Abstract
Herpes simplex virus 2 infection is characterized by cycles of viral quiescence and reactivation. CD8+ T cells persist at the site of viral reactivation, at the genital dermal-epidermal junction contiguous to neuronal endings of sensory neurons, for several months after herpes lesion resolution. To evaluate whether these resident CD8+ T cells frequently encounter HSV antigen even during times of asymptomatic viral infection, we analyzed the transcriptional output of CD8+ T cells captured by laser microdissection from human genital skin biopsy specimens during the clinically quiescent period of 8 weeks after lesion resolution. These CD8+ T cells expressed a characteristic set of genes distinct from those of three separate control cell populations, and network and pathway analyses revealed that these T cells significantly upregulated antiviral genes such as GZMB, PRF1, INFG, IL-32, and LTA, carbohydrate and lipid metabolism-related genes such as GLUT-1, and chemotaxis and recruitment genes such as CCL5 and CCR1, suggesting a possible feedback mechanism for the recruitment of CD8+ T cells to the site of infection. Many of these transcripts are known to have half-lives of <48 h, suggesting that cognate antigen is released frequently into the mucosa and that resident CD8+ T cells act as functional effectors in controlling viral spread.
INTRODUCTION
Herpes simplex virus 2 (HSV-2), the major cause of recurrent genital herpes, cycles through replicating and quiescent phases during lifelong infection of the human host. The virus occasionally causes genital lesions that are the result of virus release from sensory neurons to the mucosal epithelium, at which point HSV-2 begins replication and cell-to-cell spread (2). Recent data have shown that HSV-2 reactivation occurs frequently and wanes rapidly (24). Most reactivations of HSV-2 are subclinical, typically lasting <12 h (18, 25). A minority of reactivation episodes, less than 15%, lead to the genital lesions that are characteristic of genital herpes (2). Recent data have indicated that the clearance of infected cells and virions by the mucosal immune system is rapid, and differences in the immune response can partially explain whether a clinical or subclinical episode will occur (2; J. Zhu, presented at the Keystone Symposia, Immunologic Memory, Persisting Microbes and Chronic Disease, Banff, Canada, 2011). Antiviral cytolytic responses by the host immune system are associated with viral clearance at HSV lesion sites, with the host inflammatory response peaking during lesion resolution (13, 14, 29). These findings suggest that host innate and adaptive immune cells must be present at the time of reactivation.
After lesion resolution, a population of CD8+ T cells persists for extended periods at the dermal-epidermal junction (DEJ), contiguous to the neuronal termini of sensory nerves. These CD8+ T cells have been shown to be HSV antigen specific, and their location suggests that they may play an important role in immune surveillance. HSV-specific CD4+ T cells as well as plasmacytoid and inflammatory dendritic cells also persist for weeks after clinical reactivation (28). Recently, HSV-2 DNA and antigen have been detected in association with inflammatory CD8+ and CD4+ T cells in skin biopsy specimens with no evidence of genital ulceration, suggesting that HSV antigen may be more frequently presented into the mucosa than previously appreciated (17, 28; J. Zhu, presented at the Keystone Symposia, Immunologic Memory, Persisting Microbes and Chronic Disease, Banff, Canada, 2011).
For pathogens with chronic or “latent” phases of disease, it remains a challenge to identify when or how often the immune system is exposed to antigen. Several possibilities exist for why a viral infection would result in this phenotype: (i) the virus could be in a state of inactivity or quiescence; (ii) the virus could be actively replicating, with this replication being controlled by continuous immunosurveillance and clearance by the host immune system; or (iii) the virus may undergo only infrequent reactivation that results in clinical disease. The prevalent view of HSV pathogenesis has been the latter.
A major hurdle in studies of pathogens with dormant and activated phases is the inability to measure in vivo how the host-pathogen interactions change over the course of infection. The use of cell type-specific immunofluorescent staining and laser capture microdissection (LCM) methodologies allows one to capture and enrich for a specific cell type from a diverse and complex tissue milieu and then proceed to immediately analyze these cell populations.
To better understand the dynamic interplay between HSV-2 and the local adaptive immune response during HSV-2 infection, we specifically analyzed CD8+ T cells localized at the dermal-epidermal junction from HSV-2-affected areas of genital skin during clinically asymptomatic reactivation and evaluated the transcriptional profile of this T-cell population to determine whether these T cells exhibit effector function during periods of clinical and virological quiescence.
MATERIALS AND METHODS
Study participants.
Healthy, HSV-2-seropositive adults were recruited at the University of Washington Virology Research Clinic in Seattle, WA. The biopsy protocol was approved by University of Washington Human Subjects Review Committee, and all participants provided written consent. HSV-2 serostatus was determined by Western blotting as previously described (20); all participants were HIV seronegative. Biopsy procedures were conducted as described previously (22, 29). All samples were immediately placed on dry ice and stored at −80°C until processing.
Purification of CD8+ T cells from healed lesion biopsy specimens and matched control biopsy specimens.
We utilized a rapid immunofluorescent staining method (<15 min) to identify CD8+ T cells from lesion biopsy specimens and avoid RNA degradation, as previously described (28). We used the Zeiss PALM Microbeam LCM system to cut and catapult individual cells to designated tubes in a completely automated process. Between 50 and 100 cells were captured per sample, and 1 to 10 ng of isolated total RNA was processed for gene expression analysis via the Illumina array platform.
Three types of control cells were included: CD8+ T cells from contralateral genital biopsy specimens (CL_CD8) from an identical anatomic area that was not associated with known HSV reactivation; CD8+ T cells from arm control biopsy specimens (Arm_CD8), representing CD8+ T cells from an anatomic area in which no HSV-2 reactivation occurs; and CD1a+ cells isolated from the DEJ from biopsy specimens (CD1a) taken 8 weeks after healing for comparison to a different cell type from the same spatial area. Because the absolute number of CD8+ T cells in control tissue biopsy specimens is significantly lower than that from HSV-affected regions (28, 29), we collected CD8+ T cells from both dermal-epidermal junctions and dermal areas for these analyses.
RNA extraction, amplification, and hybridization of cDNA to Illumina bead arrays.
Total RNA from LCM-captured CD8+ T cells was extracted using PicoPure RNA isolation kits following the manufacturer's protocol (Applied Biosystems, CA). The quality of total RNA was analyzed by Agilent picochips, and RNA with a quality index (RIN) above 5 was used. Total RNA (0.5 to 1 ng) was then used for cDNA synthesis using the Ovation Pico RNA amplification system (NuGEN, CA). The size distribution of cDNA was analyzed by Agilent Technologies nanochips, and the amplified cDNA had a Gaussian distribution with an average size of 200 bp. The cDNA was biotin-labeled as per the NuGEN protocol and labeled cDNA (750 ng) was hybridized to Illumina HumanRef8_v3 bead arrays in the Shared Resource Genome Center at Fred Hutchison Cancer Research Center per the manufacturer's instructions.
Detection of HSV-2 antigen and DNA.
HSV-2 was detected as previously described (28). Briefly, we detected HSV-2 antigen by immunofluorescence staining using rabbit antibody to HSV-2 (Dako). We used a sensitive PCR assay to detect HSV-2 DNA from eight sections of each biopsy specimen. We considered one copy per reaction well a positive result (17, 18).
Quantitative RT-PCR (qRT-PCR) assay.
The cDNA synthesized from total RNA described above was used as the template for quantitative PCR (TaqMan PCR). The TaqMan probes for ACTB, PRF1, ITGAL, GLUT1, PFK, SCD, CCL5, CCL2, IL-32, CCR1, CCR5, and CCR4 were ordered from Applied Biosystems. The cDNA from NuGEN Pico Amplification Systems was diluted 1:20, and 2 μl was used in each reaction in a 96-well TaqMan PCR plate. The gene expression was normalized to that of ACTB (β-actin). The HSV DNA copy numbers were normalized to 50,000 cells by measuring the HBB (β-globin) gene copy numbers using 5′-TGAAGGCTCATGGCAAGAAA-3′ and 5′-GCTCACTCAGTGTGGCAAAGG-3′ as primers and 5′-TCCAGGTGAGCCAGGCCATCACTA-3′ as a probe. The detection limit of the assay was one copy per 50,000 cells.
Analysis of bead array data.
Raw data were imported to GenomeStudio (V2010.3; Illumina). Control summaries were generated to analyze the quality of hybridization. Data passing this initial quality control step were normalized using CubicSpline with background subtraction. Normalized data were exported to R, and genes expressed differentially between junction CD8+ T cells from samples taken 8 weeks after healing (JC_CD8) and CD8+ T cells from contralateral controls (CL_CD8) were selected using Genefilter, a Bioconductor package. A set of 987 genes were selected by the criteria that differences were found in more than five patients and showed more than a 3-fold change between experimental and control groups or that differences were found in more than four patients and showed more than a 5-fold change between the two groups. The differentially expressed genes were analyzed using an unsupervised hierarchical clustering method (clustering method, unweighted pair group method with arithmetic average [UPGMA; weighted average]; similarity measure, Euclidean distance) using SpotFire DecisionSite for functional genomics (version 9.1.2). Enriched functional categories and network analyses for the set of differentially expressed genes were performed using ingenuity pathway analysis (IPA software, version 8.8).
The GOMiner program was used to annotate all 18,401 genes on Illumina Human Ref8_v3 bead arrays. An annotation database was constructed in Microsoft Access using exported tables from GOMiner, and genes were annotated to the GO terms T-cell activation and cytolysis, cytokine activity, chemokine activity, cytokine receptor activity, and chemokine receptor activity and were exported for further analysis in SpotFire.
Table S1 in the supplemental material contains the complete list of genes differentially expressed (up- and downregulated) between control and experimental CD8+ T-cell populations.
Microarray data accession number.
Complete array data can be accessed under GEO accession number GSE39625.
RESULTS
Patients.
Nine patients were included in this study (Table 1); eight were female, and one was male. All patients had clinically and virologically documented symptomatic genital herpes and were HSV-2 seropositive; eight of nine had had genital herpes for over 2 decades. The biopsy sites studied were mostly in the buttocks region; however one was from the vulva and one was from the inner thigh, in patients 7 and 9, respectively.
Table 1.
Patient history, biopsy information, and HSV DNA levels
| Patient | Age (yrs) | Gender | No. of yrs of genital herpes | Location of biopsy | HSV DNA level ina: |
|
|---|---|---|---|---|---|---|
| 8 wph tissue | Control tissue | |||||
| 1 | 33 | F | 2 | Buttocks | ND | ND |
| 2 | 53 | M | 30 | Buttocks | ND | ND |
| 3 | 43 | F | 24 | Buttocks | 13 | ND |
| 4 | 51 | F | 32 | Buttocks | ND | ND |
| 5 | 63 | F | 26 | Buttocks | 19 | 27 |
| 6 | 56 | F | 35 | Buttocks | ND | ND |
| 7 | 48 | F | 30 | Vulva | ND | ND |
| 8 | 44 | F | 23 | Buttocks | ND | ND |
| 9 | 62 | F | 35 | Thigh | ND | ND |
Values are numbers of DNA copies per 5 × 104 cells. 8 wph, 8 weeks posthealing. ND, not detectable.
Biopsy analysis.
At the time of biopsy, the nine patients were asymptomatic and showed no evidence of genital lesions. No HSV-2 DNA or RNA was detected in eight of the nine control biopsy specimens, and none was detected in seven of the nine patients' week 8 biopsy specimens (Table 1). Patients 3 and 5 had 13 and 19 copies of HSV DNA per 5 × 104 cells in week 8 biopsy specimens, respectively, and patient 5 had 27 copies of HSV DNA per 5 × 104 cells in the biopsy specimen from the contralateral side, indicating the wide distribution of subclinical HSV-2 reactivation in these subjects at the time of biopsy. For reference, HSV DNA from lesion biopsy specimens averages 1 × 106 copies per 5 × 104 cells. Histology sections showed no evidence of genital ulcers or inflammatory infiltrates in the biopsy specimens (data not shown). Thus, clinically and histologically, HSV-2 infection was quiescent in 100% of the biopsy specimens, with undetectable levels of virus in 80% of the biopsy specimens even by the most sensitive assay.
T-cell purification and transcriptional analysis of purified JC_CD8 and CL_CD8 T cells.
We utilized a rapid immunofluorescent staining method (<15 min) to identify CD8+ T cells from biopsy specimens and proceeded with laser capture microdissection to purify selected cells from patient biopsy specimens. We obtained CD8+ T cells from biopsied tissue at the site of a prior lesion 8 weeks after healing from the dermal-epidermal junction (JC_CD8), which is where resident CD8+ T cells persist between the epithelial cells and nerve endings of sensory neurons. Control CD8+ T cells (CL_CD8) from an unaffected area were obtained at the same time point for comparison between the two CD8+ T-cell populations.
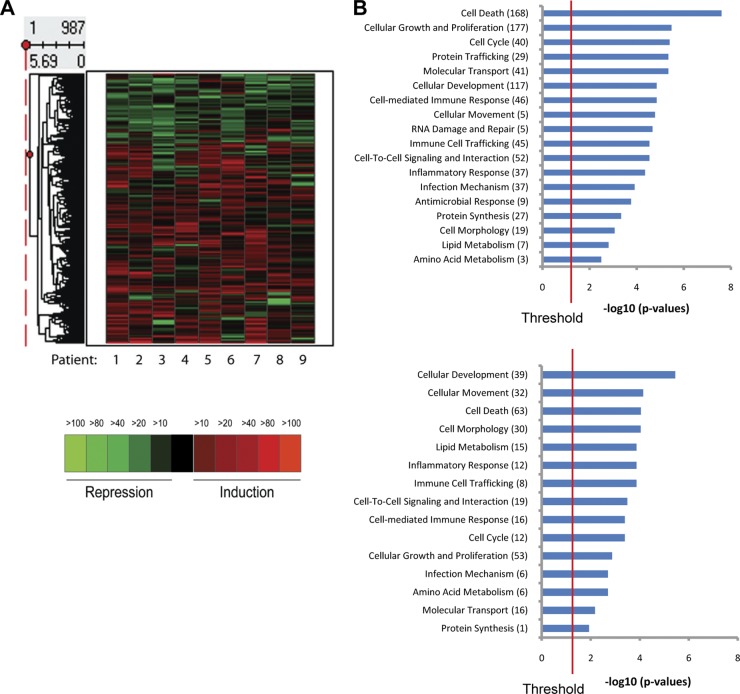
In order to compare the differential gene expression between JC_CD8 and CL_CD8 T cells, RNA was extracted from the purified T-cell populations and subjected to array analysis via Illumina human Ref8_v3 bead arrays. Using the criteria of more than a 3-fold change in expression in at least five patients or 5-fold change in at least four patients between the CD8+ T cells obtained from the HSV-affected and those from unaffected areas, we observed differential expression of 987 of the 18,401 genes on the array (737 up- and 250 downregulated; see Table S1 in the supplemental material for a complete list of genes). We generated a heat map of unsupervised hierarchical clustering of these genes and found that the pattern of up- and downregulation was uniform across all nine patients (Fig. 1A).
Fig 1.
Transcriptional profiling of laser-captured CD8+ T cells from biopsy specimens taken 8 weeks after healing and contralateral controls. (A) Heat map showing hierarchical clustering of differentially expressed genes (987 genes, 737 upregulated and 250 downregulated) between junction CD8+ cells (JC_CD8) and contralateral CD8+ T cells (CL_CD8) from nine patients. (B) Enriched functional categories in the up- and downregulated gene sets in panel A. The x axis represents the negative log10 P value calculated from the analysis of enriched functional categories using ingenuity pathway analysis.
Using ingenuity pathway analysis (IPA), we identified functional categories of genes that were significantly enriched (overrepresented) in both the upregulated and downregulated gene sets. A dominant feature of the differentially expressed gene set was a large percentage of cell death-related genes (168 [22%] genes upregulated; P = 2.49 × 10−8; 63 [25%] genes downregulated; P = 9.03 × 10−5) (Fig. 1B). Interestingly, several functional categories were overrepresented in either the upregulated or downregulated gene set compared to what would be expected by chance. A higher proportion of genes annotated to “protein synthesis” were induced compared to repressed (27 induced genes and 1 repressed gene), and genes involved in cell morphology showed a bias toward repression: 19 induced genes and 30 repressed genes (Fig. 1B).
JC_CD8+ T cells uniquely upregulated a network of genes involved in antimicrobial response.
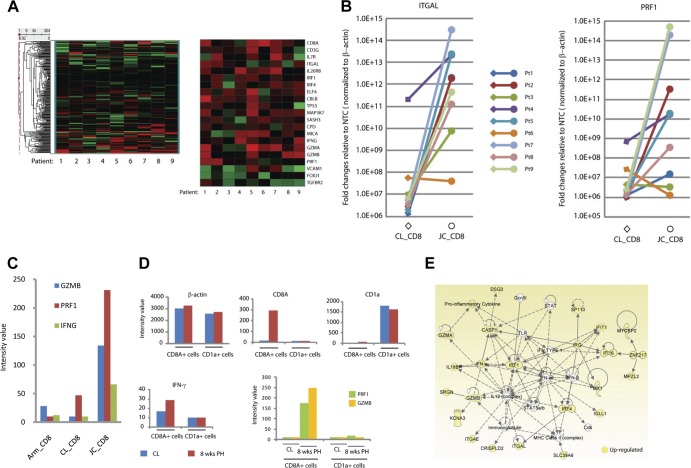
Using the GOMiner program, we annotated all 18,401 genes on Illumina human Ref8_v3 bead arrays to Gene Ontology terms (GO). Of the >18,000 genes represented on the array, 264 genes were annotated as related to T-cell activation and/or cytolysis. Unsupervised hierarchical clustering analysis of these 264 genes revealed that approximately 8% were significantly differentially expressed (Fig. 2A). Within this gene list, T-cell activation-annotated genes included CD8A and CD3G, required for T-cell receptor complex assembly; IL7R (interleukin 7 [IL-7] receptor), important for memory T-cell maturation (12); and ELF4, encoding a transcriptional regulator for perforin (PRF1) (15). In addition, three genes involved in T-cell activation were repressed in JC_CD8 cells relative to contralateral controls, including FOXJ1, encoding an NF-κB repressor (16); TGFBR2, encoding a transforming growth factor β (TGF-β) receptor; and VCAM1, encoding a vascular cell adhesion molecule, indicating a downregulation of factors involved in the inhibition of T-cell effector function. Not all genes known to be involved in T-cell activation and/or cytolysis were differentially expressed in JC_CD8 cells; the set of genes that showed no difference in expression included IL-2 and IL-4, which are known for T-cell proliferation, and BTLA, CTLA4, and PD-1 (also called PDCD1), which are known for T-cell activation inhibition (data not shown).
Fig 2.
JC_CD8+ T cells uniquely upregulated a network of genes involved in antimicrobial response. (A) Unsupervised hierarchical clustering heat maps showing the expression patterns of genes that were annotated to the GO terms T-cell activation and cytolysis. The left heat map shows the expression patterns of all 264 genes that were annotated to these two GO terms (out of the 18,401 genes on the Illumina human Ref8_v3 bead arrays); the right heat map shows the subset that are expressed differentially between JC_CD8 and CL_CD8 cells. (B) Quantitative RT-PCR (qRT-PCR) analysis shows that ITGAL and PRF1 were significantly upregulated in JC_CD8 relative to CL_CD8 cells. The y axis shows fold change of gene expression in JC_CD8 or CL_CD8 cells over that in nontemplate controls (NTC) after normalization to ACTB. Each reaction was done in duplicate. (C) JC_CD8 cells expressed GZMB, PRF1, and IFNG at significantly higher levels than Arm_CD8 and CL_CD8 cells. Data are from patient 5; similar results were seen with patient 4 (data not shown). (D) Antiviral gene expression patterns were associated with JC_CD8 cells but not with JC_CD1a cells from the same 8-week-posthealing (PH) biopsy specimens. Gene expression of ACTB, CD8A, CD1a, GZMB, PRF1, and INFG was compared in LCM-captured CD8A+ cells and CD1a+ cells from patient 1; similar results were obtained with patient 2 (data not shown). CL, contralateral. (E) JC_CD8 cells had higher levels of gene expression in an antimicrobial response network than CL_CD8 cells. A solid line between two molecules shows direct interaction; a dashed line shows indirect interaction.
To confirm the transcriptional array data, modulation of T-cell activation and cytolysis pathways was verified by quantitative RT-PCR (qRT-PCR) (Fig. 2B). We observed that the majority of patients (eight of nine) expressed significantly higher levels of ITGAL mRNA in JC_CD8 cell samples than controls (up to 107-fold). qRT-PCR also revealed that JC_CD8 cells from six of nine patients expressed notably higher levels of PRF1, a gene important for cytolytic activity, than CL_CD8 cells (up to 107-fold); the remaining three patients expressed low levels of PRF1 in both cell types.
We also performed differential transcriptional analyses of antiviral gene expression between JC_CD8 cells and Arm_CD8 cells that were captured on the same day from the axillary region (unaffected by HSV-2) in three patients (patients 3, 4, and 5). Consistent with patterns seen in CL_CD8 comparison experiments, JC_CD8 cells contained higher levels of transcripts of genes for granzymes and antiviral cytokine/chemokines than CD8+ T cells purified from the axillary area (Fig. 2C).
CD1a is a major histocompatibility complex (MHC)-like molecule expressed on the surfaces of antigen-presenting cells that presents lipid and glycolipid antigens to T cells to stimulate cell-mediated immune responses. To verify that the observed transcriptional differences were cell type specific and not representative of whole biopsy samples, we compared expression of a variety of genes between JC_CD8 and CD1a+ cells captured from the same tissue section from two patients (patients 1 and 2). CD1a+ cells expressed similar levels of housekeeping genes such as ACTB (β-actin), a significantly higher level of CD1a, and an undetectable level of CD8A. However, CD1a+ cells expressed almost undetectable levels of GZMB (granzyme B), PRF1, and IFNG (gamma interferon [IFN-γ]) (Fig. 2D). Our data therefore indicate that a set of genes for both cytolytic and noncytolytic antiviral function were specifically upregulated in JC_CD8 cells in biopsy specimens taken 8 weeks after healing.
To investigate the presence of gene interaction networks in JC_CD8 cell expression profiles, we performed a series of network analyses and subsequently selected several key genes in each network for development of qRT-PCR assays to individually quantitate differences in RNA expression between JC_CD8 cells and CL_CD8 cells. Figure 2E illustrates these data for a cellular network involved in cytolytic killing and antimicrobial recognition. Twenty-one of the 35 genes involved in this network were differentially expressed in JC_CD8 cells, including IFNG, GZMA, and GZMB; the cell-to-cell interaction genes ITGAE (human mucosal antigen 1, CD103) and ITGAL (lymphocyte function-associated antigen 1, LFA-1); the transcription factor genes IRF1 and IRF4; and several interferon-regulated genes (IFIT1 and IFI16). Integrins play critical roles in T-cell trafficking and mediating cytotoxic activities via immunological synapses between lymphocytes and target cells (8), and significant upregulation of the integrins ITGAL and ITGAE in JC_CD8 cells suggests that these cells are constantly prepared for engaging and killing infected target cells.
Differential expression of metabolism-related genes from junction CD8+ T cells.
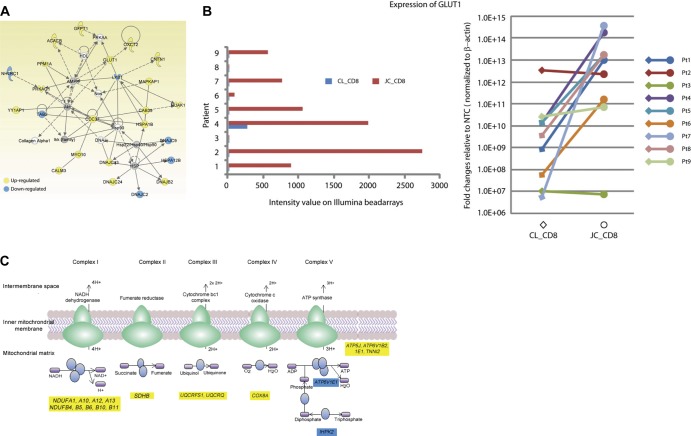
The environmental stimuli resulting in highly specific transcriptional changes in lymphocytes are known to promote global changes in cellular processes involving metabolism, and it has been well established that T cells converting from a resting to activated state require the upregulation of genes involved in nutrient metabolism. Functional analysis of biopsied T-cell mRNA identified enrichment in several metabolism-related categories from JC_CD8 cells relative to CL_CD8 cells (Fig. 1B), including a network of 18 upregulated and 6 downregulated genes related to glucose and lipid metabolism when JC_CD8 cells were compared to CD8 cells from the contralateral genital skin (Fig. 3A).
Fig 3.
JC_CD8 cells have a metabolic gene signature associated with activated T cells. (A) The differentially expressed gene set between JC_CD8 and CL_CD8 cells was significantly enriched for a network of genes involved in glucose and lipid metabolism. (B) JC_CD8 upregulation of GLUT1 relative to CL_CD8 cells. (Left) expression of GLUT1 assayed by microarray; (right) expression of GLUT1 assayed by qRT-PCR. The y axis shows fold change of gene expression in JC_CD8 or CL_CD8 cells over nontemplate controls (NTC) after normalization to β-actin (right panel). Pt, patient. Each reaction was done in duplicate. (C) JC_CD8 cells showed significant upregulation of genes in a canonical pathway for mitochondrial oxidative phosphorylation (blue ovals) relative to CL_CD8. Out of 165 genes in this pathway, JC_CD8 cells up- and downregulated 17 and 2 genes, respectively. Yellow, upregulated; blue downregulated.
Consistent with the notion that resting T cells require less sugar metabolism than activated and proliferating cells (11, 19), transcriptional profiling revealed a significant induction of GLUT1 (encoding glucose transporter 1; also called SLC2A1 [solute carrier family 2]), which encodes the primary glucose transporter in T lymphocytes (6, 9), in JC_CD8 cells from the majority of patients (six of nine) (Fig. 3B). Transcriptional expression of the gene encoding phosphofructokinase (PFKP), a key enzyme in the glycolytic cycle, was also upregulated in JC_CD8 cells (see Table S1 in the supplemental material). Together, these data suggest that CD8+ junction T cells in genital skin are metabolically activated.
In addition to alteration of the carbohydrate catabolism profile, the JC_CD8 transcriptional response was indicative of increased lipid biosynthesis and decreased lipid oxidation, another metabolic signature that is associated with T-cell activation (11). One of these gene products, acetyl coenzyme A (acetyl-CoA) carboxylase beta subunit (ACACB), catalyzes the carboxylation of acetyl-CoA to malonyl-CoA, a rate-limiting step in fatty acid synthesis, and was significantly upregulated in JC_CD8 cells (Fig. 3A); interestingly, ACACA, which encodes the alpha subunit of this enzyme, was not differentially expressed. The gene encoding stearoyl-CoA desaturase-1 (SCD), an enzyme required for unsaturated fatty acid synthesis, was also significantly induced as detected by array and qRT-PCR (data not shown).
As shown in Fig. 3A, JC_CD8 cells differentially expressed CDC37, STK11 (also known as LKB1), and PPM1A, whose protein products are known to participate in the Akt and AMP-activated protein kinase signaling pathways in metabolic control of CD8+ T-cell activation and proliferation. In addition, in canonical pathway analysis for differentially expressed genes, JC_CD8 cells exhibited a significantly higher expression level of genes involved in all five oxidative phosphorylation complexes with 17 up- and 2 downregulated genes (Fig. 3C), substantiating the active and proliferative state of this T-cell population.
JC_CD8 cells showed upregulation of antiviral cytokine/chemokine genes.
Out of 10 cytokines that were annotated by GO analyses as “response to virus” and/or “T-cell activation,” four were significantly induced in JC_CD8 T cells at the mRNA level: IFNG, IL-32, TNF (tumor necrosis factor alpha [TNF-α]), and LTA (lymphotoxin-α, also known as TNF-β) (Table 2). Several chemokines that play a role in the antiviral immune response were also transcriptionally induced, including CCL5 and CCL2 (Table 2), and the expression patterns for IL-32, CCL5, and CCL2 were verified by qRT-PCR (Fig. 4A). IFN-γ, IL-32, and TNF-α are known to have a direct antiviral effect on HSV DNA replication (3, 21, 27). As such, our transcriptional data again indicate “effector function” for CD8+ T cells located at the DEJ. Among the nine patients from whom we purified JC_CD8 T cells and analyzed their transcriptional profiles, these transcripts were elevated in four of the seven biopsy specimens in which no HSV-2 DNA was detected and one of two with detectable levels of HSV-2 DNA.
Table 2.
Cytokines and chemokines in JC_CD8 and CL_CD8 cells from biopsy samples taken 8 weeks after healing
| Gene | Avg intensity |
No. of patients with difference ofa: |
|||
|---|---|---|---|---|---|
| CL_CD8 | JC_CD8 | >3-fold | >5-fold | >10-fold | |
| IFNB | 10 | 11 | 0 | 0 | 0 |
| IFNA4 | 10 | 10 | 0 | 0 | 0 |
| IFNG | 10 | 59 | 5 | 3 | 2 |
| IL-2 | 10 | 12 | 0 | 0 | 0 |
| IL-4 | 16 | 12 | 0 | 0 | 0 |
| IL-7 | 75 | 20 | 1, 3 | 1, 3 | 0 |
| IL-32 | 31 | 126 | 5 | 2 | 1 |
| LTA | 119 | 170 | 6, 1 | 3 | 1 |
| LTB | 595 | 767 | 2, 1 | 1 | 1 |
| TNF | 50 | 85 | 5, 1 | 4, 1 | 1, 1 |
| CCL1 | 12 | 10 | 0 | 0 | 0 |
| CCL2 | 88 | 303 | 5 | 4 | 4 |
| CCL3 | 10 | 14 | 0 | 0 | 0 |
| CCL5 | 190 | 857 | 5, 1 | 3 | 3 |
| CCL13 | 75 | 25 | 3 | 1 | 1 |
| CCL19 | 65 | 55 | 3 | 2 | 0 |
| CXCL1 | 45 | 18 | 4 | 1 | 1 |
| CXCL2 | 47 | 25 | 2 | 0 | 0 |
| CXCL3 | 65 | 45 | 3 | 1 | 0 |
| CXCL9 | 23 | 40 | 2, 2 | 0 | 0 |
| CXCL10 | 20 | 27 | 1 | 0 | 0 |
| CXCL11 | 13 | 11 | 0 | 0 | 0 |
| CXCL12 | 168 | 50 | 5 | 4 | 3 |
Number of patients in which junction CD8+ T cells (JC_CD8) from 8-week-posthealing biopsy samples had either upregulated (bold) or downregulated (italic) gene expression relative to CD8+ T cells from matched contralateral biopsies (CL_CD8).
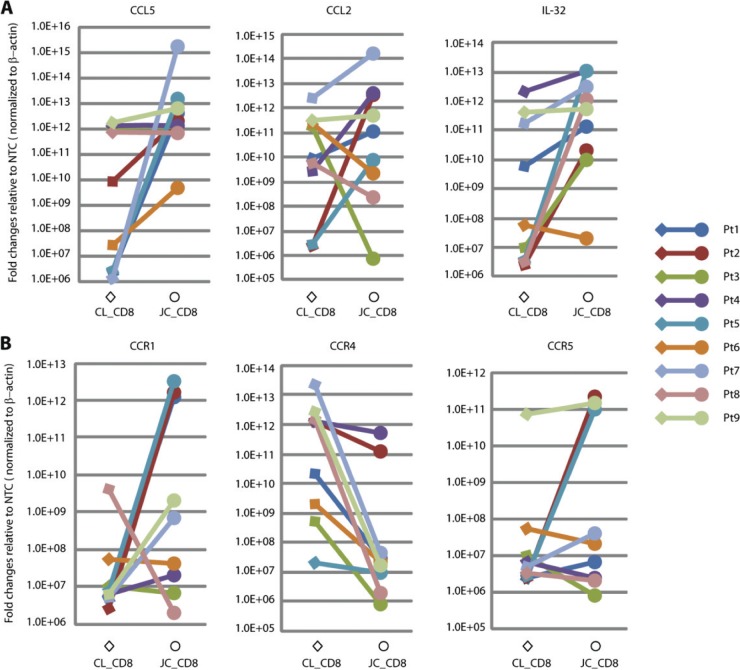
Fig 4.
Upregulation of CCL5, CCL2, IL-32, CCR1, and CCR5 and downregulation of CCR4 in JC_CD8 cells. The expression of chemokines/cytokines and chemokine receptors was assayed by qRT-PCR. The y axis shows fold change of gene expression in JC_CD8 or CL_CD8 cells over nontemplate controls (NTC) after normalization to ACTB. Pt, patient. (A) Upregulation of CCL5, CCL2, and IL-32 by JC_CD8 cells. (B) Upregulation of CCR1 and CCR5 and downregulation of CCR4 by JC_CD8 cells. Each reaction was done in duplicate.
Down-regulation of cytokines/chemokines in JC_CD8 cells.
Several chemokines were downregulated in JC_CD8 T cells, including CXCL1, CXCL3, CXCL12, CCL1, CCL13, and CCL19 (Table 2). CCL19 is known to bind CCR7 (30), a receptor involved in T-cell activation and migration to secondary lymphoid tissue, and CCR7 is significantly downregulated in JC_CD8 cells relative to CD8+ T cells in the dermal area near blood vessels (J. Zhu, T. Peng, K. Phasouk, A. Klock, L. Jin, D. M. Koelle, A. Wald, and L. Corey, submitted for publication). The proinflammatory chemokine CXCL12 was consistently downregulated in JC_CD8 cells from five of nine patients (Table 2). Interestingly, this gene was upregulated in one of four whole-tissue biopsy specimens taken 8 weeks after healing; the average array intensity was higher in 8-week-posthealing tissue than in contralateral tissue (Table 3). CCR4, the cognate receptor for the proinflammatory chemokine CXCL12, was downregulated in JC_CD8 cells in samples from all nine patients assayed by qRT-PCR (Fig. 4B) and in samples from two of nine patients tested by array (Table 4).
Table 3.
Expression patterns of chemokines in whole biopsy samples (8-week-posthealing and contralateral controls) from four patients
| Gene | Avg intensity |
No. of patients with difference ofa: |
|||
|---|---|---|---|---|---|
| Wh_CL | Wh_8wph | >3-fold | >5-fold | >10-fold | |
| CCL1 | 4 | 4 | 0 | 0 | 0 |
| CCL2 | 141 | 227 | 1 | 0 | 0 |
| CCL3 | 4 | 4 | 0 | 0 | 0 |
| CCL5 | 18 | 334 | 3 | 3 | 2 |
| CCL7 | 4 | 4 | 0 | 0 | 0 |
| CCL8 | 13 | 42 | 2 | 1 | 0 |
| CCL11 | 4 | 4 | 0 | 0 | 0 |
| CCL13 | 64 | 105 | 1 | 0 | 0 |
| CCL15 | 6 | 5 | 0 | 0 | 0 |
| CCL16 | 4 | 6 | 0 | 0 | 0 |
| CCL17 | 4 | 4 | 0 | 0 | 0 |
| CCL18 | 10 | 14 | 0 | 0 | 0 |
| CCL19 | 23 | 87 | 2 | 1 | 0 |
| CCL20 | 14 | 33 | 1 | 1 | 0 |
| CCL21 | 11 | 25 | 2 | 1 | 0 |
| CCL22 | 4 | 6 | 0 | 0 | 0 |
| CCL23 | 5 | 7 | 0 | 0 | 0 |
| CCL24 | 4 | 4 | 0 | 0 | 0 |
| CCL25 | 6 | 4 | 0 | 0 | 0 |
| CCL26 | 4 | 4 | 0 | 0 | 0 |
| CCL27 | 115 | 91 | 0 | 0 | 0 |
| CCL28 | 12 | 34 | 3 | 0 | 0 |
| CXCL1 | 5 | 6 | 0 | 0 | 0 |
| CXCL2 | 21 | 15 | 1 | 0 | 0 |
| CXCL3 | 12 | 12 | 0 | 0 | 0 |
| CXCL5 | 10 | 9 | 0 | 0 | 0 |
| CXCL6 | 4 | 4 | 0 | 0 | 0 |
| CXCL9 | 4 | 64 | 1, 1 | 1, 1 | 1, 1 |
| CXCL10 | 7 | 19 | 1 | 1 | 1 |
| CXCL11 | 4 | 4 | 0 | 0 | 0 |
| CXCL12 | 105 | 292 | 1 | 1 | 0 |
| CXCL13 | 4 | 7 | 0 | 0 | 0 |
| CXCL14 | 1,278 | 1,211 | 0 | 0 | 0 |
| CXCL16 | 44 | 27 | 0 | 0 | 0 |
| CXCL17 | 4 | 4 | 0 | 0 | 0 |
Bold, upregulated; italic, downregulated.
Table 4.
Expression patterns of chemokine receptors in JC_CD8 and CL_CD8 cells from biopsy samples taken 8 weeks after healing
| Gene | Avg intensity |
No. of patients with difference ofa: |
|||
|---|---|---|---|---|---|
| CL_CD8 | JC_CD8 | >3-fold | >5-fold | >10-fold | |
| CCR1 | 173 | 381 | 5, 3 | 5, 3 | 4, 3 |
| CCR2 | 13 | 15 | 0 | 0 | 0 |
| CCR3 | 19 | 10 | 0 | 0 | 0 |
| CCR4 | 52 | 10 | 2 | 1 | 1 |
| CCR5 | 46 | 41 | 4, 1 | 2, 1 | 1 |
| CCR7 | 34 | 43 | 1, 2 | 1, 2 | 1 |
| CCR8 | 36 | 14 | 2, 2 | 1, 2 | 2 |
| CCR9 | 33 | 24 | 0 | 0 | 0 |
| CCR10 | 17 | 16 | 0 | 0 | 0 |
| CXCR3 | 12 | 26 | 2 | 1 | 1 |
| CXCR4 | 32 | 24 | 2, 2 | 2, 1 | 1 |
| CXCR5 | 36 | 35 | 1 | 1 | 0 |
| CXCR6 | 11 | 58 | 4 | 3 | 1 |
| CXCR7 | 121 | 51 | 2, 3 | 3 | 1 |
Number of patients in which junction CD8+ T cells (JC_CD8) from 8-week-posthealing biopsy samples had either upregulated (bold) or downregulated (italic) gene expression relative to CD8+ T cells from matched contralateral biopsy samples (CL_CD8).
CCL5-CCR1/CCR5 interaction may mediate CD8+ T-cell migration to the dermal-epidermal junction.
T-cell migration into peripheral tissue is governed by the interaction between cytokine/chemokines and their receptors, which play an integral role in the regulation of lymphocyte trafficking (30). To understand which chemokines are responsible for CD8+ T-cell migration to the DEJ during reactivation episodes of HSV-2, we compared the transcriptional profiles of whole biopsy specimens from 8-week-posthealing tissue with contralateral control tissue from four patients. Among the 22 CC chemokine and 13 CXC chemokine genes represented on the Illumina Ref8_v3 bead arrays, CCL5 (RANTES) was the most significantly upregulated chemokine in the 8-week-posthealing tissue (>18-fold average upregulation) (Table 3).
Because CD8+ T cells are known to express CCL5 (5), we measured the gene expression profile of this chemokine in captured JC_CD8 cells and CL_CD8 cells by qRT-PCR. JC_CD8 cells expressed significantly higher levels of CCL5 than CL_CD8 cells in six of nine patients, similarly high levels in two patients, and modestly higher levels in one patient compared with controls (Fig. 4A). These data demonstrate that CD8+ T cells may be at least partially responsible for CCL5 expression in the DEJ.
CCR1 and CCR5, cell surface receptors known to be involved in leukocyte infiltration to sites of inflammation via interaction with CCL5 (23), showed modest induction in JC_CD8 cells from tissue taken 8 weeks after healing (Table 4). Interestingly, the gene expression patterns of CCR1 and CCR5 included both induced expression (>3-fold in five and four patients, respectively) and repressed expression (>3-fold in three and one patients, respectively) of JC_CD8 cells relative to CL_CD8 cells. CCR1 gene induction was verified with qRT-PCR, showing expression levels between 101-fold and 105-fold higher in JC_CD8 than CL_CD8 cells in five of nine patients (Fig. 4B), suggesting that expression of CCL5 by JC_CD8 cells may provide a positive feedback mechanism for CCR1+ CD8+ T-cell homing.
Differences in cytokine/chemokine receptor gene expression.
The chemokine receptors CXCR3 and CXCR6 were upregulated in two and four of nine patients, respectively, and were not downregulated in any patient samples (Table 4). CXCR3 is involved in lymphocyte recruitment, and CXCR6 is known to interact with CXCL16 as a mechanism for inflammation. CXCR7 was downregulated at least fivefold in JC_CD8 cells compared to CL_CD8 cells from three patients, although this receptor was upregulated threefold in two of nine patients. We found inconsistent up- and downregulation in the receptor genes CCR1, CCR5, CCR7, CCR8, and CXCR4. No other significant variation in expression of genes encoding other receptors involved in T-cell inflammation, such as CCR4 and CCR10 (10) (Table 4 and Fig. 4), within the two groups of cells was detected.
DISCUSSION
Previously we showed that CD8+ T cells persist at the dermal-epidermal junction at a high density for several months upon complete healing of herpes lesions (29). In this report, we provide evidence that the CD8+ T cells that are adjacent to the neuronal nerve endings are not quiescent but exhibit the metabolic and functional pattern of effector T cells, despite the lack of HSV-2 DNA or inflammation in these areas. The transcriptional induction of genes for T-cell activation in this cell population is consistent with an immunosurveillance function, as many of the signature transcripts identified have half-lives of <48 h. Thus, the uniform and consistent pattern of a wide variety of RNA transcripts in biopsy specimens from areas of known HSV-2 reactivation compared to areas with infrequent or undetectable reactivation during periods of clinical quiescence suggests that these T cells likely see HSV-2 antigen frequently without evidence of clinical reactivation.
One salient feature of our experimental approach was the lack of any in vitro manipulation of cells from human skin biopsy specimens, which allowed direct analysis of T-cell gene expression profiles indicative of the cells' in vivo physiological state. In addition, by comparing JC_CD8 cells to three types of controls (CL_CD8 cells, axillary cells, and CD1a+ cells) from the same patients, we were able to define the distinctive transcriptional response of T cells persisting at the site of previous herpes lesions.
During lymphocyte activation and proliferation, transcriptional regulation of a variety of cellular processes, including cell cycle initiation and glucose metabolism, adapts in response to antigenic and inflammatory stimuli. GLUT1, the primary glucose transporter in T lymphocytes, is transcriptionally expressed at low levels in resting T cells and both transcriptional induction and posttranslational trafficking are necessary for GLUT1-dependent glucose uptake during T-cell activation (6, 9), a process largely dependent on T-cell receptor (TCR) stimulation (19). In 89% of patients (8/9), GLUT1 was expressed at low levels in CL_CD8 cells (Fig. 3B), suggesting that this cell population was largely quiescent. In contrast, JC_CD8 cells in these patients expressed high levels of the transporter, indicative of an activated phenotype, with values ranging from 10- to 270-fold over control samples. Due to the short half-life of GLUT1 mRNA (between 10 and 12 h) (4), its detection indicates active transcription of the gene and suggests that resident CD8+ T cells likely encounter HSV-2 during periods in which HSV-2 DNA can be detected. Similarly, the half-life of IFNG mRNA is approximately 3 h in plasma T cells (7), and as with GLUT1, the expression pattern of IFNG was low in CL_CD8 cells but much higher in JC_CD8 cells. The detection of these short-lived transcripts is consistent with the model that HSV-2 frequently leaks from sacral neurons into the genital tract, where it subsequently stimulates resident T cells (2).
JC_CD8 cells express a variety of antiviral cytokines and chemokines, and the expression pattern we observed corresponds to known antiviral mechanisms of the cytokines IFN-γ, IL-32, and TNF-α on virus DNA replication of HSV-2 and vaccinia virus (3, 21, 27). In addition, IFN-γ can act synergistically with type I IFN or TNF-α in the production of antiviral immune responses. The IL-32-mediated antiviral cellular pathway effects only partially overlap those of type I IFN; it will be interesting to investigate whether IFN-γ, IL-32, and TNF-α can have cooperative effects to inhibit HSV-2 replication at peripheral sites.
One of the interesting observations of our study was that CD8+ T cells at the junction site express a substantial amount of CCL5/CCR1. These data suggest a positive feedback mechanism for the recruitment of CD8+ T cells to this region during the clinically quiescent phase of HSV-2 reactivation. It is not clear whether keratinocytes in 8-week-posthealing tissue express high levels of CCL5, although these cells can secret large quantities of this chemokine in response to Th1 cytokine (including IFN-γ and TNF-α) stimulation in vitro (1). Expression of CCL5 as measured by qRT-PCR was significantly higher in JC_CD8 cells relative to CL_CD8 cells in five of the nine patients, similarly high in three patients in both cell populations, and modestly higher in JC_CD8 cells in one patient (Fig. 4A). Expression of CCR1 (by qRT-PCR) was higher in JC_CD8 cells than in CL_CD8 cells in six of the nine patients, slightly lower in JC_CD8 cells in two patients, and significantly downregulated in JC_CD8 cells in one patient (Fig. 4B). Interestingly, the patients with increased levels of CCL5 also had increased levels of CCR1, which suggests that this pathway may be an important chemotactic mechanism in these patients. These results are partially consistent with numerous studies that have shown that CCL5 is responsible for chemotaxis of CCR1+ and CCR5+ lymphocytes (23, 26). The correlation of upregulation of both genes in the same patient may be due to the location of the biopsy. Studies have shown that microenvironments of the genital dermis and DEJ separated by just millimeters can have diverse immune cell density. Therefore, we hypothesize that the CCL5-CCR1 interaction may play a role in mediating the migration of CD8 cells to the dermal-epidermal junction area.
One limitation of this study is variance in the specificity and sensitivity of both microarray and qRT-PCR techniques for each gene. In most cases, both methods produced similar expression patterns. However, there were instances in which expression was inconsistent between the two methods. For example, expression of CCL2 was upregulated >3-fold in JC_CD8 cells in five of nine patients and >10-fold in four of nine patients as measured by microarray (Table 2). In contrast to array data, CCL2 was downregulated in three of nine patients as measured by qRT-PCR (patients 3, 6, and 8) (Fig. 4A). Expression of CCL2 in patient 6 was near basal level and the limit of detection in microarray studies; qPCR detected modest expression of CCL2 in both cell types and a downregulation in JC_CD8 relative to CL_CD8 cells. The low level of CCL2 expression in patients 3 and 8 can be potentially explained by experimental design: cDNA synthesized from laser captured cells (approximately 100 cells) has a mean size of 300 to 500 bp, and the Illumina single 50-bp probes and the PCR product (about 60 bp) detected by ABI TaqMan probes do not cover the same area of the cDNA.
Thus, while validation of gene expression studies via microarray and qRT-PCR is essential for making conclusions regarding expression patterns, the dynamic range and similarities in detection sensitivity and specificity may be complex. It should be noted that we observed concurrence in the two methods with a large number of genes.
In summary, transcriptional analysis of resident CD8+ T cells of the genital mucosal junction epithelium uncovered highly expressed genes in the functional categories of activation, proliferation, antiviral activity, and metabolism. High expression of chemokines and their receptors in these cells, as well as mucosal tissue, provides possible evidence for a positive regulatory loop in the homing of T cells to the sites of herpes lesions. This study will help further elucidate the complicated dynamic interplay between the adaptive host immune system and HSV-2 during frequent reactivation episodes over the course of this chronic infection.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, grants R37AI042528 and P01 AI030731-18 and by the James B. Pendleton Charitable Trust.
We thank Mindy Miner for technical editing. We also thank all study participants who contributed to this work.
Footnotes
Published ahead of print 18 July 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Albanesi C, et al. 2001. A cytokine-to-chemokine axis between T lymphocytes and keratinocytes can favor Th1 cell accumulation in chronic inflammatory skin diseases. J. Leukoc. Biol. 70:617–623 [PubMed] [Google Scholar]
- 2. Alsallaq RA, et al. 2010. Population level impact of an imperfect prophylactic vaccine for herpes simplex virus-2. Sex. Transm. Dis. 37:290–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bartee E, Mohamed MR, Lopez MC, Baker HV, McFadden G. 2009. The addition of tumor necrosis factor plus beta interferon induces a novel synergistic antiviral state against poxviruses in primary human fibroblasts. J. Virol. 83:498–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bruckner BA, Ammini CV, Otal MP, Raizada MK, Stacpoole PW. 1999. Regulation of brain glucose transporters by glucose and oxygen deprivation. Metabolism 48:422–431 [DOI] [PubMed] [Google Scholar]
- 5. Catalfamo M, et al. 2004. Human CD8+ T cells store RANTES in a unique secretory compartment and release it rapidly after TcR stimulation. Immunity 20:219–230 [DOI] [PubMed] [Google Scholar]
- 6. Chakrabarti R, Jung CY, Lee TP, Liu H, Mookerjee BK. 1994. Changes in glucose transport and transporter isoforms during the activation of human peripheral blood lymphocytes by phytohemagglutinin. J. Immunol. 152:2660–2668 [PubMed] [Google Scholar]
- 7. Curiel R, Garcia C, Farooq L, Aguero M, Espinoza-Delgado I. 2001. Bryostatin-1 and IL-2 synergize to induce IFN-γ expression in human peripheral blood T cells: implications for cancer immunotherapy. J. Immunol. 167:4828–4837 [DOI] [PubMed] [Google Scholar]
- 8. Evans R, et al. 2009. Integrins in immunity. J. Cell Sci. 122:215–225 [DOI] [PubMed] [Google Scholar]
- 9. Frauwirth KA, Thompson CB. 2004. Regulation of T lymphocyte metabolism. J. Immunol. 172:4661–4665 [DOI] [PubMed] [Google Scholar]
- 10. Homey B, et al. 2002. CCL27-CCR10 interactions regulate T cell-mediated skin inflammation. Nat. Med. 8:157–165 [DOI] [PubMed] [Google Scholar]
- 11. Jones RG, Thompson CB. 2007. Revving the engine: signal transduction fuels T cell activation. Immunity 27:173–178 [DOI] [PubMed] [Google Scholar]
- 12. Kaech SM, et al. 2003. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat. Immunol. 4:1191–1198 [DOI] [PubMed] [Google Scholar]
- 13. Koelle DM, et al. 2002. Expression of cutaneous lymphocyte-associated antigen by CD8(+) T cells specific for a skin-tropic virus. J. Clin. Invest. 110:537–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Koelle DM, et al. 1998. Clearance of HSV-2 from recurrent genital lesions correlates with infiltration of HSV-specific cytotoxic T lymphocytes. J. Clin. Invest. 101:1500–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lacorazza HD, et al. 2002. The ETS protein MEF plays a critical role in perforin gene expression and the development of natural killer and NK-T cells. Immunity 17:437–449 [DOI] [PubMed] [Google Scholar]
- 16. Lin L, Spoor MS, Gerth AJ, Brody SL, Peng SL. 2004. Modulation of Th1 activation and inflammation by the NF-kappaB repressor Foxj1. Science 303:1017–1020 [DOI] [PubMed] [Google Scholar]
- 17. Magaret A, Wald A, Huang ML, Selke S, Corey L. 2007. Optimizing PCR positivity criterion for detection of herpes simplex virus DNA on skin and mucosa. J. Clin. Immunol. 45:1618–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mark KE, et al. 2008. Rapidly cleared episodes of herpes simplex virus reactivation in immunocompetent adults. J. Infect. Dis. 198:1141–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Michalek RD, Rathmell JC. 2010. The metabolic life and times of a T-cell. Immunol. Rev. 236:190–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Morrow R, Friedrich D, Meier A, Corey L. 2005. Use of “Biokit HSV-2 rapid assay” to improve the positive predictive value of Focus HerpeSelect HSV-2 ELISA. BMC Infect. Dis. 5:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Peng T, Zhu J, Hwangbo Y, Corey L, Bumgarner RE. 2008. Independent and cooperative antiviral actions of beta interferon and gamma interferon against herpes simplex virus replication in primary human fibroblasts. J. Virol. 82:1934–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Peng T, et al. 2009. Evasion of the mucosal innate immune system by herpes simplex virus type 2. J. Virol. 83:12559–12568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sato K, Kawasaki H, Morimoto C, Yamashima N, Matsuyama T. 2002. An abortive ligand-induced activation of CCR1-mediated downstream signaling event and a deficiency of CCR5 expression are associated with the hyporesponsiveness of human naive CD4+ T cells to CCL3 and CCL5. J. Immunol. 168:6263–6272 [DOI] [PubMed] [Google Scholar]
- 24. Schiffer JT, Wald A, Selke S, Corey L, Magaret A. 2011. The kinetics of mucosal herpes simplex virus-2 infection in humans: evidence for rapid viral-host interactions. J. Infect. Dis. 204:554–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wald A, et al. 1997. Frequent genital herpes simplex virus 2 shedding in immunocompetent women. Effect of acyclovir treatment. J. Clin. Invest. 99:1092–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wu L, et al. 1997. Interaction of chemokine receptor CCR5 with its ligands: multiple domains for HIV-1 gp120 binding and a single domain for chemokine binding. J. Exp. Med. 186:1373–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zepp JA, Nold-Petry CA, Dinarello CA, Nold MF. 2011. Protection from RNA and DNA viruses by IL-32. J. Immunol. 186:4110–4118 [DOI] [PubMed] [Google Scholar]
- 28. Zhu J, et al. 2009. Persistence of HIV-1 receptor-positive cells after HSV-2 reactivation is a potential mechanism for increased HIV-1 acquisition. Nat. Med. 15:886–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhu J, et al. 2007. Virus-specific CD8+ T cells accumulate near sensory nerve endings in genital skin during subclinical HSV-2 reactivation. J. Exp. Med. 204:595–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zlotnik A, Yoshie O. 2000. Chemokines: a new classification system and their role in immunity. Immunity 12:121–127 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.