Abstract
A variety of cellular and viral processes are coordinately regulated during adenovirus (Ad) infection to achieve optimal virus production. The Ad late gene product L4-22K has been associated with disparate activities during infection, including the regulation of late gene expression, viral DNA packaging, and infectious virus production. We generated and characterized two L4-22K mutant viruses to further explore L4-22K functions during viral infection. Our results show that L4-22K is indeed important for temporal control of viral gene expression not only because it activates late gene expression but also because it suppresses early gene expression. We also show that the L4-22K protein binds to viral packaging sequences in vivo and is essential to recruit two other packaging proteins, IVa2 and L1-52/55K, to this region. The elimination of L4-22K gave rise to the production of only empty virus capsids and not mature virions, which confirms that the L4-22K protein is required for Ad genome packaging. Finally, L4-22K contributes to adenovirus-induced cell death by regulating the expression of the adenovirus death protein. Thus, the adenovirus L4-22K protein is multifunctional and an integral component of crucial aspects of infection.
INTRODUCTION
Adenoviruses (Ad) have a nonenveloped, icosahedral virion that contains the linear, double-stranded DNA genome. Ad infect a wide range of animals, and over 50 different serotypes of human Ad have been identified. Human Ad infection can lead to respiratory tract, gastrointestinal, and ophthalmologic diseases. Most of these infections are self-limiting but can be severe and even fatal in immunocompromised hosts and occasionally in healthy young children and adults (8). During Ad infection, the virus enters the cell through receptor-mediated endocytosis and the partially degraded virus capsid escapes from the endosome and travels to the nuclear pore complex, where the viral genome is translocated into the nucleus (36). E1A, the immediate early gene product, is expressed and transactivates the Ad early genes (E1B, E2, E3, and E4) whose products function to optimize the cellular milieu for viral replication, counteract a variety of antiviral defenses, and promote viral DNA replication (4). With the onset of viral DNA replication, the major late promoter (MLP) is fully activated and late gene products (L1 to L5) are synthesized to produce progeny viruses (1). The MLP is activated by a number of cellular transcription factors and a viral protein, IVa2, which bind to upstream elements (UE) and downstream elements (DE) of the MLP, respectively (46). Mutations in DE that abolish IVa2 binding or mutation of IVa2 coding sequences, however, have only a modest effect on Ad late gene expression (30, 48). These and other results indicate a redundancy of MLP elements (46). The viral protein L4-33K facilitates late gene expression by enhancing the alternative splicing of Ad late transcripts (39) and facilitating the early-to-late switch during infection (10, 23). During the final stage of infection, the Ad death protein (ADP) induces cell lysis, which leads to the release of progeny viruses from the nucleus (37).
The mature Ad virion consists of major capsid proteins (hexon, penton, and fiber), minor capsid proteins (IIIa, VI, VIII, and IX), and core proteins (V, VII, Mu, and TP) that associate with the viral genome (36). In addition, the Ad protease (AVP) is incorporated in the virion and cleaves precursor proteins (IIIa, pVI, pVII, pVIII, pX, pTP, and L1-52/55K) to produce the fully infectious mature virion (22). From the results of a number of studies, it is believed that Ad empty capsids (EC) are first assembled in the nucleus and that the viral genome is subsequently encapsidated (27). The EC produce bands at a lighter density (1.29 to 1.30 g/cc) than the mature virions (MV) (1.34 g/cc) in a CsCl density equilibrium gradient. MV contain proteolytically processed forms of proteins VI, VII, and VIII; precursor forms of pVI and pVIII are present in EC. In contrast to the MV, EC also contain the L1-52/55K protein (15). Core proteins V and VII, which are associated with the viral genome, are present in MV but are absent or present at lower levels in EC.
The Ad genome is packaged into the capsid, utilizing cis-acting packaging sequences (PS) located at the left end of the viral genome (27). The PS overlap with the E1A transcriptional enhancer region and contain seven AT-rich motifs, termed A repeats, which have the conserved, bipartite sequence motif 5′-TTTG-N8-CG-3′ (27). Deletion of the PS results in no progeny virus production (17). Chromatin immunoprecipitation (ChIP) experiments have shown that Ad proteins IVa2, L1-52/55K, and IIIa bind to the PS in vivo (21, 28, 32). Analyses of mutant viruses demonstrate that these three proteins are required for Ad packaging, since only EC are produced in their absence (7, 13, 16, 24). In vitro DNA-protein binding analyses have shown that the IVa2 protein binds to the CG motif of the PS and that another Ad late gene product, L4-22K, binds to the TTTG motif of the PS; binding of L4-22K to the PS is dependent on the IVa2 protein (9, 28, 40, 44, 45, 47). While the L1-52/55K protein does not bind to the PS in vitro, it does interact with the IVa2 protein in Ad-infected cells and with protein IIIa in vitro (14, 21). The IVa2 protein binds ATP (26) and is located at a single virion vertex (6). A working model of Ad DNA packaging suggests that the IVa2 and L4-22K proteins bind directly to the PS and recruit the L1-52/55K and IIIa proteins to promote viral DNA packaging; the IVa2 protein may function as a packaging motor by analogy to bacteriophage ATPases (12).
The L4-22K protein has been suggested to have distinct functions during viral infection. Transfection into cells of an Ad5 infectious clone which contains a stop codon in L4-22K results in no infectious virus production (25), suggesting that L4-22K is required for genome packaging and consistent with the observation that L4-22K binds to the PS in vitro. By transfection analyses performed using the same Ad5 L4-22K mutant infectious clone, Morris and Leppard showed that L4-22K promotes Ad late gene expression (23). Because these two analyses indicated completely different roles for the L4-22K protein during Ad infection and due to the limitations of experiments involving transient-transfection assays, we sought to study L4-22K function in the context of virus infection. In the current study, we generated two Ad5 L4-22K mutant viruses, Δ22K (a null mutant) and 22K− (an amber mutant), using an L4-22K-complementing cell line, to further explore the functions of L4-22K in vivo. Here we demonstrate that the L4-22K protein is important for appropriate temporal control of viral early and late gene expression. The effects of L4-22K in the activation of Ad late gene expression differed for different late genes. In addition, the L4-22K mutants produced only EC and not MV in noncomplementing cells, demonstrating that the L4-22K protein is required for genome packaging. ChIP assays demonstrated that L4-22K and IVa2 are dependent on each other for binding to the PS and that both of these proteins are required to recruit L1-52/55K to the PS in vivo. Finally, the L4-22K protein is required for expression of ADP at late times after infection and the induction of cellular cytopathic effect. We conclude that the Ad L4-22K protein is multifunctional and an integral component in crucial aspects of infection.
MATERIALS AND METHODS
Cells and viruses.
A549 (ATCC) and N52.E6 (35) cells were maintained in Dulbecco's modified Eagle medium (DMEM) containing penicillin and streptomycin and supplemented with 10% bovine calf serum and FetalClone III serum (HyClone), respectively. N52.E6-Cre (a kind gift from Gudrin Schiedner and Stefan Kochanek, Ulm University) and 293-L1 (13) cells were maintained in DMEM containing penicillin and streptomycin and 200 μg/ml Geneticin and supplemented with 10% FetalClone III and bovine calf serum, respectively.
To make the L4-22K-ΔC cell line, the N-terminal 173 amino acids of Ad5 L4-22K, with a stop codon at the end, were subjected to PCR amplification and cloned into pUHD 10-3 (kindly provided by H. Bujard, Zentrum für Molekulare Biologie, Germany) under the control of the tetracycline-response element and a minimal cytomegalovirus (CMV) promoter using EcoRI and XbaI sites. The resulting plasmid, pUHD-22K-ΔC, was cotransfected with pSuper-Blasticidin (Invitrogen) into TET-C4 (24), a cell line that constitutively expresses the tTA tetracycline-controlled transactivator in the background of N52.E6-Cre cells. An L4-22K-ΔC-expressing cell line (termed TetC4-22K-ΔC) was selected and maintained in DMEM containing penicillin and streptomycin, 200 μg/ml Geneticin, 200 μg/ml hygromycin B, 10 μg/ml Blasticidin S, and 1 μg/ml doxycycline and supplemented with 10% fetal bovine serum (HyClone). The same culture conditions were used for maintaining cell lines TetC4-IVa2 (24) and TetC4-33K. The generation of TetC4-33K is to be described elsewhere.
An Ad5-wild-type (Ad5-WT) virion stock was generated by transfecting PacI-linearized infectious clone pTG3602 (5) into N52.E6-Cre cells; a cell lysate was prepared and purified virions were produced using A549 cells. IVa2 mutant viruses pm8002 (48) and ΔIVa2 (24) were propagated using TetC4-IVa2 cells minus doxycycline in the culture medium. L1-52/55K mutant virus pm8001 (13) and an Ad5 virus containing loxP sites flanking the PS (termed Ad5-Ψ-loxP [unpublished data]) were propagated in 293-L1 and N52.E6 cells, respectively. All virions were purified using CsCl equilibrium density gradient centrifugation, as described previously (24). Throughout this study, A549 cells were infected at a multiplicity of infection (MOI) of 12.5 PFU/cell; N52.E6, N52.E6-Cre, TetC4-IVa2, TetC4-22K-ΔC, and TetC4-33K cells were infected at an MOI of 4 PFU/cell.
The strategy used for making pTG3602-Δ22K was to delete the L4-33K intron (Fig. 1A) so that the C terminus of L4-22K would be deleted without interfering with L4-33K expression. Cloning details are available upon request. The Δ22K sequence corresponds to Ad5 nucleotides (nt) 26195 to 27590 with the L4-33K intron (nt 26511 to 26712) deleted. This fragment was cotransformed with SpeI-digested pTG3602 into Escherichia coli BJ5183 cells for homologous recombination (5), and pTG3602-Δ22K was isolated and confirmed by nucleotide sequencing. The construction of pTG3602-22K− was described previously (25).
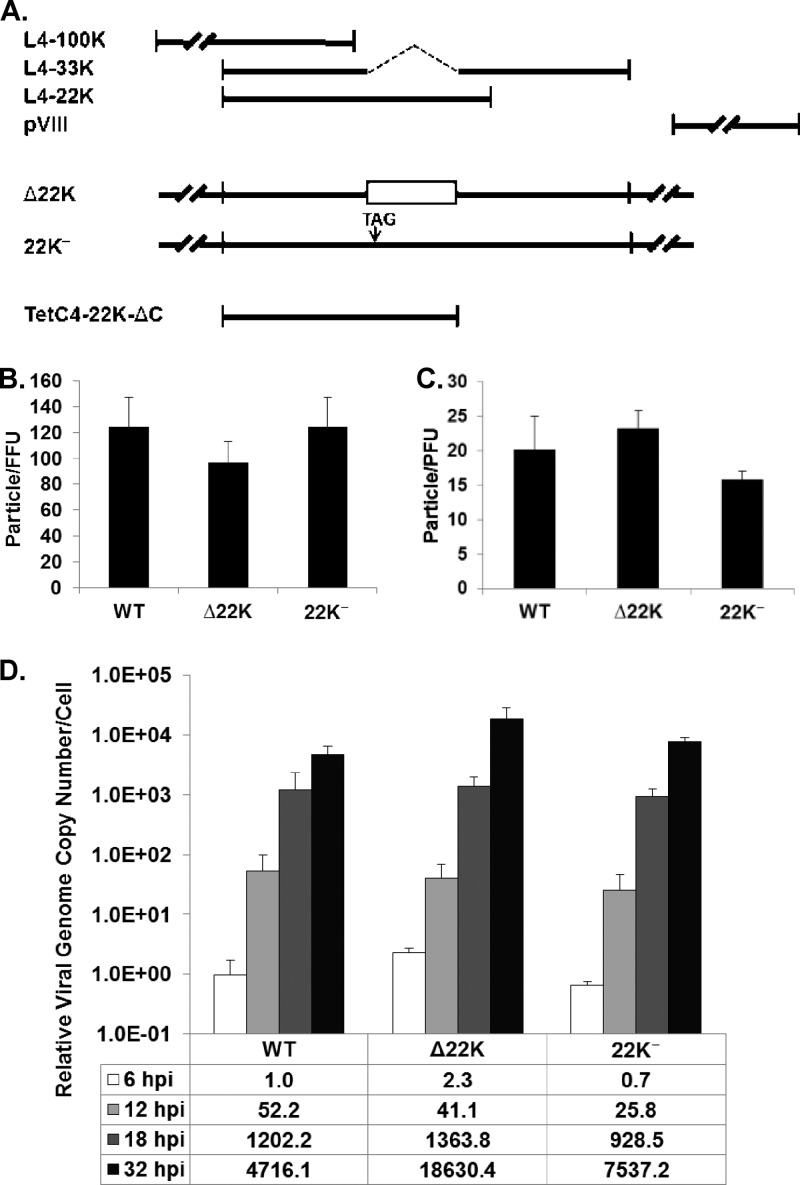
Fig 1.
L4-22K mutant viruses. (A) Schematic diagram of the Ad5 L4 region. Coding sequences for L4-100K, L4-33K, L4-22K, and pVIII are shown. L4-22K mutants Δ22K and 22K− are depicted along with the L4-22K-ΔC protein expressed in TetC4-22K-ΔC cells. (B) The particle/FFU ratios of Ad5-WT, Δ22K, and 22K− viruses determined in A549 cells are presented. (C) The particle/PFU ratios of Ad5-WT, Δ22K, and 22K− viruses determined in TetC4-22K-ΔC cells are presented. (D) Viral genome replication in Ad5-WT-, Δ22K-, or 22K−-infected A549 cells. The genome copy number was normalized to the GAPDH level, and this value was compared to that of the WT at 6 hpi for presentation of the calculations.
To make Δ22K and 22K− viruses, PacI-linearized pTG3602-Δ22K and pTG3602-22K− plasmids were transfected using NanoJuice (EMD Millipore) into TetC4-22K-ΔC cells which had been seeded on the plates without doxycycline for 2 days. Virus stocks were subjected to passage twice on TetC4-22K-ΔC cells without doxycycline. The titer of the passage 2 lysate of each mutant virus was determined on TetC4-22K-ΔC cells without doxycycline and used to generate purified virions.
Fluorescent focus assay.
A549 cells grown on glass coverslips were infected with different numbers of viral particles in 24-well plates. Cells were fixed using methanol at 18 h postinfection (hpi) and subjected to indirect immunofluorescence analysis using a monoclonal antibody (MAb) against the Ad DNA binding protein (DBP) (MAb-B6; provided by Arnold Levine, Princeton University) and a secondary goat anti-mouse antibody conjugated with fluorescein isothiocyanate (FITC) (Invitrogen). Coverslips were examined under a Zeiss Axiovert 200 M digital deconvolution microscope, and 10 random fields of each coverslip were counted for DBP-positive cells. The average number of DBP-positive cells (quantified as fluorescent focus units [FFU]) per field was used to calculate total FFU in the well, and the particle/FFU ratio was calculated as (number of particles)/(total FFU in the well).
Measurement of viral genome replication by quantitative PCR.
A549 cells were infected with Ad5-WT, Δ22K, or 22K− viruses and harvested at 6, 12, 18, and 32 hpi. Cell pellets were washed once with phosphate-buffered saline (PBS) and resuspended in 500 μl isotonic NP-40 buffer (150 mM NaCl, 10 mM Tris [pH 7.5], 1.5 mM MgCl2, 0.6% NP-40). Cell lysates were kept on ice for 5 to 10 min, and nuclei were pelleted by centrifugation at 2,000 × g at 4°C for 5 min and then resuspended in 200 μl PBS for DNA extraction using a DNeasy blood and tissue kit (Qiagen) and following the manufacturer's instructions. The final DNA sample was resuspended in Tris-EDTA (TE) buffer and then subjected to quantitative PCR using an Applied Biosystems 7500 real-time PCR system. The two sets of primers used for quantitative PCR were primer pair PACK-4 (Ad5 nt 44 to 63 [TAATGAGGGGGTGGAGTTTG]) and PACK-5 (Ad5 nt 280 to 261 [GCGAAAATGGCCAAATGTTA]) and primer pair GAPDH-5 (CCCCACACACATGCACTTACC) and GAPDH-6 (CCTAGTCCCAGGGCTTTGATT). Genome replication was calculated as the value of the viral genome copy number divided by the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) copy number. For comparison, the value of each virus at each time point was normalized to that of WT virus at 6 hpi, shown as the relative viral genome copy number per cell.
Western blot analysis.
Mock- or virus-infected cells were lysed in 2× Laemmli sample buffer (0.15 M Tris [pH 6.8], 4% sodium dodecyl sulfate [SDS]), and protein concentrations were determined using a bicinchoninic acid (BCA) protein assay kit (Pierce). Whole-cell extract (40 μg) was loaded for SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to nitrocellulose (Whatman) or polyvinylidene difluoride (PVDF) (GE Healthcare) membranes. Membranes were blocked with 3% bovine serum albumin (BSA)–Tris-buffered saline (TBS) for 1 h at room temperature and then probed with primary antibody for 1 h at room temperature or 4°C overnight. Primary antibodies included rabbit polyclonal E1A antibody (SC430; Santa Cruz Biotechnology) (1:1,000 dilution), rabbit polyclonal DBP antibody (Peter van der Vliet, University of Utrecht) (1:2,000 dilution), rabbit polyclonal IVa2 antibody (28) (1:500 to 1:1,000 dilution), rabbit polyclonal L1-52/55K antibody (28) (1:1,000 dilution), rabbit polyclonal IIIa antibody (21) (1:1,000 dilution), rabbit polyclonal penton antibody (Carl Anderson, Brookhaven National Laboratory) (1:1,000 dilution), rabbit polyclonal VII antibody (Daniel Engel, University of Virginia) (1:2,000 dilution), rabbit polyclonal V antibody (David Matthews, University of Bristol) (1:1,000 dilution), rabbit polyclonal VI antibody (Christopher Wiethoff, Loyola University Chicago) (1:10,000 dilution), rabbit polyclonal hexon antibody (GTX44240; GeneTex) (1:1,000 dilution), rabbit polyclonal L4-100K antibody (1:1,000 dilution), rabbit polyclonal L4-22K antibody (against the distinct C terminus of L4-22K [25]) (1:2,000 dilution), rabbit polyclonal L4-33K antibody (against the distinct C terminus of L4-33K) (1:5,000 dilution), rabbit polyclonal VIII and ADP antibodies (Ann Tollefson and William Wold, St. Louis University) (1:400 dilution), rabbit polyclonal fiber antibody (Carl Anderson) (1:1,000 dilution), and rabbit polyclonal γ-tubulin antibody (T5192; Sigma) (1:1,000 dilution). After primary antibody incubation, the membrane was washed 3 times with 5% nonfat milk–TBS containing 0.05% Tween 20 and then subjected to analysis with IRDye CW800-conjugated goat (polyclonal) anti-rabbit antibody (catalog no. 926-32211; Li-COR) for 45 min at room temperature. The final membrane was washed again 3 times with 5% nonfat milk–TBS buffer containing 0.05% Tween 20 and examined by the use of an Odyssey infrared imaging system (Li-COR).
For virus particle analyses, ∼2 × 108 A549, N52.E6-Cre, N52.E6, TetC4-22K-ΔC, and TetC4-33K cells were infected by Ad5-WT, Δ22K, 22K−, or loxP viruses. At 48 to 72 hpi, infected cells were harvested for preparation of virus particles by a CsCl density gradient. Isolated MV or EC from the CsCl gradient were subsequently analyzed by silver staining and Western blotting as described previously (24).
Northern blot analysis.
A549 cells infected by Ad5-WT, Δ22K, or 22K− viruses were harvested at 12, 24, and 48 hpi for preparation of cytoplasmic mRNA using an RNeasy Mini Kit (Qiagen) following the manufacturer's instructions. A 12-μg volume of RNA of each sample was separated on a 1% formaldehyde–agarose gel and transferred to a positively charged nylon membrane (GE Healthcare). The probe for detecting E1A mRNAs corresponds to Ad5 nt 147 to 1334, which was amplified by PCR. The purified probe was labeled with [32P]dATP by random primer labeling using exo− Klenow DNA polymerase (NEB).
ChIP.
A549 cells were infected with Ad5-WT, Δ22K, 22K−, pm8002, ΔIVa2, or pm8001, and TetC4-IVa2 or TetC4-22K-ΔC cells were infected with Ad5-WT, Δ22K, 22K−, pm8002, or ΔIVa2. At 18 hpi, infected cells were cross-linked with formaldehyde and sonicated, and the supernatants from the sonication were subjected to a preclearing process with protein A-agarose beads and then to overnight incubation with the corresponding antibodies at 4°C, as described previously (43). After overnight incubation with the antibody, the manufacturer's instructions for the use of protein A-agarose/salmon sperm DNA (Millipore) were followed for the washing and DNA precipitation processes. The final DNA sample was dissolved in 50 μl TE buffer and subjected to quantitative PCR analysis using two sets of primers: the PACK-4/-5 primer set described above was used to assess the number of copies of the PS that had been specifically immunoprecipitated by the antibodies, and an E4-ORF6 primer set (Ad5 nt 33597 to 33578 [TACCGGGAGGTGGTGAATTA] and Ad5 nt 33428 to 33447 [TTCAAAATCCCACAGTGCAA]) was used to assess the copy number of an Ad5 right-end DNA fragment that had been nonspecifically immunoprecipitated or reflected incomplete sonication products. The capacity of each viral protein (L4-22K, IVa2, or L1-52/55K) to bind the PS in virus-infected cells was calculated as the value of the PS copy number divided by the E4-ORF6 copy number. For comparison, the PACK/E4-ORF6 ratio for each antibody in a particular virus infection was normalized using ChIP with preimmune serum. To confirm viral protein expression in ChIP experiments, 20 μl of 600 μl of supernatant remaining after the preclearing was boiled for 10 min in 0.1 M dithiothreitol (DTT) to reverse the cross-links and analyzed by Western blotting as the input control.
Propidium iodide (PI) staining.
Mock- or virus-infected A549 cells (including detached ones) were collected from 100-mm-diameter plates and washed with PBS once at 24, 48, or 72 hpi. A fraction of each set of cells were spun down and resuspended in culture medium with 1 μg/ml PI (Invitrogen). A total of 10,000 cells of each sample were analyzed for cell viability as determined by PI exclusion using a FACSCalibur flow cytometer (BD Biosciences).
RESULTS
Generation, infectivity, and viral genome replication of L4-22K mutant viruses.
The Ad L4 region encodes four proteins: L4-100K, L4-33K, L4-22K, and pVIII (Fig. 1A). In order to assess the role of L4-22K during Ad infection, two L4-22K mutant viruses were generated and characterized in this study: Δ22K (a null mutant) and 22K− (an amber mutant). A Δ22K infectious clone (pTG3602-Δ22K) was made by deleting the L4-33K intron, leaving the L4-33K coding region intact but truncating the L4-22K protein at amino acid 105 (of 194). Deletion of the L4-33K intron shifts the reading frame of the remaining C terminus of L4-22K and might generate a novel L4 product with the same N terminus as L4-22K and L4-33K. However, Western blot analysis of Δ22K-infected cell lysate did not detect such a product using an antibody against the shared N-terminal region of L4-22K and L4-33K (9) (data not shown). A 22K− infectious clone (pTG3602-22K−) which contains a stop codon (TAG) at amino acid 114 of L4-22K was previously constructed without affecting the L4-33K open reading frame (ORF) (25). Two different L4-22K mutant viruses were analyzed in these studies to verify the specificity of the results for the L4-22K mutations and because of uncertain effects that these different mutations may have on L4-33K expression. Our previous observation that transfection of pTG3602-22K− resulted in no infectious virus production (25) suggested that an L4-22K-complementing cell line would be needed to propagate the L4-22K mutant viruses. To reduce the chance of homologous recombination that would give rise to revertants, we made a tetracycline-inducible cell line expressing L4-22K-ΔC (amino acids 1 to 173 of Ad5 L4-22K) termed TetC4-22K-ΔC (Fig. 1A). Earlier work from our laboratory had shown that Ad17 L4-22K was able to complement the growth of pTG3602-22K− in a complementation assay (42); Ad5 and Ad17 L4-22K proteins are well conserved except for an extra 23 C-terminal amino acids in Ad5 L4-22K. We reasoned that the extra 23 C-terminal amino acids of Ad5 L4-22K would not be required to complement Ad5 L4-22K mutant viruses and that the removal of these sequences would eliminate or reduce the chance of generating revertants of the Δ22K and 22K− mutant viruses. Indeed, TetC4-22K-ΔC cells efficiently complemented the growth of Δ22K and 22K− mutant viruses. Southern blot analysis of DNA isolated from Δ22K- or 22K−-infected A549 cells confirmed that both mutant virus stocks were pure (data not shown).
To test the infectivity of Δ22K and 22K− viruses, virus particles were used in a plaque assay using the L4-22K-complementing cell line and in a fluorescent focus assay performed using A549 cells. The particle/PFU and particle/FFU ratios for each virus were calculated, and the results showed no significant differences between Ad5-WT and the two L4-22K mutant viruses (Fig. 1B and C). Next, we evaluated viral DNA replication (Fig. 1D). From the onset of infection to 18 hpi, Ad5-WT and both L4-22K mutant viruses exhibited similar input levels (6 hpi) and DNA replication levels (12 and 18 hpi). At 32 hpi, however, the L4-22K mutant viruses replicated at levels 2-to-4-fold higher than those seen with Ad5-WT. In contrast to these results, neither L4-22K mutant produced detectable infectious virus in A549 cells as measured by titer determinations in a growth curve (data not shown). We conclude that the two L4-22K mutant viruses had levels of infectivity and viral genome replication comparable to those of Ad5-WT and that the L4-22K protein is critical for the production of infectious progeny.
L4-22K mutant virus gene expression.
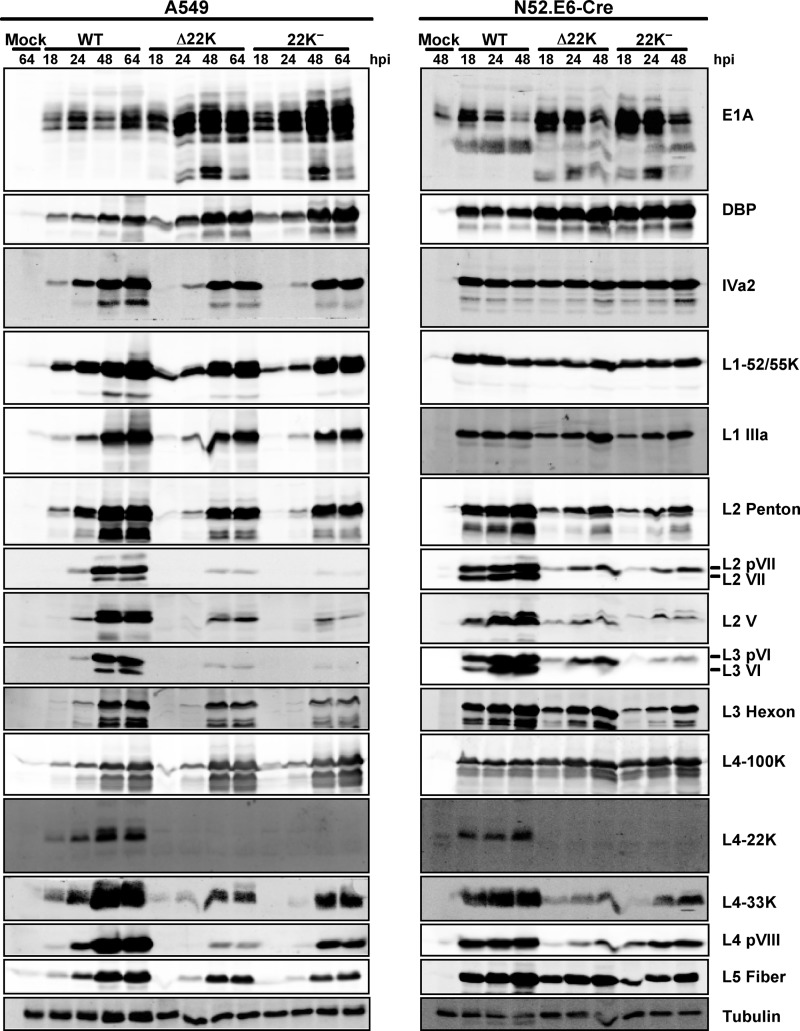
To examine viral gene expression with the L4-22K mutant viruses, A549 cells were infected with Ad5-WT, Δ22K, or 22K− viruses and total cell extracts were prepared at different time points and analyzed by Western blotting. With early gene expression, both L4-22K mutant viruses had significantly higher E1A expression levels than Ad5-WT during the late phase of infection (Fig. 2, left panel [24 to 64 hpi]). In addition, DBP levels were also increased 2-to-4-fold at late times after infection, which corresponded to the higher viral genome replication observed at 32 hpi with the L4-22K mutant viruses (Fig. 1D). The analysis of intermediate (IVa2) and late (L1 to L5) gene expression showed that there were delays and decreases to various extents with respect to gene production with the L4-22K mutant viruses (Fig. 2, left panel). Among the gene products whose expression was delayed and decreased, expression of major capsid proteins (hexon, penton, and fiber) and one minor capsid protein (IIIa) was modestly decreased, and expression of IVa2 and L1-52/55K was delayed. Other minor capsid proteins (VI and VIII), core proteins (V and VII), and L4-33K exhibited a significant decrease in expression compared with Ad5-WT. In contrast, levels of expression of L4-100K with Ad5-WT and the L4-22K mutant viruses were quite similar. Full-length L4-22K protein was not detected with the mutant viruses, as expected, and stable truncated products were not observed. Similar results were found following infection of N52.E6-Cre cells (Fig. 2, right panel), but the extent of delay and decrease of expression for the late gene products was less significant with the L4-22K mutant viruses than with the infection in A549 cells. In addition, the increases in E1A and DBP expression in L4-22K mutant virus infections were also observed in N52.E6-Cre cells during the late phase of infection (Fig. 2, right panel). These results demonstrated that mutation of the L4-22K gene had pleiotropic effects on viral early and late gene expression; increased early gene expression and decreased late gene expression indicate that the L4-22K protein is important to drive the early-to-late phase switch during Ad infection.
Fig 2.
Western blot analysis of Ad early and late proteins. Representative early (E1A and DBP), intermediate (IVa2), and late (L1 to L5) gene products were analyzed using whole-cell extracts isolated from Ad5-WT-, Δ22K-, and 22K−-infected A549 cells (left) and N52.E6-Cre cells (right) at different times postinfection (indicated at the top in hours). Protein designations are indicated on the right; tubulin was analyzed as a loading control.
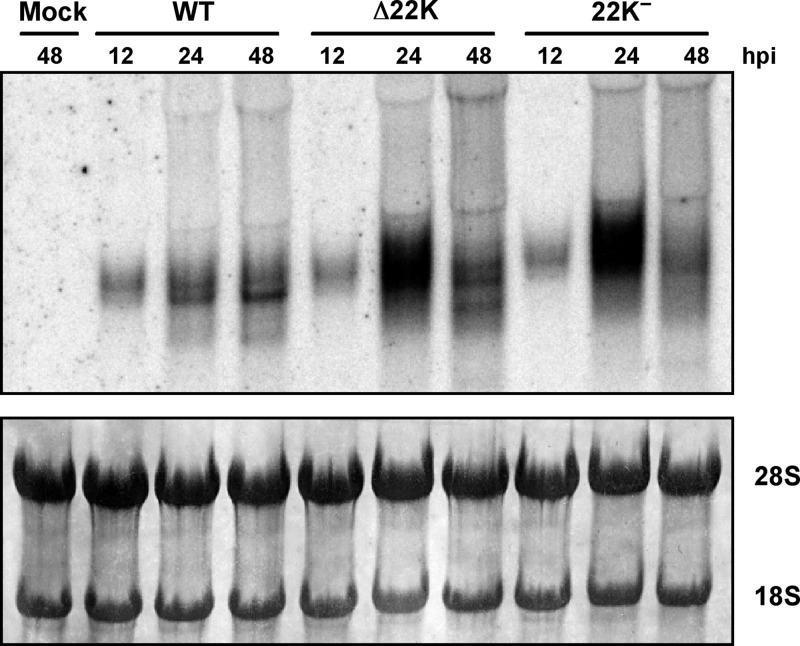
To further explore the increased levels of E1A observed with L4-22K mutant viruses during the late phase of infection, Northern blot analysis was performed to measure E1A mRNA levels. These results showed that E1A mRNA levels were significantly elevated in both Δ22K- and 22K−-infected cells at 24 hpi compared to Ad5-WT-infected cells (Fig. 3). This increase suggests that the L4-22K protein plays a role in suppressing early gene expression.
Fig 3.
Northern blot analysis of E1A mRNA levels. Total cytoplasmic mRNAs isolated from Ad5-WT-, Δ22K-, and 22K−-infected A549 cells at 12, 24, and 48 hpi were analyzed by Northern blotting using a 32P-labeled E1A probe (top panel). The blot was stained with methylene blue as a loading control (bottom panel).
L4-22K mutant viruses produce only empty capsids.
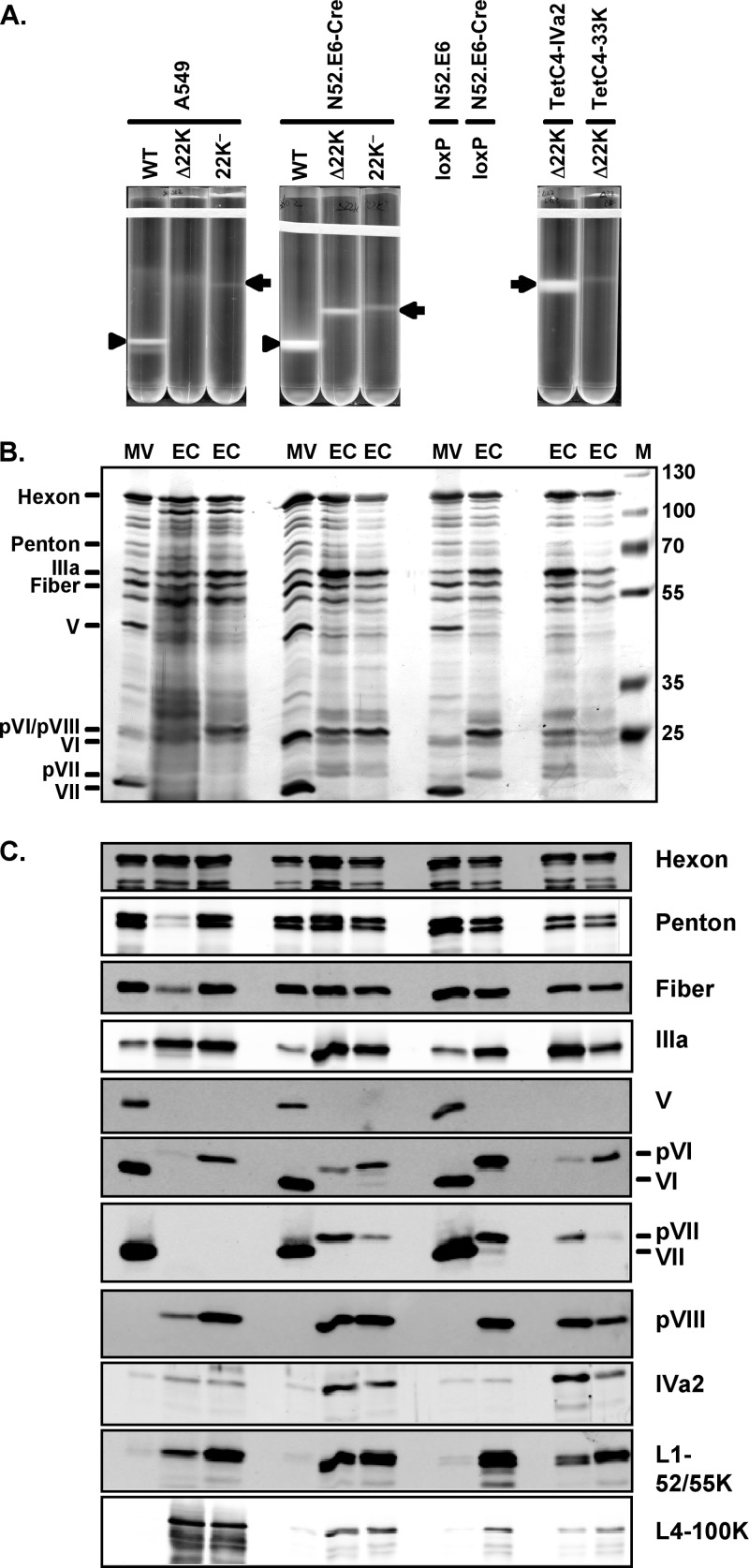
To determine if L4-22K is required for Ad genome packaging in vivo, A549 and N52.E6-Cre cells were infected with the L4-22K mutant viruses and virus particle production was analyzed using CsCl equilibrium density gradient centrifugation. In both A549 and N52.E6-Cre cells, only EC were observed with Δ22K- or 22K−-infected cells, which produced bands at a density of 1.29 g/cc (Fig. 4). The yield of empty capsids from both L4-22K mutant viruses was lower in A549 than in N52.E6-Cre cells, which might reflect the more permissive nature of N52.E6-Cre cells compared to A549 cells, since elevated levels of Ad5-WT were also observed. This phenotype of the L4-22K mutant viruses strongly indicates that the L4-22K protein is required for genome packaging. However, expression of many of the late viral gene products, including IVa2 and L4-33K, which regulate late gene expression on the transcriptional and posttranscriptional levels, respectively, was moderately or significantly delayed and decreased (Fig. 2) (1, 30, 39). In order to examine the contribution of delayed IVa2 and reduced L4-33K gene expression to the L4-22K mutant virus phenotype, we infected IVa2- or L4-33K-expressing cell lines with the Δ22K mutant virus to investigate if virion production was altered when IVa2 or L4-33K was supplemented in trans. These results showed that supplementation of IVa2 or L4-33K to Δ22K infection did not augment mature virion production (Fig. 4). Curiously, Δ22K infection of the IVa2-expressing cell line did increase empty capsid production, although we do not know if this represents complementation or a property of this particular cell subclone. Since empty capsids were produced in L4-22K mutant virus infections that had not resulted in encapsidation of the viral genome, we conclude that the L4-22K protein is required for viral DNA packaging.
Fig 4.
Virus particle production with L4-22K mutant viruses. (A) CsCl equilibrium density gradient profiles of virus particles produced from Ad5-WT-, Δ22K-, and 22K−-infected A549 and N52.E6-Cre cells and Δ22K-infected TetC4-IVa2 and TetC4-33K cells. Arrows and arrowheads indicate mature virions (MV) and empty capsids (EC), respectively. (B) The results of silver staining of protein components of MV and EC isolated from the virus particles indicated in panel A are shown; in addition, MV and EC isolated from Ad5-Ψ-loxP-infected N52.E6 (MV) and N52.E6-Cre (EC) cells are shown. Protein designations are indicated on the left. M, molecular weight markers. (C) Western blot analysis of protein components of MV and EC isolated from the virus particles indicated in panel B. Protein designations are indicated on the right.
We used silver staining and Western blot analysis to analyze the EC generated from Δ22K- and 22K−-infected cells in comparison to Ad5-WT MV as well as EC generated by floxing of the PS using Cre recombinase (31). These results revealed that EC generated from Δ22K or 22K− mutant viruses in A549, N52.E6-Cre, TetC4-IVa2, or TetC4-33K cells had protein composition similar to that of the EC of the Ad5-Ψ-loxP virus grown in N52.E6-Cre cells (Fig. 4B and C), although reduced levels of penton, fiber, and pVI were evident in EC isolated in some experiments from Δ22K-infected cells compared to 22K−-infected cells. The basis for these differences is not known but may reflect slight leakiness of the 22K− mutant virus. All of these EC lacked core protein V and the proteolytically processed form of VII, both characteristic of MV, and they all contained precursor forms of pVI and pVIII, as well as readily evident levels of L1-52/55K and L4-100K, which are present at very low abundance in MV. In addition, the Δ22K or 22K− EC had slightly elevated levels of IVa2 in comparison to MV or EC from Ad5-Ψ-loxP. Thus, the EC generated by the L4-22K mutant viruses were bona fide empty capsids. We were surprised to detect pVII in EC isolated from N52.E6-Cre cells, since core protein VII is associated with viral DNA. We do not know if this reflects true association of pVII within EC or minor contamination of EC with free pVII or nonpackaged viral DNA associated with pVII.
The L4-22K protein binds to PS in vivo only in the presence of IVa2.
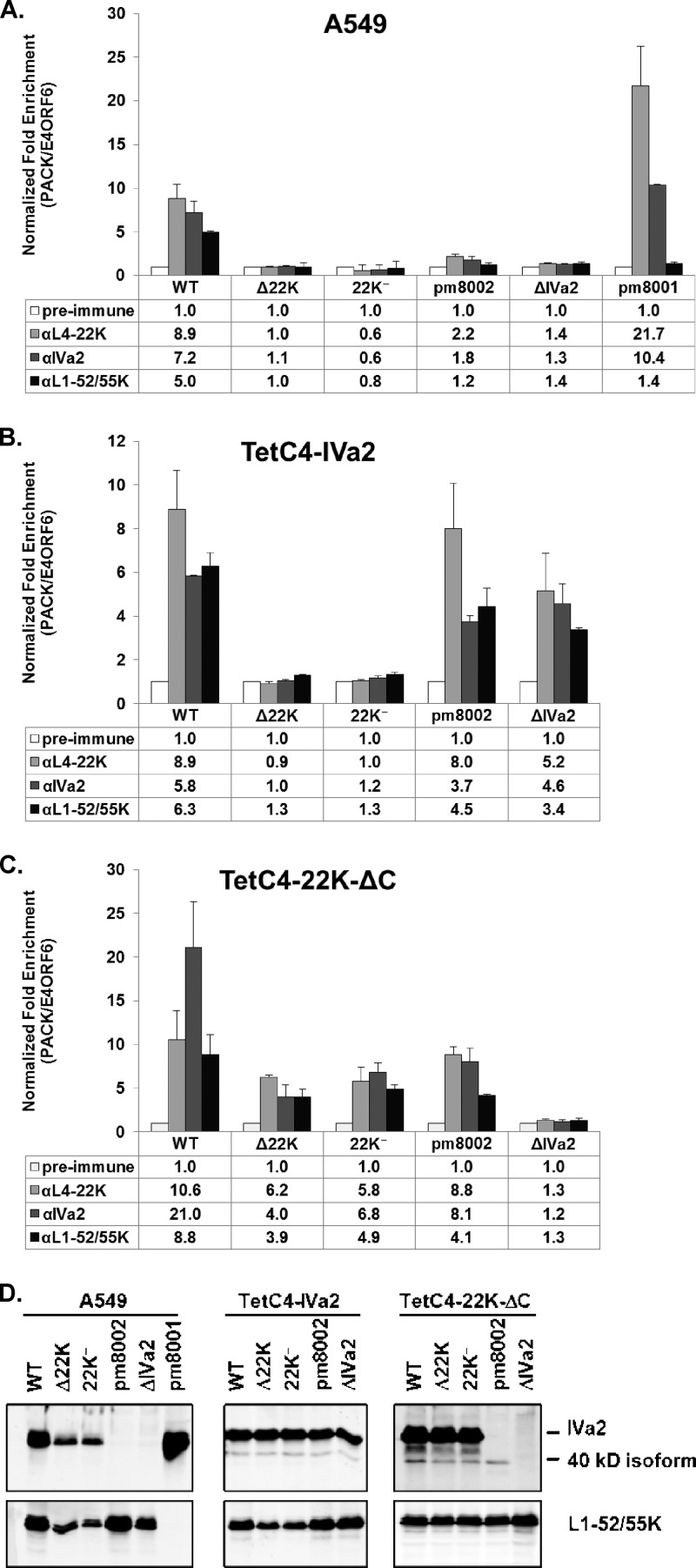
To explore packaging in vivo, we performed ChIP assays to decipher the relationship between the viral packaging proteins IVa2, L4-22K, and L1-52/55K and the PS in the genome. A549 cells infected with Ad5-WT, Δ22K, 22K−, pm8002 (a IVa2 mutant virus [48]), ΔIVa2 (24), or pm8001 (an L1-52/55K mutant virus [13]) were analyzed by ChIP at 18 hpi using rabbit preimmune serum or rabbit serum directed against L4-22K, IVa2, or L1-52/55K. These results showed that the L4-22K protein bound to the PS of Ad5-WT in vivo, but this was not observed in Δ22K- or 22K−-infected cells, as expected (Fig. 5A). In addition, L4-22K did not bind to the PS in pm8002- or ΔIVa2-infected cells, indicating that the IVa2 protein is required for L4-22K to bind to the PS in vivo, consistent with in vitro DNA-protein binding studies (9, 25, 44). Results from pm8001-infected cells demonstrated that in the absence of L1-52/55K, the interaction between L4-22K or IVa2 and the PS was not reduced. IVa2 bound to the PS in Ad5-WT- or pm8001-infected cells but not in pm8002- or ΔIVa2-infected cells, consistent with previous reports (28, 32). However, IVa2 did not bind to the PS in Δ22K- or 22K−-infected cells, implying that the L4-22K protein is essential for IVa2 to bind to the PS in vivo, in contrast to the results of previous in vitro binding analyses (28, 40, 45). In addition, in the absence of L4-22K (Δ22K, 22K−) or IVa2 (pm8002, ΔIVa2), the binding of L1-52/55K to the PS in vivo was completely abolished, indicating that both the L4-22K and IVa2 proteins are required for L1-52/55K to bind to the PS. The loss of IVa2 and L1-52/55K binding to the PS in the absence of the L4-22K protein may be due, at least in part, to decreased IVa2 and L1-52/55K expression in Δ22K- or 22K−-infected cells (Fig. 2 and 5D). Taken together, the ChIP experiments indicate that the IVa2 protein is essential for L4-22K and L1-52/55K to bind to the PS, the L4-22K protein is likely required for IVa2 and L1-52/55K to bind the PS, and the L1-52/55K protein is dispensable for IVa2 or L4-22K binding to the PS in vivo.
Fig 5.
ChIP assays for packaging protein binding to the PS in vivo. A549 (A), TetC4-IVa2 (B), and TetC4-22K-ΔC (C) cells were infected with Ad5-WT, Δ22K, 22K−, pm8002, ΔIVa2, or pm8001 viruses, and L4-22K-, IVa2-, and L1-52/55K-specific antibodies were used for ChIP analysis to quantify binding to the PS. The readout is presented as normalized fold enrichment as calculated by dividing the number of copies of PS pulled down specifically by the antibody-antigen complex by the number of copies of the E4-ORF6 fragment pulled down nonspecifically by the antibody. For each individual virus infection, the fold enrichment value of each antibody was normalized to that of the preimmune serum negative control. (D) Western blot analysis of ChIP input samples for determination of IVa2 and L1-52/55K protein expression levels.
The L4-22K protein is required for IVa2 to bind to the PS in vivo.
To examine the possibility that the loss of IVa2 binding to the PS in Δ22K- or 22K−-infected A549 cells was due to the decreased level of IVa2 expression (Fig. 2 and 5D), we infected TetC4-IVa2 cells with Ad5-WT, Δ22K, 22K−, pm8002, or ΔIVa2 for ChIP analyses; this provided similar levels of the IVa2 protein in the different virus infections (Fig. 5D). These results demonstrated that complementation of IVa2 protein levels in IVa2 mutant virus infections restored not only the binding of IVa2 but also the binding of the L4-22K and L1-52/55K proteins to the PS in vivo (Fig. 5B). However, the supplementation of IVa2 protein levels did not restore the binding of IVa2 or L1-52/55K to the PS in Δ22K- or 22K−-infected cells (Fig. 5B), even though IVa2 protein levels were the same among the different virus-infected cells (Fig. 5D). These results ruled out the possibility that the decreased IVa2 protein expression was responsible for the loss of IVa2 binding to the PS in Δ22K- or 22K−-infected cells and confirmed that the L4-22K protein is required for IVa2 and L1-52/55K to bind to the PS in vivo. We note that there were similar levels of L1-52/55K expression among TetC4-IVa2 cells infected with the different viruses (Fig. 5D).
To further confirm the relationship between L4-22K, IVa2, and L1-52/55K and binding to the PS in vivo, the L4-22K-expressing cell line, TetC4-22K-ΔC, was employed in ChIP analyses. Complementation of L4-22K fully restored the expression levels of IVa2 and L1-52/55K in cells infected with the L4-22K mutant viruses (Fig. 5D) and partially restored the binding of L4-22K-ΔC, IVa2, and L1-52/55K to the PS in Δ22K- or 22K−-infected cells (Fig. 5C). The different extents of binding of IVa2 and L1-52/55K to the PS between Ad5-WT and the two L4-22K mutant viruses may have been due to the fact that the WT virus had additional full-length WT L4-22K expressed in addition to the L4-22K-ΔC provided by the cell line. Surprisingly, in pm8002-infected TetC4-22K-ΔC cells, where there was no full-length IVa2 expression (Fig. 5D), the binding of the IVa2, L4-22K, and L1-52/55K proteins to the PS was partially restored; this was not observed with the ΔIVa2 mutant virus (Fig. 5C). Western blot analysis of the input samples showed that the 40-kDa IVa2 isoform (29) was expressed in pm8002-infected TetC4-22K-ΔC cells, even though the full-length IVa2 protein was not evident, as expected (Fig. 5D). Since it is known that the 40-kDa IVa2 isoform binds to the PS (29, 45), its presence may be responsible for the partially restored binding of the IVa2, L4-22K, and L1-52/55K proteins to the PS in pm8002-infected TetC4-22K-ΔC cells.
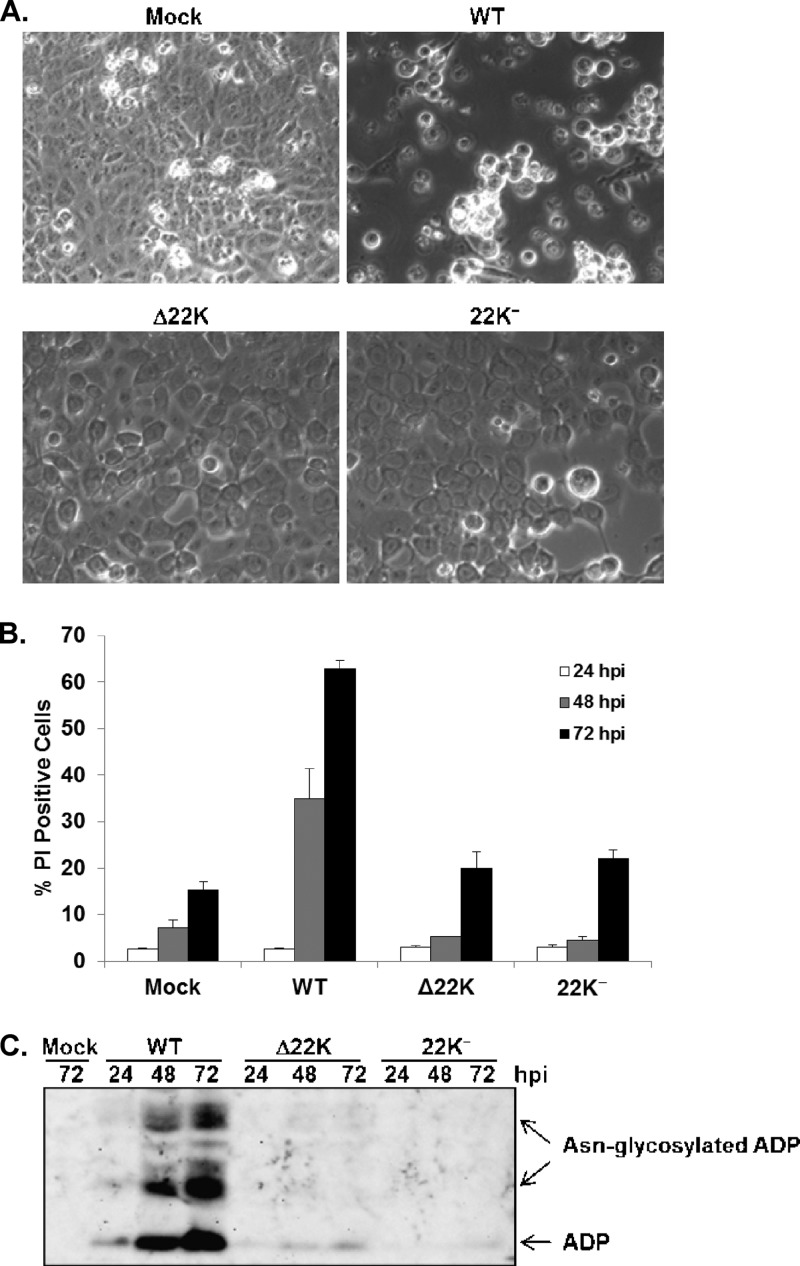
The L4-22K protein promotes Ad-induced cell lysis.
During the course of these studies, we noted that there was a remarkable difference in the morphologies of A549 cells infected with Ad5-WT versus the L4-22K mutant viruses (Fig. 6A). With Ad5-WT-infected cells, significant cytopathic effect was observed at 48 hpi, with most of the cells rounded up and detached from the plate. In contrast, Δ22K- or 22K−-infected cells had enlarged nuclei but were still confluent and attached to the plate (Fig. 6A). In order to determine if the L4-22K protein influenced cell viability, PI staining was used to monitor cell death in Ad5-WT-, Δ22K-, or 22K−-infected cells at different time points after infection. The results showed that Ad5-WT showed significant cell killing by 48 to 72 hpi whereas both L4-22K mutant viruses showed very modest cell killing compared with mock-infected cell results (Fig. 6B). This finding indicated that the L4-22K protein contributes to cell killing during Ad infection. ADP is known to be responsible for Ad-induced cell lysis (37). We examined ADP expression levels in Ad5-WT-, Δ22K- or 22K−-infected A549 cells. Western blot analysis showed that ADP expression levels were dramatically decreased in cells infected with the L4-22K mutant viruses compared to those infected with Ad5-WT (Fig. 6C). ADP is transcribed from the MLP during the late phase of infection (38); thus, the L4-22K protein likely regulates ADP synthesis in parallel with other Ad late genes. These data suggest that the L4-22K protein promotes Ad-induced cell lysis by regulating ADP expression.
Fig 6.
L4-22K promotes Ad-induced cell lysis by regulating ADP expression. (A) Light-microscopy images of mock-, Ad5-WT-, Δ22K-, or 22K−-infected A549 cells at 48 hpi. (B) Propidium iodide staining of mock-, Ad5-WT-, Δ22K-, or 22K−-infected A549 cells at 24, 48, and 72 hpi. (C) Western blot analysis of ADP expression levels in total cell extracts prepared from mock-, Ad5-WT-, Δ22K-, or 22K−-infected A549 cells at 24, 48, and 72 hpi. Free and Asn-glycosylated ADP forms (34) are indicated on the right.
DISCUSSION
We previously demonstrated that L4-22K is an essential viral protein and that it binds to the TTTG motif of the PS in vitro in the presence of IVa2, suggesting a role in viral genome packaging (25). Morris and Leppard have shown that the L4-22K protein is important for the temporal control of viral gene expression by regulating intermediate and late gene expression (23). We generated and characterized two L4-22K mutant viruses, Δ22K and 22K−, to further explore L4-22K functions during Ad infection. Our results showed that L4-22K is indeed important for the temporal control of viral gene expression not only because it activates late gene expression but also because it suppresses early gene expression. We also showed that the L4-22K protein binds to the PS in vivo and is essential to recruit two other packaging proteins, IVa2 and L1-52/55K, to this region. The elimination of L4-22K gave rise only to the production of EC and not MV, which confirms that the L4-22K protein is required for Ad genome packaging. Finally, L4-22K contributes to Ad-induced cell death by regulating expression of ADP. Thus, the Ad L4-22K protein is multifunctional and an integral component of crucial aspects of infection.
The L4-22K mutant viruses behaved similarly to Ad5-WT through the onset of infection to viral DNA replication (Fig. 1 and 2). Alterations in both early and late gene expression were observed with the L4-22K mutant viruses as the late phase progressed, consistent with the findings by Morris and Leppard (23). The mechanism by which the L4-22K protein influences late gene expression has been suggested to be at the level of late gene transcription and/or RNA processing (3, 23). The overall delay and reduction in expression of late gene products observed with both L4-22K mutant viruses may relate to its role in activating MLP transcription (3). However, the extents of reduction of expression of different late gene products were quite different, which supports a role for L4-22K in the posttranscriptional processing of certain viral mRNAs (23). Interestingly, both L4-22K mutant viruses showed significantly increased E1A expression and mRNA levels late after infection (Fig. 2 and 3) and moderately increased DBP expression at this time (Fig. 2). This may explain the modestly higher genome replication level that was observed with both L4-22K mutant viruses at late times after infection (Fig. 1D). These results also suggest that L4-22K suppresses early gene expression during the late stage of infection. A role for an Ad late trans-acting component in the downregulation of viral early gene expression was previously proposed (11).The increased E1A protein level that we observed correlated well with E1A mRNA levels, suggesting that L4-22K suppresses early gene expression. The Ad PS overlaps significantly with the E1A enhancer region (17), suggesting that the binding of packaging proteins to this region might suppress E1A transcription during the late phase of infection. However, this model is open to question, since the elimination of IVa2 expression with mutant pm8002, and consequently the binding of both IVa2 and L4-22K to the PS (Fig. 5A), did not enhance E1A expression at 24 hpi (48) or at later times of infection (data not shown) in A549 cells. But it is still plausible that, independently of its binding to the PS, L4-22K indirectly suppresses E1A expression by affecting the activity of an E1A transcription factor(s) or that L4-22K regulates E1A expression posttranscriptionally. Increased E1A transcript levels with the L4-22K mutant viruses also could reflect a failure to remove viral genomes from the transcription pool due to the packaging defect. The significance of suppressing E1A expression during the late phase of infection might be to redistribute the cellular transcriptional-translational machinery from early genes to late genes. Overall, our results and those published by others cited above indicate that the L4-22K protein plays an important role in regulating the temporal control of viral gene expression during the course of Ad infection.
The L4-22K mutant viruses produced only empty capsids, indicating that L4-22K is essential for Ad genome packaging (Fig. 4), but other aspects of mutant virus infection might contribute to this phenotype. First, the IVa2 and L4-33K proteins regulate Ad late gene expression at the transcriptional and posttranscriptional levels (2, 10, 30, 39) and the L4-22K mutant viruses exhibited decreased expression of both of these proteins, particularly with L4-33K (Fig. 2). Supplementation of IVa2 or L4-33K using complementing cell lines, however, did not rescue the ability of the L4-22K mutants to produce MV (Fig. 4). Second, the significantly reduced levels of V, pVII/pVII, and pVIII (Fig. 2) might also underlie the L4-22K mutant phenotype with respect to EC production. However, protein VII is not required for Ad genome packaging (P. Ostapchuk and P. Hearing, unpublished data) and a temperature-sensitive mutant in protein VIII did not display a packaging defect (20), but this possibility has not been fully resolved. Third, pVI promotes hexon import into the nucleus (41) and the reduction of pVI in L4-22K mutant virus infections (Fig. 2) may contribute to the defect in MV formation. However, the yield of EC from Δ22K-infected N52.E6-Cre cells (Fig. 4) indicates that pVI is not limiting in the absence of the L4-22K protein. While we cannot exclude the possibility of combinatorial effects of reduction of expression of different Ad late proteins, the binding of the L4-22K protein to the PS in vitro (9, 25, 44) and in vivo (Fig. 5) and the lack of MV production with both L4-22K mutant viruses (Fig. 4) lead us to conclude that the L4-22K protein is required for Ad genome packaging. This conclusion is strengthened by the analyses of viruses carrying mutations in the PS, where a precise correlation was observed between the binding of the L4-22K protein to PS in vitro and virus viability in vivo (28).
Our results demonstrate that the L4-22K and IVa2 proteins are dependent upon each other for binding to the PS in vivo and that both of these proteins are required to recruit the L1-52/55K protein to this region of the Ad genome (Fig. 5). These results are consistent with previous studies that showed that IVa2 is required for L4-22K to bind to the PS in vitro (9, 25, 44). However, in vitro binding studies also have shown that purified recombinant IVa2 can bind to the PS alone (28, 40, 45). A possible explanation for this discrepancy is that in virus-infected cells, the IVa2 protein needs L4-22K to remove other factors that might anchor on or close to the IVa2 binding site and impede IVa2 DNA binding. Perhaps more likely is the observation that the IVa2 and L4-22K proteins bind to the PS cooperatively (44); we anticipate that weak DNA-protein interactions can be identified using in vitro binding studies but that the infected cell nucleus in vivo provides a more rigorous environment where only the more stable DNA-protein interactions are maintained. It has been reported that L1-52/55K binds to the PS in pm8002-infected cells in the absence of IVa2 (32). However, we could not detect such binding in our experimental setting (Fig. 5). Different L1-52/55K antibodies and cells were used in these two studies. These differences also may reflect the use of semiquantitative PCR (32) versus quantitative PCR (Fig. 5) in these analyses. We observed partially restored binding of L4-22K, IVa2, and L1-52/55K with the PS in pm8002-infected TetC4-22K-ΔC cells (Fig. 5C). Western blot analysis confirmed the existence of a 40-kDa IVa2 isoform in pm8002-infected TetC4-22K-ΔC cells but not in pm8002-infected A549 cells at 18 hpi (Fig. 5D). In this context, the 40-kDa IVa2 isoform appears to have partially restored the binding of L4-22K, IVa2, and L1-52/55K to the PS in vivo (Fig. 5).
The L4-22K protein contributes significantly to expression of ADP (Fig. 6), which in turn contributes to the lysis of Ad-infected cells (37). Even though the ADP coding region is located in the E3 region, ADP is transcribed from the MLP and not expressed in great abundance until the late phase of infection (38). The link between Ad genome packaging and augmented ADP expression to induce cell lysis, and the involvement of the L4-22K protein in both of these processes, makes perfect sense with respect to the temporal coordination of the Ad infection cycle. A number of recent studies also have shown that Ad infection induces autophagy, which also contributes to the killing of infected cells (18, 19, 33). It should be interesting to determine if the autophagy process is altered by mutation of L4-22K and if ADP, L4-22K, or other viral genes regulated by L4-22K are responsible for induction of autophagy during Ad infection.
ACKNOWLEDGMENTS
We thank our colleagues, including Gudrin Schiedner and Stefan Kochanek, Michael Imperiale, Arnold Levine, Peter van der Vliet, Carl Anderson, Daniel Engel, David Matthews, Christopher Wiethoff, Ann Tollefson, and William Wold, for providing important reagents used in this study. We thank Philomena Ostapchuk for critical reading of the manuscript and laboratory members for informative discussions throughout the project. We thank Wei-Xing Zong, Ji-An Pan, and Zhixun Dou for informative discussions and technical help. We thank Ilana Shoshani for excellent technical assistance.
This work was supported by NIH grant AI041636.
Footnotes
Published ahead of print 18 July 2012
REFERENCES
- 1. Akusjarvi G. 2008. Temporal regulation of adenovirus major late alternative RNA splicing. Front. Biosci. 13:5006–5015 [DOI] [PubMed] [Google Scholar]
- 2. Ali H, LeRoy G, Bridge G, Flint SJ. 2007. The adenovirus L4 33-kilodalton protein binds to intragenic sequences of the major late promoter required for late phase-specific stimulation of transcription. J. Virol. 81:1327–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Backström E, Kaufmann KB, Lan X, Akusjarvi G. 2010. Adenovirus L4-22K stimulates major late transcription by a mechanism requiring the intragenic late-specific transcription factor-binding site. Virus Res. 151:220–228 [DOI] [PubMed] [Google Scholar]
- 4. Berk AJ. 2005. Recent lessons in gene expression, cell cycle control, and cell biology from adenovirus. Oncogene 24:7673–7685 [DOI] [PubMed] [Google Scholar]
- 5. Chartier C, et al. 1996. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J. Virol. 70:4805–4810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Christensen JB, et al. 2008. Presence of the adenovirus IVa2 protein at a single vertex of the mature virion. J. Virol. 82:9086–9093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. D'Halluin JC, Milleville M, Boulanger PA, Martin GR. 1978. Temperature-sensitive mutant of adenovirus type 2 blocked in virion assembly: accumulation of light intermediate particles. J. Virol. 26:344–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Echavarría M. 2008. Adenoviruses in immunocompromised hosts. Clin. Microbiol. Rev. 21:704–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ewing SG, Byrd SA, Christensen JB, Tyler RE, Imperiale MJ. 2007. Ternary complex formation on the adenovirus packaging sequence by the IVa2 and L4 22-kilodalton proteins. J. Virol. 81:12450–12457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Farley DC, Brown JL, Leppard KN. 2004. Activation of the early-late switch in adenovirus type 5 major late transcription unit expression by L4 gene products. J. Virol. 78:1782–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fessler SP, Young CS. 1998. Control of adenovirus early gene expression during the late phase of infection. J. Virol. 72:4049–4056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guo P, Lee TJ. 2007. Viral nanomotors for packaging of dsDNA and dsRNA. Mol. Microbiol. 64:886–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gustin KE, Imperiale MJ. 1998. Encapsidation of viral DNA requires the adenovirus L1 52/55-kilodalton protein. J. Virol. 72:7860–7870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gustin KE, Lutz P, Imperiale MJ. 1996. Interaction of the adenovirus L1 52/55-kilodalton protein with the IVa2 gene product during infection. J. Virol. 70:6463–6467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hasson TB, Ornelles DA, Shenk T. 1992. Adenovirus L1 52- and 55-kilodalton proteins are present within assembling virions and colocalize with nuclear structures distinct from replication centers. J. Virol. 66:6133–6142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hasson TB, Soloway PD, Ornelles DA, Doerfler W, Shenk T. 1989. Adenovirus L1 52- and 55-kilodalton proteins are required for assembly of virions. J. Virol. 63:3612–3621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hearing P, Shenk T. 1983. The adenovirus type 5 E1A transcriptional control region contains a duplicated enhancer element. Cell 33:695–703 [DOI] [PubMed] [Google Scholar]
- 18. Ito H, et al. 2006. Autophagic cell death of malignant glioma cells induced by a conditionally replicating adenovirus. J. Natl. Cancer Inst. 98:625–636 [DOI] [PubMed] [Google Scholar]
- 19. Jiang H, et al. 2011. Human adenovirus type 5 induces cell lysis through autophagy and autophagy-triggered caspase activity. J. Virol. 85:4720–4729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu GQ, Babiss LE, Volkert FC, Young CS, Ginsberg HS. 1985. A thermolabile mutant of adenovirus 5 resulting from a substitution mutation in the protein VIII gene. J. Virol. 53:920–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ma HC, Hearing P. 2011. Adenovirus structural protein IIIa is involved in the serotype specificity of viral DNA packaging. J. Virol. 85:7849–7855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mangel WF, Toledo DL, Ding J, Sweet RM, McGrath WJ. 1997. Temporal and spatial control of the adenovirus proteinase by both a peptide and the viral DNA. Trends Biochem. Sci. 22:393–398 [DOI] [PubMed] [Google Scholar]
- 23. Morris SJ, Leppard KN. 2009. Adenovirus serotype 5 L4-22K and L4-33K proteins have distinct functions in regulating late gene expression. J. Virol. 83:3049–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ostapchuk P, Almond M, Hearing P. 2011. Characterization of empty adenovirus particles assembled in the absence of a functional adenovirus IVa2 protein. J. Virol. 85:5524–5531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ostapchuk P, Anderson ME, Chandrasekhar S, Hearing P. 2006. The L4 22-kilodalton protein plays a role in packaging of the adenovirus genome. J. Virol. 80:6973–6981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ostapchuk P, Hearing P. 2008. Adenovirus IVa2 protein binds ATP. J. Virol. 82:10290–10294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ostapchuk P, Hearing P. 2005. Control of adenovirus packaging. J. Cell. Biochem. 96:25–35 [DOI] [PubMed] [Google Scholar]
- 28. Ostapchuk P, Yang J, Auffarth E, Hearing P. 2005. Functional interaction of the adenovirus IVa2 protein with adenovirus type 5 packaging sequences. J. Virol. 79:2831–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pardo-Mateos A, Young CS. 2004. A 40 kDa isoform of the type 5 adenovirus IVa2 protein is sufficient for virus viability. Virology 324:151–164 [DOI] [PubMed] [Google Scholar]
- 30. Pardo-Mateos A, Young CS. 2004. Adenovirus IVa2 protein plays an important role in transcription from the major late promoter in vivo. Virology 327:50–59 [DOI] [PubMed] [Google Scholar]
- 31. Parks RJ, et al. 1996. A helper-dependent adenovirus vector system: removal of helper virus by Cre-mediated excision of the viral packaging signal. Proc. Natl. Acad. Sci. U. S. A. 93:13565–13570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Perez-Romero P, Tyler RE, Abend JR, Dus M, Imperiale MJ. 2005. Analysis of the interaction of the adenovirus L1 52/55-kilodalton and IVa2 proteins with the packaging sequence in vivo and in vitro. J. Virol. 79:2366–2374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rodriguez-Rocha H, et al. 2011. Adenoviruses induce autophagy to promote virus replication and oncolysis. Virology 416:9–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Scaria A, Tollefson AE, Saha SK, Wold WS. 1992. The E3-11.6K protein of adenovirus is an Asn-glycosylated integral membrane protein that localizes to the nuclear membrane. Virology 191:743–753 [DOI] [PubMed] [Google Scholar]
- 35. Schiedner G, Hertel S, Kochanek S. 2000. Efficient transformation of primary human amniocytes by E1 functions of Ad5: generation of new cell lines for adenoviral vector production. Hum. Gene Ther. 11:2105–2116 [DOI] [PubMed] [Google Scholar]
- 36. Smith JG, Wiethoff CM, Stewart PL, Nemerow GR. 2010. Adenovirus. Curr. Top. Microbiol. Immunol. 343:195–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tollefson AE, et al. 1996. The adenovirus death protein (E3-11.6K) is required at very late stages of infection for efficient cell lysis and release of adenovirus from infected cells. J. Virol. 70:2296–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tollefson AE, Scaria A, Saha SK, Wold WS. 1992. The 11,600-MW protein encoded by region E3 of adenovirus is expressed early but is greatly amplified at late stages of infection. J. Virol. 66:3633–3642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Törmänen H, Backstrom E, Carlsson A, Akusjarvi G. 2006. L4-33K, an adenovirus-encoded alternative RNA splicing factor. J. Biol. Chem. 281:36510–36517 [DOI] [PubMed] [Google Scholar]
- 40. Tyler RE, Ewing SG, Imperiale MJ. 2007. Formation of a multiple protein complex on the adenovirus packaging sequence by the IVa2 protein. J. Virol. 81:3447–3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wodrich H, et al. 2003. Switch from capsid protein import to adenovirus assembly by cleavage of nuclear transport signals. EMBO J. 22:6245–6255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wohl BP, Hearing P. 2008. Role for the L1-52/55K protein in the serotype specificity of adenovirus DNA packaging. J. Virol. 82:5089–5092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yang J, Hearing P. 2007. Chromatin immunoprecipitation to study the binding of proteins to the adenovirus genome in vivo. Methods Mol. Med. 131:113–121 [DOI] [PubMed] [Google Scholar]
- 44. Yang TC, Maluf NK. 2012. Cooperative heteroassembly of the adenoviral L4-22K and IVa2 proteins onto the viral packaging sequence DNA. Biochemistry 51:1357–1368 [DOI] [PubMed] [Google Scholar]
- 45. Yang TC, Yang Q, Maluf NK. 2009. Interaction of the adenoviral IVa2 protein with a truncated viral DNA packaging sequence. Biophys. Chem. 140:78–90 [DOI] [PubMed] [Google Scholar]
- 46. Young CS. 2003. The structure and function of the adenovirus major late promoter. Curr. Top. Microbiol. Immunol. 272:213–249 [DOI] [PubMed] [Google Scholar]
- 47. Zhang W, Imperiale MJ. 2000. Interaction of the adenovirus IVa2 protein with viral packaging sequences. J. Virol. 74:2687–2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang W, Imperiale MJ. 2003. Requirement of the adenovirus IVa2 protein for virus assembly. J. Virol. 77:3586–3594 [DOI] [PMC free article] [PubMed] [Google Scholar]