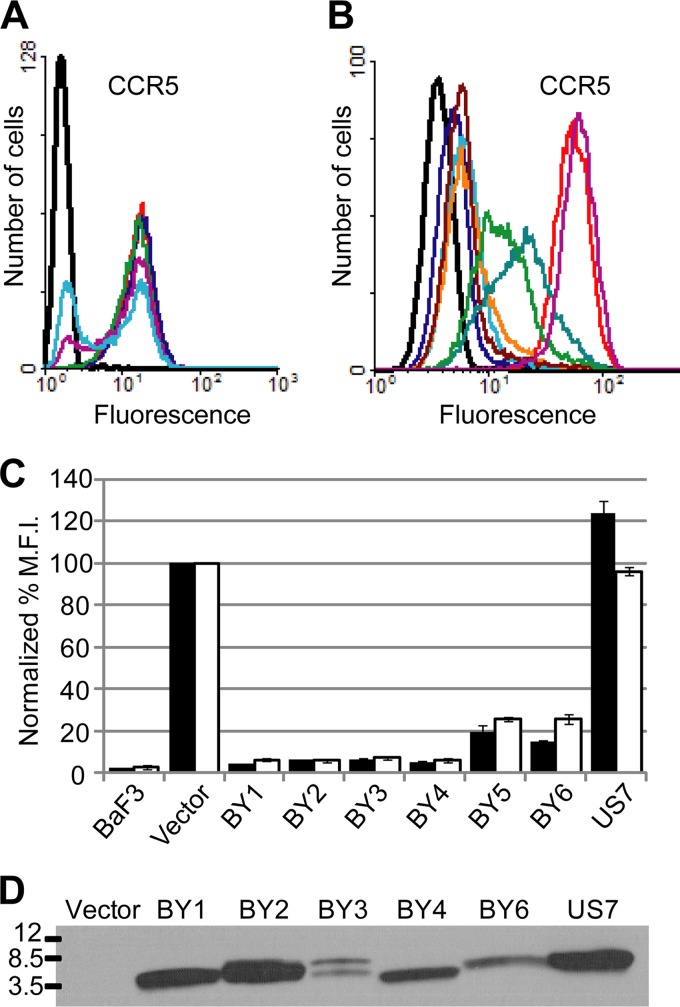
Several independent pools of BaF3-CCR5 cells were infected with the YX4 traptamer expression library and stained without permeabilization with a conformation-specific anti-CCR5 antibody (Fig. 1B). The library-infected cells displayed flow cytometry profiles similar to those of uninfected cells (Fig. 2A), consistent with our expectation that traptamers targeting CCR5 are rare in the library. FACS was used to isolate the 10% of cells from each infected pool that displayed the lowest binding to the CCR5 antibody to enrich for cells in which a traptamer inhibited the expression of CCR5 or affected its folding or localization. Library sequences were recovered by PCR from genomic DNA isolated from the sorted cells, cloned back into the retroviral vector to generate a secondary library, packaged into retrovirus particles, and used to infect naïve BaF3-CCR5 cells. The 2.5% of infected cells that displayed the lowest binding to the anti-CCR5 antibody were isolated by FACS, and library sequences were again recovered and used to generate tertiary libraries, which were used to infect naïve BaF3-CCR5 cells. In this round, approximately 10% of the infected cells showed a clear reduction in anti-CCR5 antibody binding (Fig. 2A). These cells were recovered by FACS, and the library sequences amplified from genomic DNA were used to generate quaternary libraries. Strikingly, 30 to 40% of BaF3-CCR5 cells infected with the quaternary libraries displayed dramatically reduced CCR5 antibody binding (Fig. 2A). These cells were recovered, and inserts encoding the TM proteins were amplified, cloned, and sequenced. A number of clones were repeated multiple times (Fig. 1A, clones BY1 to BY6). The TM sequences of the repeated clones were not related to one another, to the wild-type BPV E5 sequence, or to other known proteins, and all of them were truncated after the randomized TM domain, suggesting a strong selection against disulfide-linked dimers.
Fig 2.
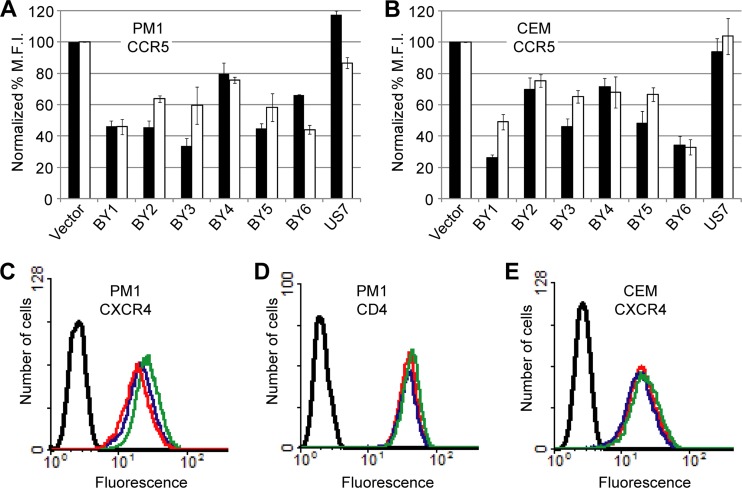
Identification of traptamers that inhibit cell surface expression of CCR5. (A) BaF3-CCR5 cells were infected with the empty vector (red), the YX4 library (dark blue), or the secondary, tertiary, or quaternary libraries generated after each round of the screen (green, purple, and light blue, respectively). Data for parental BaF3 cells lacking CCR5 are shown in black. Nonpermeabilized cells were incubated with a conformation-specific, anti-CCR5 antibody (45523) and analyzed by flow cytometry. (B) Parental BaF3 cells (black) and BaF3-CCR5 cells expressing vector (red), clone BY1 (dark blue), clone BY2 (light blue), clone BY3 (orange), clone BY4 (dark red), clone BY5 (blue-green), clone BY6 (green), or unselected clone US7 (purple) were stained and analyzed as described for panel A. (C) The geometric mean fluorescence intensity (MFI) calculated for each sample in the experiment shown in panel B (black bars) and in a similar experiment using the non-conformation-specific anti-CCR5 antibody CTC5 (white bars) is plotted as a percentage of that observed for the vector-only control cells. Results shown are the averages of two independent trials, with error bars representing standard error. (D) RIPA-MOPS extracts were prepared from BaF3-CCR5 cells expressing empty vector, the indicated selected clone, or US7. Samples were immunoprecipitated and immunoblotted with anti-HA antibodies. Molecular mass markers in kDa are shown on the left. Clone BY5 had a mutation in the HA tag (Fig. 1A) and was therefore not included here. Correction of the mutation allowed detection of this traptamer but did not affect the ability of BY5 to downregulate cell surface CCR5 expression (data not shown).
Retroviral vectors encoding BY1 to BY6, as well as a truncated sequence from the unselected YX4 library (clone US7), were packaged and used to infect clonal BaF3-CCR5 cells. Expression of cell surface CCR5 was assessed by flow cytometry of nonpermeabilized cells. Each of the selected proteins reduced the binding of conformation-specific (45523, 2D7, and 45531) and non-conformation-specific (CTC5) anti-CCR5 antibodies to cells, but the unselected protein did not (Fig. 2B and C; also data not shown). Notably, all of the antibodies displayed similar patterns of binding, in which expression of proteins BY1 to BY4 resulted in nearly complete elimination of antibody binding, whereas there was greater residual antibody binding to cells expressing BY5 and BY6. As shown in Fig. 2D, the traptamers were readily detectable by immunoblotting.
Effects of traptamers on CCR5 mRNA and protein levels.
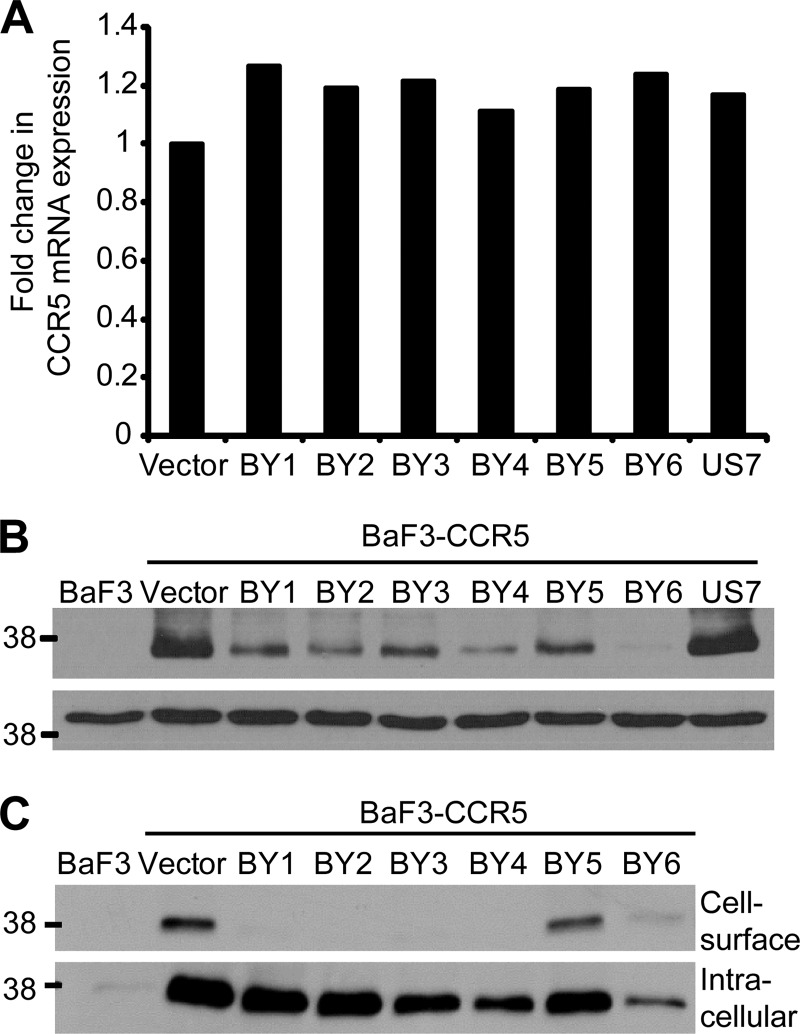
To determine whether the traptamers affected CCR5 transcription, total RNA was harvested from BaF3-CCR5 cells expressing the empty vector, an active clone, or US7, and qRT-PCR was used to quantify CCR5 mRNA. None of the traptamers affected CCR5 mRNA levels (Fig. 3A), indicating that they did not repress CCR5 transcription or destabilize CCR5 mRNA. Because the traptamers inhibited binding of a non-conformation-specific antibody that recognizes a linear epitope in CCR5 (Fig. 2C), they did not appear to simply alter CCR5 conformation at the cell surface. Indeed, immunoblotting of whole-cell lysates showed that the active traptamers caused a marked reduction in total CCR5 protein expression compared to the samples obtained from cells expressing the empty vector or the unselected clone (Fig. 3B). Notably, cells expressing BY6 displayed the most dramatic reduction in total levels of CCR5 protein even though BY6 was not as active as the other traptamers at reducing the amount of cell surface CCR5, as measured by flow cytometry (Fig. 2C).
Fig 3.
Traptamers reduce total CCR5 protein levels but not mRNA levels. (A) Total RNA was isolated from BaF3-CCR5 cells expressing empty vector, the indicated active clone, or an unselected clone (US7). CCR5 mRNA was measured by qRT-PCR and normalized to GAPDH expression. Fold change in mRNA expression is plotted compared to levels in vector-infected cells. Three independent experiments yielded similar results, and data from one such experiment are shown. (B) RIPA-MOPS extracts were prepared from parental BaF3 cells and from BaF3-CCR5 cells expressing empty vector, the indicated traptamer, or an unselected clone (US7) and then immunoblotted with a CCR5-specific antibody (CKR-5) (top panel). The membrane was stripped and reprobed for actin (bottom panel). Molecular mass markers in kDa are shown on the left. Similar results were obtained in multiple independent experiments. (C) Intact cells were biotinylated as described in Materials and Methods, and biotinylated proteins were recovered using streptavidin beads. The pellet fraction was immunoblotted directly to detect cell surface CCR5 (top panel), while the supernatant fraction was immunoprecipitated with an antibody specific for CCR5 (CKR-5) and then immunoblotted with the same antibody to detect intracellular CCR5 (bottom panel). The two panels are at the same exposure. Molecular mass markers in kDa are shown on the left. Similar results were obtained in multiple independent experiments.
To explore the effects of the traptamers in more detail, cell surface biotinylation and immunoblotting were used to measure cell surface and intracellular pools of CCR5. Nonpermeabilized cells were treated with biotin, and biotinylated proteins in whole-cell lysates were separated from nonbiotinylated proteins by streptavidin affinity pulldown. Both fractions were then analyzed by immunoblotting with a non-conformation-specific anti-CCR5 antibody (CKR-5). As shown in Fig. 3C, the majority of CCR5 in control cells expressing empty vector was intracellular. Furthermore, the effects of the traptamers on cell surface CCR5 expression (top panel) detected by this method were consistent with the flow cytometry results; namely, the traptamers markedly reduced cell surface CCR5 expression, but more cell surface CCR5 persisted in cells expressing BY5 and BY6 than in cells expressing the other traptamers. In contrast, most traptamers caused little reduction in intracellular (i.e., nonbiotinylated) CCR5 (Fig. 3C, bottom panel). However, cells expressing BY6 contained markedly reduced levels of intracellular CCR5. Thus, the low level of CCR5 detected in total lysates of cells expressing BY6 is due to the preferential depletion of intracellular CCR5 by this traptamer. These results confirmed that the active traptamers inhibited cell surface CCR5 expression and suggested that they decreased CCR5 stability. In addition, BY6 appears to act differently than the other active traptamers because it efficiently reduced intracellular as well as cell surface CCR5.
Traptamers downregulate CCR5 and inhibit HIV in human cells.
We also determined the effect of the traptamers on expression of CCR5 in two human T-cell lines: PM1 cells, which express endogenous CCR5, CXCR4, and CD4, and CEM.NKR-CCR5 cells, which express endogenous CXCR4 and CD4 and exogenous human CCR5. Expression of individual traptamers reduced the binding of conformation-specific (2D7) and non-conformation-specific (CTC5) anti-CCR5 antibodies in both cell lines (Fig. 4A and B), but CCR5 downregulation was less dramatic in either human cell line than in BaF3-CCR5 cells. In addition, the relative potencies of the various traptamers differed in different cells (e.g., BY6 was one of the most active traptamers in downregulating cell surface CCR5 in CEM.NKR-CCR5 cells, while it was one of the least active traptamers in BaF3-CCR5 cells). Stable expression of the traptamers did not inhibit cell growth (data not shown), consistent with the expectation that our cellular selection method would eliminate toxic traptamers.
Fig 4.
Traptamers downregulate CCR5 in human T-cell lines. Flow cytometry was performed on nonpermeabilized PM1 (A) and CEM.NKR-CCR5 (B) cells expressing empty vector, the indicated active clone, or an unselected clone (US7). Bar graphs show the geometric mean fluorescence intensity (MFI) for each sample, calculated as a percentage of that observed for the vector-only control in cells stained with the conformation-specific anti-CCR5 antibody 2D7 (black bars) or the non-conformation-specific anti-CCR5 antibody CTC5 (white bars). Results shown are the averages of at least two independent trials, with error bars indicating standard error. Flow cytometry was also performed with PM1 or CEM.NKR-CCR5 cells, as indicated, expressing vector (red), clone BY1 (blue), or clone BY6 (green) and stained with the anti-CXCR4 antibody 12G5 (C and E) or the anti-CD4 antibody B4 (D). Data for unstained cells are shown in black.
We then tested the effects of the traptamers on transduction by pseudotyped, single-cycle HIV reporter viruses expressing enhanced yellow fluorescent protein (eYFP). The HIV reporter was complemented in trans with CCR5-tropic (R5-tropic) or CXCR4-tropic (X4-tropic) HIV envelope proteins or with the vesicular stomatitis virus (VSV) G envelope protein, which does not depend on CCR5 for entry. These reporter viruses were used to infect PM1 cells stably expressing individual traptamers (the reporter viruses did not efficiently infect CEM.NKR-CCR5 cells). After 72 h, the cells were analyzed by flow cytometry for eYFP fluorescence. In this assay, all of the traptamers recovered from the library reduced transduction by reporter viruses pseudotyped with R5-tropic envelope proteins (ADA and BaL), while the unselected clone had no effect (Fig. 5 and data not shown). BY1 and BY6 were the most active, inhibiting transduction by up to 80%. As expected, none of the traptamers inhibited infection by a VSV-G pseudotyped reporter virus. Clones BY2 to BY6 did not affect transduction by an X4-tropic pseudovirus (Fig. 5, Lai; also data not shown). In contrast, clone BY1 had a modest inhibitory effect on this pseudovirus (Fig. 5) even though it did not reduce cell surface CXCR4 levels (Fig. 4C). None of the traptamers altered cell surface CD4 levels in either human cell line (Fig. 4D and data not shown). Thus, traptamers isolated on the basis of their ability to reduce cell surface expression of CCR5 in murine cells preferentially inhibited transduction of human T cells by HIV reporter viruses dependent on CCR5 for entry.
Interestingly, BY6 reproducibly increased the binding of multiple, different anti-CXCR4 antibodies to PM1 cells (Fig. 4C and data not shown) even though it did not affect transduction by the X4-tropic pseudovirus (Fig. 5). This increase in antibody binding did not occur in CEM.NKR-CCR5 (Fig. 4E) or in BaF3-CXCR4 (data not shown) cells, and none of the other traptamers increased CXCR4 staining (data not shown), again suggesting that BY6 acts differently than the other traptamers.
Optimization of anti-HIV activity in human cell lines.
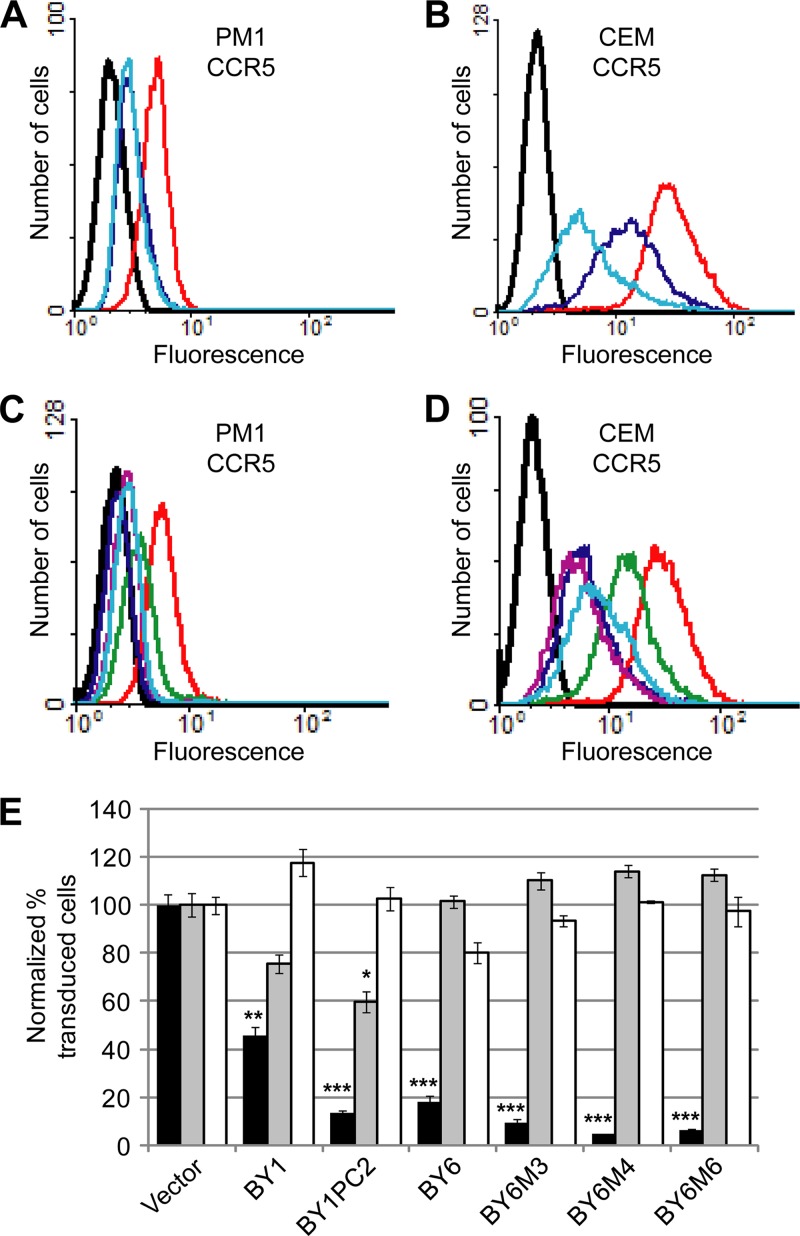
We subjected the TM segment of BY1 and BY6 to limited random mutagenesis (LRM) and screened the resulting mutant libraries for variants with increased ability to downregulate CCR5 in human T cells. Serial FACS-based isolation of cells displaying the lowest CCR5 antibody binding, recovery of retroviral inserts, and reinfection of naïve PM1 and/or CEM.NKR-CCR5 cells were conducted as described in Materials and Methods, and individual, recovered clones were tested for activity. The most active clone isolated from the BY1 LRM library, designated BY1PC2, had three amino acid substitutions in its TM domain (Fig. 1A). Three active clones were isolated from the BY6 LRM library and had either three (BY6M3 and BY6M6) or four (BY6M4) amino acid substitutions in the TM domain (Fig. 1A).
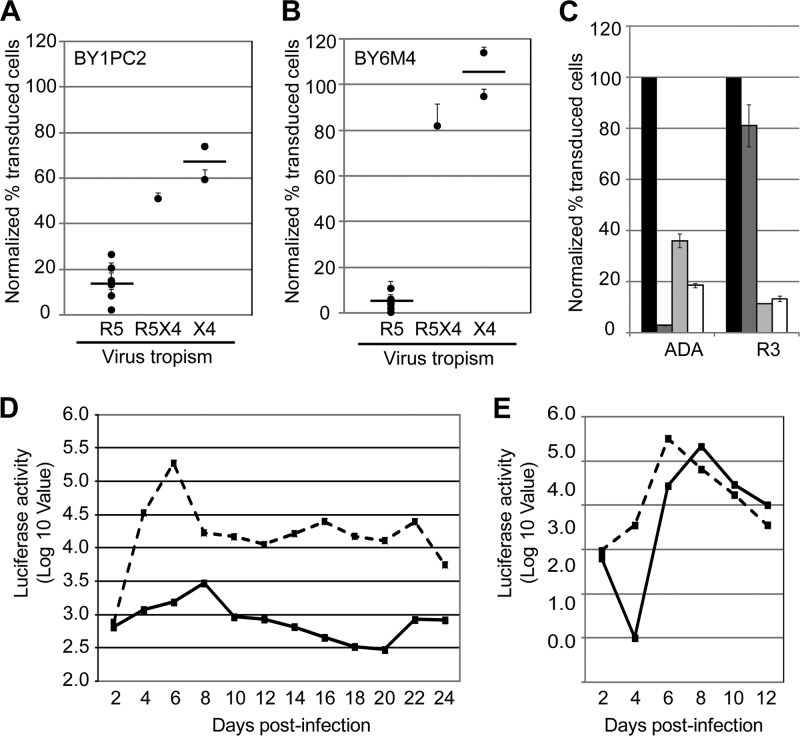
In CEM.NKR-CCR5 cells, the mutant traptamers reduced CCR5 cell surface levels approximately 3-fold better than the parent clones (Fig. 6B and D). In PM1 cells, these traptamers appeared to downregulate CCR5 only slightly better than or, in the case of BY1PC2, as well as the parent clones (Fig. 6A and C), but because of the relatively low levels of CCR5 on the surface of the PM1 cells, it is difficult to reliably detect slight differences in CCR5 expression in these cells. The mutant traptamers did not affect cell surface expression of CXCR4 or CD4 (data not shown). Notably, BY1PC2 was reproducibly 3-fold more active than BY1 in the HIV pseudovirus transduction assay in PM1 cells, inhibiting transduction by an R5-tropic reporter virus by almost 90% compared to cells expressing the empty vector (Fig. 6E). BY1PC2 also inhibited an X4-tropic pseudotype by 40% but was inactive against the VSV-G pseudotype. Similarly, the BY6 mutants were reproducibly 2- to 4-fold more active than BY6, with BY6M4 inhibiting R5-tropic reporter virus transduction by greater than 95% (Fig. 6E). BY6 and the BY6 mutants were inactive against the X4-tropic and VSV-G pseudotypes. BY1PC2 and BY6M4 also dramatically inhibited transduction by several additional R5-tropic pseudotypes, including three packaged with envelope genes cloned directly from patient isolates of various HIV subtypes (Fig. 7A and B). Furthermore, BY1PC2 and BY6M4 displayed intermediate activity against an R5X4-tropic pseudotype, 89.6, which can use either CCR5 or CXCR4. These results indicated that BY1PC2 and BY6M4 are active against a broad range of R5-tropic pseudotypes derived from various HIV subtypes and that this activity is dictated by the dependence of a given Env strain on CCR5.
Fig 6.
BY1 and BY6 mutants with increased activity in human T-cell lines. Flow cytometry was performed on PM1 (A and C) and CEM.NKR-CCR5 (B and D) cells expressing empty vector or traptamers as indicated below. Nonpermeabilized cells were stained with the anti-CCR5 antibody 2D7 (A through D). In panels A to D, black represents unstained cells, and red represents cells expressing empty vector. In panels A and B, values for cells expressing BY1 (dark blue) and BY1PC2 (light blue) are shown. In panels C and D, values for cells expressing BY6 (green), BY6M3 (dark blue), BY6M4 (purple), and BY6M6 (light blue) are shown. (E) PM1 cells expressing empty vector or the indicated clones were infected with HIV-based eYFP reporter viruses pseudotyped with an R5-tropic HIV Env (ADA; black bars), an X4-tropic HIV Env (Lai; gray bars), or VSV-G (white bars), and reporter virus transduction was determined as described in the legend for Fig. 5. Data shown are the average of at least two independent trials (three for Lai and five for ADA) with error bars indicating standard error. Significance relative to cells expressing vector only was determined by a two-tailed t test: *, P < 0.05; **, P < 0.005; ***, P < 0.001.
Fig 7.
Traptamer mutants are active against multiple R5-tropic pseudotypes and against replication-competent virus. PM1 cells expressing BY1PC2 (panel A) or BY6M4 (panel B) were infected with HIV-based eYFP reporter viruses pseudotyped with CCR5-specific (R5: ADA, BaL, JRFL, SF162, B1206, BK184, and MG505), dual-tropic (R5X4: 89.6), or CXCR4-specific (X4: Lai and HXB2) envelope proteins. HIV reporter virus transduction was assessed by flow cytometry as described in the legend of Fig. 5 and is shown as the percentage of cells that expressed eYFP, normalized to transduction of cells expressing empty vector. Results shown are the average of at least two independent trials, with error bars representing standard error. The difference between transduction by R5- and X4-tropic Envs (means indicated by the horizontal bars) is statistically significant in cells expressing either traptamer (P < 0.001, two-tailed t test). (C) TZM-bl cells stably expressing empty vector were treated with maraviroc (dark gray bars) or left untreated (black bars), or cells expressing BY1PC2 (light gray bars) or BY6M4 (white bars) were left untreated. Cells were then infected with reporter viruses pseudotyped with ADA (maraviroc sensitive) and R3 (maraviroc resistant) R5-tropic Env proteins. After 72 h, reporter virus transduction was assessed by flow cytometry as described in the legend for Fig. 5. Results shown are the average of at last two independent trials with error bars indicating standard error. BY1PC2 and BY6M4 inhibited transduction by the maraviroc-resistant R3 pseudotype significantly better than maraviroc itself (P < 0.001). (D and E) PM1 cells expressing clone BY1PC2 (solid line) or the unselected clone US7 (dashed line) were infected with R5-tropic pNL-BaL (D) or X4-tropic pNL4-3 (E) virus, and supernatants harvested from the infected cells at the indicated time points were used to infect TZM-bl reporter cells. Viral titer was quantified by luciferase activity in extracts from infected TZM-bl cells. Data shown are from experiments performed in triplicate with the background from mock-infected cells subtracted. Similar results were obtained in two independent experiments.
Because the traptamers appeared to act by reducing expression of CCR5 rather than by altering its conformation, we reasoned that they might retain activity against viruses resistant to small-molecule CCR5 inhibitors. To test whether BY1PC2 and BY6M4 were effective against a drug-resistant Env, we generated an eYFP reporter virus pseudotyped with R3, an R5-tropic, maraviroc-resistant HIV Env. The R3 Env was isolated from a patient exhibiting virologic failure while on a maraviroc-containing treatment regimen (42). Because the R3-pseudotyped virus did not efficiently infect PM1 cells, presumably because of the relatively low CCR5 expression levels in these cells, we tested the ability of traptamers to inhibit transduction of TZM-bl cells, which were engineered to overexpress exogenous CD4 and CCR5. As assessed by flow cytometry for eYFP fluorescence, treatment with maraviroc almost completely inhibited transduction by reporter viruses pseudotyped with the R5-tropic envelope protein ADA, but the pseudotype with the maraviroc-resistant R3 envelope was markedly resistant to the drug, as expected (Fig. 7C). Strikingly, BY1PC2 and BY6M4 were highly active against the R3-pseudotyped virus (∼90% inhibition). Thus, env mutations that confer high-level resistance to maraviroc did not inhibit the action of these traptamers.
We also tested the activity of BY1PC2 in a multicycle HIV replication assay. PM1 cells expressing this traptamer were exposed to an infectious R5-tropic HIV strain, pNL-BaL, and a control X4-tropic strain, pNL4-3. Virus produced by these cells was harvested at various time points, and HIV replication was assayed by infecting TZM-bl cells with the viral supernatants and measuring luciferase activity in response to Tat-mediated induction of the integrated HIV promoter in these reporter cells. In PM1 cells expressing the unselected clone US7, HIV titers rose almost 300-fold following infection, peaked at 6 days after infection, and then declined to a steady-state level approximately 1.5 logs higher than the day 2 time point, as expected (Fig. 7D). Strikingly, expression of BY1PC2 delayed HIV replication and inhibited peak virus production by approximately 100-fold, and HIV titers returned to baseline levels approximately 2 weeks after the initial infection. In contrast, peak production of X4-tropic pNL4-3 was not inhibited by BY1PC2 (Fig. 7E). We note that pNL4-3 replication was delayed in the presence of BY1PC2, consistent with our finding that this traptamer caused a modest inhibition of transduction by a reporter virus pseudotyped with an X4-tropic HIV envelope (Fig. 6E).
Activity of traptamers against CCR2b and chimeric receptors.
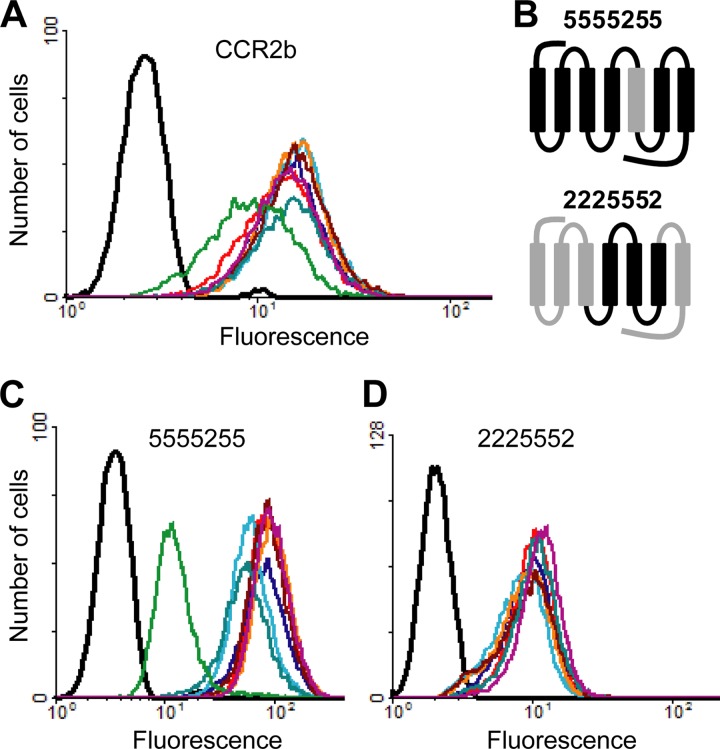
To begin to explore the mechanism of action of the traptamers, we first tested their activity against human CCR2b, a chemokine receptor which is nearly 90% identical to CCR5 in the TM regions (8). Despite this extensive similarity, most of the traptamers did not affect cell surface expression of exogenous CCR2b in BaF3 cells (Fig. 8A). Expression of clone BY6 caused a modest reduction in CCR2b expression. In order to determine whether specific TM domains of CCR5 were required for the activity of the traptamers, two receptor chimeras were constructed and tested (Fig. 8B). In one chimera (denoted 5555255), the fifth TM domain of CCR5 (TM5) was replaced with the corresponding CCR2b sequence, which differs by only 4 amino acids. Cell surface 5555255 expression was completely refractory to inhibition by three of the selected proteins (BY1, BY3, and BY4), suggesting that they directly interact with TM sequences of CCR5, likely including residue(s) within TM5 (Fig. 8C). Clones BY2 and BY5 caused a slight decrease in cell surface expression of this chimera, far less than their effect on wild-type CCR5. Clone BY6 effectively downregulated 5555255, indicating that this chimera was not intrinsically resistant to traptamer-mediated downregulation (Fig. 8C) and that BY6 recognized CCR5 differently than the other traptamers. In the second chimera (denoted 2225552), the region encompassing CCR5 TM domains 4, 5, and 6 was introduced into CCR2b (35). None of the tested traptamers inhibited 2225552 cell surface expression (Fig. 8D), showing that TM domains 4, 5, and 6 of CCR5 were not sufficient for traptamer activity.
Fig 8.
Activity of traptamers against CCR2b and chimeric receptors. (A) Nonpermeabilized parental BaF3 cells (black) and BaF3-CCR2b cells expressing vector (red), clone BY1 (dark blue), clone BY2 (light blue), clone BY3 (orange), clone BY4 (dark red), clone BY5 (blue-green), clone BY6 (green), or clone US7 (purple) were incubated with a CCR2b-specific antibody (48607) and analyzed by flow cytometry. The same color scheme applies to panels C and D. (B) Schematic diagram of CCR5/CCR2b chimeric receptors, with the N termini at the left. Segments from CCR5 and CCR2b are shown in black and gray, respectively. Numbers in the name of each chimera refer to the origin of each TM domain. (C) BaF3 cells expressing 5555255 and individual traptamers were stained with a non-conformation-specific anti-CCR5 antibody that recognizes the N terminus of the chimeric receptor (CTC5). (D) BaF3 cells expressing 2225552 and individual traptamers were stained with an antibody specific for the N terminus of CCR2b (48607).
DISCUSSION
We developed a FACS-based screening approach to identify small, artificial TM proteins that specifically downregulated cell surface expression of the HIV coreceptor CCR5. The most active traptamers almost completely blocked expression of CCR5 and dramatically inhibited R5-tropic HIV reporter viruses and infectious R5-tropic HIV. These results highlight the power of a FACS-based biological screen of a completely randomized TM protein library to isolate rare clones encoding biologically active proteins from among hundreds of thousands of inactive sequences.
We previously isolated traptamers that stimulated cell growth by activating the platelet-derived growth factor β receptor (PDGFβR) or the human erythropoietin receptor (hEPOR), each of which contains single membrane-spanning segments (7, 13, 14). The traptamers reported here specifically inhibited expression of a multipass TM protein, CCR5, by affecting its translation or stability. Posttranscriptional downregulation of a target is distinct from the mechanisms employed by the traptamers we isolated previously. Traptamers that activate the PDGFβR induce receptor dimerization, and the activator of the hEPOR is thought to cause an activating conformational change in the receptor. Thus, despite the superficial similarity of these small, hydrophobic proteins, they can have diverse effects on their targets. In addition, covalent dimerization of the hEPOR activator and most of the PDGFβR activators is required for activity, but all of the traptamers active against CCR5 were truncated, indicating that neither the C-terminal cysteines nor covalent dimerization is required for anti-CCR5 activity.
The very hydrophobic nature of the traptamers suggests that they reside within cellular membranes, and CCR5, with seven TM domains, is mostly embedded in the membrane as well. Furthermore, amino acid differences in a single CCR5 TM domain blocked the activity of most of the traptamers, and certain amino acid substitutions in the TM domains of BY1 and BY6 (in the optimized traptamers) increased their activity. Based on these results and previous results with the well-studied E5 protein (40), we hypothesize that the traptamers engage in lateral, hydrophobic interactions with the TM domains of CCR5. We further hypothesize that binding of the traptamer disrupts the intramolecular interactions that normally stabilize the hydrophobic core of CCR5, thereby affecting its ability to fold into the proper three-dimensional conformation. This in turn is likely to destabilize CCR5 directly or result in its aberrant trafficking, which might indirectly result in accelerated degradation. Attempts to demonstrate a physical interaction between the traptamers and CCR5 are complicated by the low receptor levels resulting from traptamer action and have so far been unsuccessful. Additional experiments are required to determine the mechanism by which the traptamers downregulate CCR5.
The traptamers that cause CCR5 downregulation have diverse TM sequences, and they are highly specific. Only BY6 affected expression of CCR2b, demonstrating that this traptamer has different and possibly less specific sequence requirements for activity. Most traptamers had minimal effects on the 5555255 chimera, implying that TM5 is a target domain common to several traptamers. Alternatively, TM5 may be important for maintaining a conformation of CCR5 required for traptamer activity or contain a signal required for traptamer-mediated downregulation. However, the 5555255 chimera traffics to the cell surface, is recognized by the conformation-specific antibody 2D7, and can support infection by R5-tropic HIV pseudoviruses (unpublished results), implying that it is not grossly misfolded. Furthermore, TM5 is not required for BY6-mediated downregulation of 5555255. CCR5 TM5 in the context of the 2225552 chimera is not sufficient for traptamer-mediated downregulation, implying that additional segments of CCR5 are required for traptamer recognition or for subsequent steps of traptamer action.
The traptamers downregulated expression of CCR5 in human T cells, but they were more active in mouse cells, where the initial selection was performed, perhaps because of species- or cell-type-specific differences in the cellular machinery that mediates CCR5 downregulation by these proteins. We increased the activity of two of the traptamers by limited random mutagenesis and additional cycles of screening in human cells. The most active traptamers isolated so far specifically inhibited transduction of PM1 T cells by a panel of pseudotyped R5-tropic HIV reporter viruses by 95% or more and dramatically inhibited replication of an R5-tropic HIV strain, demonstrating that traptamers are active in both single-cycle and multicycle HIV infectivity assays. The traptamers were inactive or showed markedly lower activity against X4-tropic HIV reporter viruses or X4-tropic HIV. This specificity implies that the traptamers block virus entry, the step mediated by the gp120-coreceptor interaction.
These traptamers may inform our understanding of how the TM domains of CCR5 interact with one another and contribute to its proper folding, trafficking, cell surface expression, metabolism, and activity, and they may help identify the TM segments of CCR5 that are most suitable as therapeutic targets. For example, although the small-molecule inhibitor maraviroc is thought to interact with residues in CCR5 TM domains 2, 3, and 7 (15, 26), our results suggest that TM5 should also be considered in inhibitor design. Indeed, other studies have demonstrated that residues in TM5 play an important role in stabilizing an HIV-resistant conformation of CCR5 that is recognized by small-molecule inhibitors (5, 15).
Because the traptamers have diverse TM sequences, they are likely to have different effects on CCR5 conformation and activity. Thus, different traptamers may uniquely probe different aspects of CCR5 structure and function. For example, only BY6 showed strong activity against 5555255, implying that it interacted with this chimera differently than the other traptamers. In fact, BY6 downregulated 5555255 more effectively than CCR5 itself, implying that TM5 of CCR5, which is required by the other traptamers, may actually restrain the activity of BY6. BY6 is also unusual because it increased antibody binding to CXCR4 in PM1 cells. BY6 may alter the heterodimerization of CCR5 and CXCR4, a phenomenon which is well documented in the literature (9, 22), and thus promote the exposure of CXCR4 epitopes that are otherwise hidden. Additionally, BY6 has the unique ability to deplete the internal pool of CCR5, which may contribute to its strong activity against HIV. BY1 also appears to engage CCR5 in a different manner than the other traptamers. BY1 and its mutant BY1PC2 were the only traptamers that inhibited transduction by an X4-tropic reporter virus in PM1 cells, which express both CXCR4 and CCR5, even though these traptamers did not downregulate CXCR4 expression in these cells (Fig. 5 and 6E). The ability of these traptamers to inhibit an X4-tropic virus may be due to cross talk between CCR5 and CXCR4, mediated by coreceptor heterodimerization (9, 22). It is likely that traptamers can be further optimized and diversified by additional rounds of mutagenesis and selection, thereby generating a wide variety of sequences with subtly different activities. Indeed, the BY6 mutants have lost the ability to upregulate CXCR4 expression in PM1 cells (data not shown) but have acquired even stronger anti-HIV activity. Furthermore, by screening additional libraries and by using different selection strategies, we recently isolated additional CCR5-specific traptamers with different TM sequences (unpublished results). Thus, traptamers are likely to provide a new and highly diverse set of reagents to probe TM domain structure and function.
Our results showed that traptamers are highly active against reporter viruses pseudotyped with HIV envelope proteins from clinical isolates, including a maraviroc-resistant envelope protein. Thus, traptamers that downregulate CCR5 may lead to new gene therapy or peptide antagonist approaches to inhibit HIV infection. Traptamers may also serve as the basis for the design of peptidomimetic drugs. Rational design of drugs targeting TM domains is often challenging because minimal structural information about the target is available. Our screening approach is advantageous in this regard as it allows for the genetic selection of active starting points for inhibitor design. Although each of these approaches would require a major development effort, our isolation of multiple, diverse, highly potent anti-HIV proteins that have never occurred in nature suggests that we have created a large and untapped pool of structures with potential clinical utility.
Our success in isolating artificial proteins that modulate single-pass and multipass TM proteins points to the generality of this method. In contrast to a previous study, where peptides were designed based on the predicted TM regions of CCR5 and CXCR4 (41), we used biological selection to identify multiple different proteins with the desired activity. Thus, our approach did not require previous knowledge of the sequence or structure of the target protein and is potentially applicable to numerous TM protein targets for which the appropriate antibodies or labeled ligands exist. This approach seems ideally suited for identifying agents that activate or inhibit GPCRs as each of their seven TM domains constitutes a potential binding site for a traptamer. Because GPCRs comprise a large class of drug targets, traptamers may be a rich source of important new bioactive compounds.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the NCI to D.D. (CA37157) and from NIAID to R.E.S. (AI067034), by postdoctoral fellowships from the American Cancer Society (PF-11-273-01-TBE) and the James Hudson Brown-Alexander Brown Coxe Foundation to E.H.S., by an NIH predoctoral Genetics Training grant (T32 GM007499) to S.A.M., and by the on-going generous support of Laurel Schwartz.
We thank Robert Doms, Ned Landau, Dan Littman, Peter Glazer, Robert Means, Priti Kumar, Susan Baserga, Erica Schleifman, and Kristina Talbert-Slagle for advice and reagents and acknowledge the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, for essential reagents. We also thank Edward Barbieri, Jill Hagey, and Shuang Shao for help with preliminary experiments and the Yale School of Medicine Cell Sorter Core Facility. We also thank Jan Zulkeski for assistance in preparing the manuscript.
Yale University has filed a provisional patent covering the use of traptamers to inhibit HIV infection (application number 61668736).
Footnotes
Published ahead of print 18 July 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. An DS, et al. 2007. Stable reduction of CCR5 by RNAi through hematopoietic stem cell transplant in non-human primates. Proc. Natl. Acad. Sci. U. S. A. 104:13110–13115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anderson J, Akkina R. 2007. Complete knockdown of CCR5 by lentiviral vector-expressed siRNAs and protection of transgenic macrophages against HIV-1 infection. Gene Ther. 14:1287–1297 [DOI] [PubMed] [Google Scholar]
- 3. Bai J, et al. 2000. Characterization of anti-CCR5 ribozyme-transduced CD34+ hematopoietic progenitor cells in vitro and in a SCID-hu mouse model in vivo. Mol. Ther. 1:244–254 [DOI] [PubMed] [Google Scholar]
- 4. Berger EA, Murphy PM, Farber JM. 1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 17:657–700 [DOI] [PubMed] [Google Scholar]
- 5. Billick E, et al. 2004. The differential sensitivity of human and rhesus macaque CCR5 to small-molecule inhibitors of human immunodeficiency virus type 1 entry is explained by a single amino acid difference and suggests a mechanism of action for these inhibitors. J. Virol. 78:4134–4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Burns JC, Friedmann T, Driever W, Burrascano M, Yee JK. 1993. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc. Natl. Acad. Sci. U. S. A. 90:8033–8037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cammett TJ, et al. 2010. Construction and genetic selection of small transmembrane proteins that activate the human erythropoietin receptor. Proc. Natl. Acad. Sci. U. S. A. 107:3447–3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Charo IF, et al. 1994. Molecular cloning and functional expression of two monocyte chemoattractant protein 1 receptors reveals alternative splicing of the carboxyl-terminal tails. Proc. Natl. Acad. Sci. U. S. A. 91:2752–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Contento RL, et al. 2008. CXCR4-CCR5: a couple modulating T cell functions. Proc. Natl. Acad. Sci. U. S. A. 105:10101–10106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Coskun AK, van Maanen M, Nguyen V, Sutton RE. 2006. Human chromosome 2 carries a gene required for production of infectious human immunodeficiency virus type 1. J. Virol. 80:3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Doms RW, Peiper SC. 1997. Unwelcomed guests with master keys: how HIV uses chemokine receptors for cellular entry. Virology 235:179–190 [DOI] [PubMed] [Google Scholar]
- 12. Doranz BJ, et al. 1996. A dual-tropic primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3, and CKR-2β as fusion cofactors. Cell 85:1149–1158 [DOI] [PubMed] [Google Scholar]
- 13. Freeman-Cook LL, DiMaio D. 2005. Modulation of cell function by small transmembrane proteins modeled on the bovine papillomavirus E5 protein. Oncogene 24:7756–7762 [DOI] [PubMed] [Google Scholar]
- 14. Freeman-Cook LL, et al. 2004. Selection and characterization of small random transmembrane proteins that bind and activate the platelet-derived growth factor β receptor. J. Mol. Biol. 338:907–920 [DOI] [PubMed] [Google Scholar]
- 15. Garcia-Perez J, et al. 2011. An allosteric model of maraviroc binding to CCR5. J. Biol. Chem. 286:33409–33421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. George SR, et al. 1998. A transmembrane domain-derived peptide inhibits D1 dopamine receptor function without affecting receptor oligomerization. J. Biol. Chem. 273:30244–30248 [DOI] [PubMed] [Google Scholar]
- 17. Gulick RM, et al. 2008. Maraviroc for previously treated patients with R5 HIV-1 infection. N. Engl. J. Med. 359:1429–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Haas DL, Case SS, Crooks GM, Kohn DB. 2000. Critical factors influencing stable transduction of human CD34+ cells with HIV-1-derived lentiviral vectors. Mol. Ther. 2:71–80 [DOI] [PubMed] [Google Scholar]
- 19. Hebert TE, et al. 1996. A peptide derived from a β2-adrenergic receptor transmembrane domain inhibits both receptor dimerization and activation. J. Biol. Chem. 271:16384–16392 [DOI] [PubMed] [Google Scholar]
- 20. Holt N, et al. 2010. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat. Biotechnol. 28:839–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hütter G, et al. 2009. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N. Engl. J. Med. 360:692–698 [DOI] [PubMed] [Google Scholar]
- 22. Isik N, Hereld D, Jin T. 2008. Fluorescence resonance energy transfer imaging reveals that chemokine-binding modulates heterodimers of CXCR4 and CCR5 receptors. PLoS One 3:e3424 doi:10.1371/journal.pone.0003424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ji TH, Grossman M, Ji I. 1998. G protein-coupled receptors. I. Diversity of receptor-ligand interactions. J. Biol. Chem. 273:17299–17302 [DOI] [PubMed] [Google Scholar]
- 24. Kondru R, et al. 2008. Molecular interactions of CCR5 with major classes of small-molecule anti-HIV CCR5 antagonists. Mol. Pharmacol. 73:789–800 [DOI] [PubMed] [Google Scholar]
- 25. Kuhmann SE, Hartley O. 2008. Targeting chemokine receptors in HIV: a status report. Annu. Rev. Pharmacol. Toxicol. 48:425–461 [DOI] [PubMed] [Google Scholar]
- 26. Labrecque J, et al. 2011. HIV-1 entry inhibition by small-molecule CCR5 antagonists: A combined molecular modeling and mutant study using a high-throughput assay. Virology 413:231–243 [DOI] [PubMed] [Google Scholar]
- 27. Lee B, et al. 1999. Epitope mapping of CCR5 reveals multiple conformational states and distinct but overlapping structures involved in chemokine and coreceptor function. J. Biol. Chem. 274:9617–9626 [DOI] [PubMed] [Google Scholar]
- 28. Li W, Yu M, Bai L, Bu D, Xu X. 2006. Downregulation of CCR5 expression on cells by recombinant adenovirus containing antisense CCR5, a possible measure to prevent HIV-1 from entering target cells. J. Acquir. Immune Defic. Syndr. 43:516–522 [DOI] [PubMed] [Google Scholar]
- 29. Liu R, et al. 1996. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 86:367–377 [DOI] [PubMed] [Google Scholar]
- 30. Mansky LM. 2002. HIV mutagenesis and the evolution of antiretroviral drug resistance. Drug Resist. Updat. 5:219–223 [DOI] [PubMed] [Google Scholar]
- 31. Marlatt SA, et al. 2011. Construction and maintenance of randomized retroviral expression libraries for transmembrane protein engineering. Protein Eng. Des. Sel. 24:311–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Moore JP, Kuritzkes DR. 2009. A piece de resistance: how HIV-1 escapes small molecule CCR5 inhibitors. Curr. Opin. HIV AIDS 4:118–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Naviaux RK, Costanzi E, Haas M, Verma I. 1996. The pCL vector system: rapid production of helper-free, high titer, recombinant retroviruses. J. Virol. 70:5701–5705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rana S, et al. 1997. Role of CCR5 in infection of primary macrophages and lymphocytes by macrophage-tropic strains of human immunodeficiency virus: resistance to patient-derived and prototype isolates resulting from the Δccr5 mutation. J. Virol. 71:3219–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rucker J, et al. 1996. Regions in β-chemokine receptors CCR5 and CCR2b that determine HIV-1 cofactor specificity. Cell 87:437–446 [DOI] [PubMed] [Google Scholar]
- 36. Samson M, et al. 1996. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR5-chemokine receptor gene. Nature 382:722–725 [DOI] [PubMed] [Google Scholar]
- 37. Sattentau QJ, Weiss RA. 1988. The CD4 antigen: physiological ligand and HIV receptor. Cell 52:631–633 [DOI] [PubMed] [Google Scholar]
- 38. Schleifman EB, et al. 2011. Targeted disruption of the CCR5 gene in human hematopoietic stem cells stimulated by peptide nucleic acids. Chem. Biol. 18:1189–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shimizu S, et al. 2010. A highly efficient short hairpin RNA potently down-regulates CCR5 expression in systemic lymphoid organs in the hu-BLT mouse model. Blood 115:1534–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Talbert-Slagle K, DiMaio D. 2009. The bovine papillomavirus E5 protein and the PDGF β receptor: it takes two to tango. Virology 384:345–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tarasova NI, Rice WG, Michejda CJ. 1999. Inhibition of G-protein-coupled receptor function by disruption of transmembrane domain interactions. J. Biol. Chem. 274:34911–34915 [DOI] [PubMed] [Google Scholar]
- 42. Tilton JC, et al. 2010. A maraviroc-resistant HIV-1 with narrow cross-resistance to other CCR5 antagonists depends on both N-terminal and extracellular loop domains of drug-bound CCR5. J. Virol. 84:10863–10876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. van't Wout AB, et al. 1994. Macrophage-tropic variants initiate human immunodeficiency virus type 1 infection after sexual, parenteral, and vertical transmission. J. Clin. Invest. 94:2060–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yee JK, et al. 1994. A general method for the generation of high-titer, pantropic retroviral vectors: highly efficient infection of primary hepatocytes. Proc. Natl. Acad. Sci. U. S. A. 91:9564–9568 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.