Abstract
LGP2, a member of the RIG-I-like receptor family, lacks the amino-terminal caspase activation recruitment domains (CARDs) required for initiating the activation of interferon regulatory factor 3 (IRF3) and interferon (IFN) transcription. The role of LGP2 in virus infection is controversial, and the only LGP2 experiments previously carried out with mammalian influenza A viruses employed an attenuated, mouse-adapted H1N1 A/PR/8/34 (PR8) virus that does not encode the NS1 protein. Here we determine whether LGP2 has a role during infection with wild-type, nonattenuated influenza A viruses that have circulated in the human population, specifically two types of seasonal influenza A viruses: (i) H3N2 and H1N1 viruses that activate IRF3 and IFN transcription and (ii) recent H1N1 viruses that block these two activations. In human cells infected with an H3N2 virus that activates IRF3, overexpression of LGP2 or its repressor domain decreased STAT1 activation and IFN-β transcription approximately 10-fold. Overexpression of LGP2 also caused a 10-fold decrease of STAT1 activation during infection with other seasonal influenza A viruses that activate IRF3. Using LGP2+/+ and LGP2−/− mouse cells, we show that endogenous LGP2 decreased IFN production during H3N2 virus infection 3- to 4-fold. In contrast, in both mouse and human cells infected with H1N1 viruses that do not activate IRF3, LGP2 had no detectable role. These results demonstrate that LGP2 downregulates IFN production during infection by seasonal influenza A viruses that activate IRF3 and IFN transcription. It is intriguing that LGP2, a host protein induced during influenza A virus infection, downregulates the host antiviral IFN response.
INTRODUCTION
Innate immunity against many viruses is initiated by the binding of viral RNA species to the repressor and helicase domains of RIG-I, a cytosolic protein that also contains two amino-terminal caspase activation recruitment domains (CARDs) (23). The RIG-I CARDs then associate with the CARDs of the mitochondrion-associated adaptor MAVS protein, thereby triggering the signaling pathway that leads to the activation of the interferon regulatory factor 3 (IRF3) and NF-κB transcription factors and the activation of interferon (IFN) transcription (6, 11, 16, 21). LGP2 has repressor and helicase domains similar to those of RIG-I but lacks CARDs and, hence, cannot trigger the signaling pathway that leads to the activation of IFN transcription (13, 22). The role of LGP2 in virus infection is controversial: it has been reported to either positively or negatively affect the RIG-I-mediated activation of IFN transcription and the production of IFN in virus-infected cells (14, 15, 20).
Influenza A virus, which utilizes the RIG-I-initiated pathway (4, 10), was reported to be unique in that the production of IFN in infected mammalian cells was not affected by LGP2, unlike with other viruses (15). These experiments used an attenuated, mouse-adapted H1N1 A/PR/8/34 (PR8) virus that does not encode the NS1 protein. A major role of the multifunctional NS1 protein is to counter the host antiviral response, including the synthesis of IFN (2). Consequently, infection with this attenuated PR8 virus results in robust activation of IFN synthesis (1), unlike with the wild-type (wt) PR8 virus. It was reported that the same amounts of IFN were produced in wt and LGP2−/− mouse embryo fibroblasts (MEFs) infected with this attenuated PR8 virus, indicating that LGP2 has no role during infection with this attenuated virus (15).
Our goal was to determine whether LGP2 has a role during infection with wt, nonattenuated influenza A viruses that have circulated in the human population. Currently circulating H1N1 and H3N2 viruses differ in the ability of their NS1 proteins to inhibit the activation of IRF3 and IFN transcription (9). The NS1 proteins of H1N1 viruses that have circulated since 1991, including the 2009 H1N1 pandemic virus, block the activation of IRF3 and IFN transcription, whereas the NS1 proteins of all tested H3N2 viruses do not inhibit these activations. Interestingly, the NS1 proteins of most H1N1 viruses in two earlier time periods, 1940 to 1957 and 1977 to 1990, function like the NS1 proteins of H3N2 viruses, i.e., they do not inhibit IRF3 activation. We determined the role of LGP2 in two types of seasonal influenza A viruses, those that activate IRF3 and those that block IRF3 activation. The NS1 proteins of the H1N1 and H3N2 viruses that we analyzed share the property of binding the 30-kDa subunit of the cellular cleavage and polyadenylation specificity factor (CPSF30), a protein that is required for 3′-end processing of cellular pre-mRNAs, and hence, these NS1 proteins inhibit the 3′-end processing of IFN-β pre-mRNA (7, 12, 19).
Here we show that LGP2 has strikingly different roles during infection of MEFs and human cells with these two types of seasonal influenza A viruses. With influenza A viruses that activate IRF3 and IFN transcription, LGP2, which is induced during infection, downregulates the synthesis of IFN. Hence, counterintuitively, the induced LGP2 host protein downregulates the host antiviral IFN response against many influenza A viruses. In contrast, with seasonal influenza A viruses that block IRF3 activation, LGP2 has no detectable role.
MATERIALS AND METHODS
Cells and viruses.
MDCK, A549, and HeLa cells were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS). LGP2 knockout (LGP2−/−) mice were generated on a pure C57BL/6 background (17) and did not exhibit the defects noted in other LGP2−/− mouse lines (15). LGP2−/− and LGP2 wt (LGP2+/+) mouse embryo fibroblasts (MEFs) were grown in DMEM with 10% heat-inactivated FBS. Influenza A viruses were amplified in 10-day-old fertilized eggs, and virus titers were determined by plaque assays in MDCK cells. Vesicular stomatitis virus (VSV) Indiana strain was amplified in Vero cells, which were also used for plaque assays. Cells (A549, HeLa, and MEFs) were infected with 5 PFU/cell of the indicated virus. After 1 h of adsorption at 37°C, cells were washed once with phosphate-buffered saline (PBS), replenished with DMEM containing 2% FBS, and incubated at 37°C for the indicated times.
RNA assays.
RNA was isolated from cells at the indicated times after infection using the TRIzol reagent. Quantitative real-time-PCR (RT-PCR) to measure the levels of human IFN-β pre-mRNA and IFN-β mRNA produced in infected HeLa cells was carried out as previously described (9). The reactions were carried out in triplicate and were normalized to the levels of β-actin mRNA. Similar procedures were used to measure the levels of LGP2 mRNAs in infected A549 cells and the levels of mouse IFN-β mRNA in infected MEFs.
Immunoblots.
For the assays for activated (phosphorylated) IRF3 and STAT1, cells were suspended in the PhosphoSafe extraction reagent (Novagen) at the indicated times after infection. After vortexing, the mixture was maintained at 4°C for 10 min and then centrifuged at 10,000 × g for 5 min. An aliquot of the supernatant was subjected to SDS-polyacrylamide gel electrophoresis, followed by immunoblots using an antibody against (serine 396-phosphorylated) IRF3 or (tyrosine 701-phosphorylated) STAT1 (Cell Signaling). For the assays of other proteins, cells collected at the indicated times after infection were lysed in RIPA buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) supplemented with complete protease inhibitor (Roche). Immunoblots were probed using antibodies against the viral NS1 protein, total STAT1 (Cell Signaling), total IRF3 (Cell Signaling), LGP2 (IBL-America), or β-tubulin (Cell Signaling). Quantitation of immunoblots was carried out by scanning the films and analyzing the scans using ImageJ software (NIH).
Transfection/infection assays.
The pOTB7 plasmid containing the cDNA for LGP2 was purchased from Thermo Scientific. The LGP2 cDNA was subcloned into a pcDNA3 vector that had been modified by inserting a V5 coding region adjacent to the cytomegalovirus (CMV) promoter, resulting in a cDNA encoding N-terminal V5-tagged LGP2. The cDNA sequence encoding the C-terminal repressor domain (RD) of LGP2 (amino acids 537 to 678) was also subcloned into the modified pcDNA3 vector, resulting in a cDNA encoding the N-terminal V5-tagged LGP2 RD. HeLa cells were transfected with either of these pcDNA3 vectors for 24 h using the Mirus TransIT-1 transfection reagent, followed by infection with the indicated virus or mock infection for 6 or 9 h. The amounts of full-length LGP2 or the LGP2 RD were determined using immunoblots probed with anti-V5 antibody.
RESULTS
Role of LGP2 in influenza A virus-infected human cells.
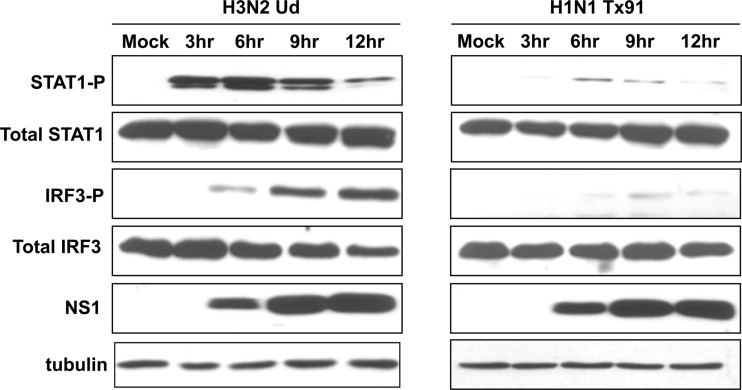
For most experiments, we used the following two influenza A viruses: H3N2 influenza A/Udorn/72 (Ud) virus, which activates IRF3 and IFN transcription, and H1N1 influenza A/Texas/91 (Tx91) virus, which inhibits these activations (9). Here we measured the production of IFN by assaying for activation (phosphorylation) of STAT1, the transcription factor activated by IFN (3, 5). STAT1 was strongly activated in H3N2 Ud virus-infected HeLa cells at 3 and 6 h after infection (Fig. 1, left panels). In fact, detectable STAT1 activation preceded the detection of IRF3 activation, presumably because the STAT1-P antibody works better than the IRF3-P antibody in immunoblots. The synthesis of the NS1 protein was detected starting at 6 h after infection, and the amount of the NS1 protein substantially increased at later times (9 and 12 h). Concomitant with the increase in the amount of the NS1 protein, the level of STAT1 activation decreased. This decrease is attributable to the increased levels of the NS1 protein, which would effectively inhibit the 3′-end processing of IFN pre-mRNA (12). In contrast, in H1N1 Tx91 virus-infected cells, little or no activation of either STAT1 or IRF3 was detected (Fig. 1, right panels), even though the time course of NS1 protein synthesis was similar to that in Ud virus-infected cells.
Fig 1.
H3N2 Ud and H1N1 Tx91 viruses have different effects on innate immune signaling. Levels of phosphorylated STAT1 and phosphorylated IRF3 at the indicated times after infection of HeLa cells with either H3N2 Ud (left panel) or H1N1 Tx91 (right panel) virus, as assayed by immunoblots.
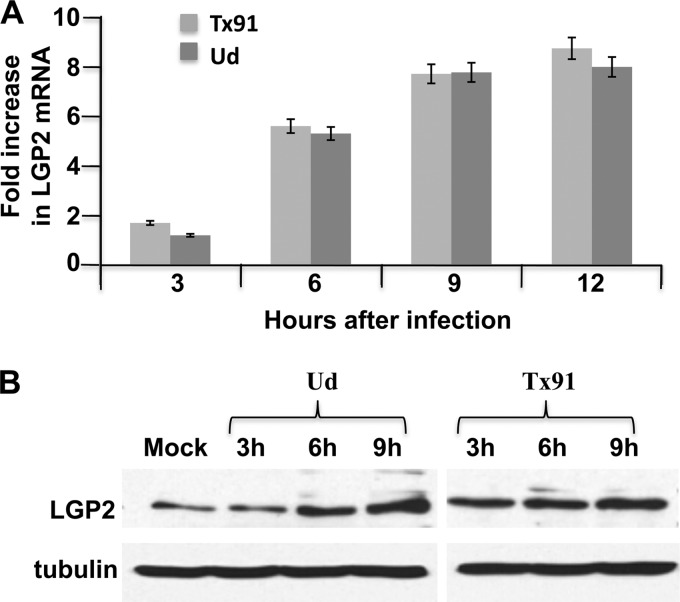
Unexpectedly, increasing amounts of LGP2 mRNA, as measured by quantitative RT-PCR, were produced in A549 cells infected with either of these influenza A viruses (Fig. 2A). By 12 h after infection with either virus, the amount of LGP2 mRNA increased approximately 8-fold. Because little or no activation of IRF3 and IFN transcription occurs during Tx91 infection, these results show that these activations are not required for the synthesis of LGP2 mRNA in influenza A virus-infected cells. Because these LGP2 mRNAs are derived from LGP2 pre-mRNAs that have escaped the inhibition of their 3′-end processing resulting from the binding of the NS1 protein to CPSF30 (12), it was likely that transcription of the LGP2 gene increased substantially more than 8-fold. This was, in fact, the case, as shown by carrying out an infection with a Ud virus that expresses an NS1 protein with a mutated CPSF30 binding site (G replaced by R at position 184). At 12 h after infection, the increase in LGP2 mRNA was approximately 30-fold. The increase in mature LGP2 mRNA resulted in an increase in LGP2 protein of approximately 5-fold by 9 h after infection by the Ud and Tx91 viruses (Fig. 2B).
Fig 2.
LGP2 is induced after infection with the H3N2 Ud and H1N1 Tx91 viruses. (A) Increase in LGP2 mRNA at the indicated times of infection of A549 cells with either H3N2 Ud or H1N1 Tx91 virus. The quantitative RT-PCRs were carried out in triplicate. Error bars represent the standard deviations from the mean values of the three independent assays. (B) LGP2 protein at the indicated times of infection of A549 cells with either H3N2 Ud or H1N1 Tx91 virus, as measured by an immunoblot probed with anti-LGP2 antibody.
To determine the role of this increased amount of LGP2 protein in infected cells, we attempted to deplete the LGP2 protein in infected cells using small interfering RNAs (siRNAs). We identified two siRNAs that effectively inhibited the synthesis of LGP2 mRNA and protein in experiments in which cells were transfected with a plasmid expressing LGP2 mRNA for 24 h, followed by transfection of the siRNAs for an additional 24 h (data not shown). However, when these siRNAs were transfected into cells 24 h prior to infection, little or no inhibition of the amount of LGP2 protein produced in Ud-infected cells was found (data not shown). A plausible explanation for this lack of inhibition is that most of the LGP2 mRNA and protein was synthesized during virus infection, at the tail end of siRNA treatment, so that there was not sufficient time for the siRNA to deplete the newly synthesized LGP2 mRNA and protein.
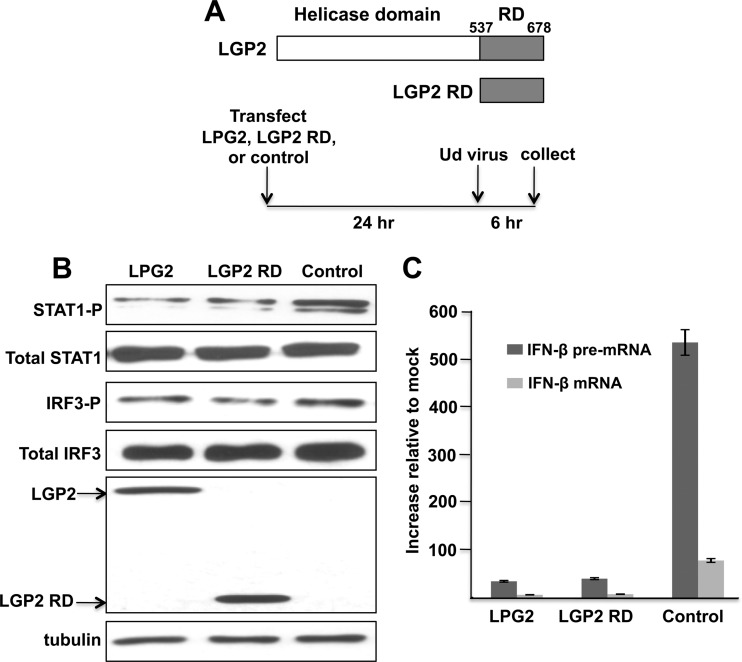
Our alternative approach was to overexpress LGP2. HeLa cells were transfected for 24 h with a plasmid expressing either full-length LGP2 or the LGP2 repressor domain (RD) or with an empty plasmid as a control, followed by infection with the H3N2 Ud virus for 6 h (Fig. 3A). Overexpression of LGP2 or its RD domain suppressed the antiviral response induced by the H3N2 Ud virus. STAT1 activation was reduced approximately 10-fold, as determined by an immunoblot (Fig. 3B), reflecting a similar 10-fold reduction in the production of both IFN-β pre-mRNA and IFN-β mRNA, as measured by quantitative RT-PCR (Fig. 3C). The latter results also document the strong inhibition of the 3′-end processing of IFN-β pre-mRNA by the NS1 protein at 6 h after infection: the amount of IFN-β mRNA was only 10 to 15% of the amount of IFN-β pre-mRNA. The reduction in IRF3 activation, as determined by an immunoblot, was smaller, at approximately 2- to 4-fold (Fig. 3B). These results demonstrate that LGP2 overexpression downregulates the antiviral response and the production of IFN in cells infected by the H3N2 Ud virus. In contrast, LGP2 overexpression had no detectable effect on the antiviral response induced by the H1N1 Tx91 virus (data not shown) because the antiviral response, e.g., STAT1 and IRF3 activation, was already so minimal in the absence of LGP2 overexpression (Fig. 1, right panels).
Fig 3.
LGP2 inhibits STAT1 activation in human cells infected with the H3N2 Ud virus. (A) HeLa cells were transfected with plasmids expressing LGP2 or the LGP2 RD or with an empty plasmid for 24 h, followed by infection with the H3N2 Ud virus. (B) Levels of phosphorylated STAT1, phosphorylated IRF3, and V5-tagged LGP2 and LGP2 RD were assayed by immunoblots. (C) Increase in IFN-β pre-mRNA and IFN-β mRNA after 6 h of infection of HeLa cells with the H3N2 Ud virus, as measured by quantitative RT-PCR carried out in triplicate.
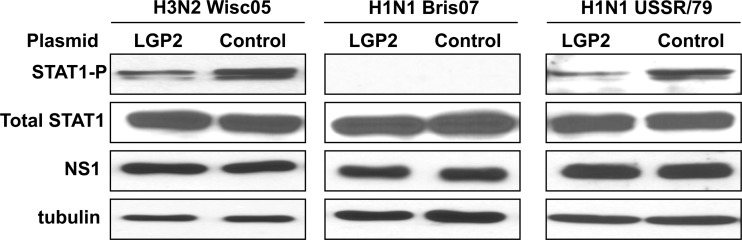
In contrast to the H1N1 Tx91 virus, the H1N1 influenza A/USSR/46/79 (USSR/79) virus activates IRF3 and IFN transcription (9). As shown in Fig. 4, LGP2 overexpression reduced the STAT1 activation induced by the H1N1 USSR/79 virus approximately 10-fold, similar to the reduction seen with both the H3N2 influenza A/Wisconsin/67/05 (Wisc05) virus and the H3N2 Ud virus. In contrast, because the H1N1 influenza A/Brisbane/59/07 (Bris07) virus does not induce detectable activation of STAT1, no effect of LGP2 overexpression on STAT1 activation was detected. Consequently, LGP2 overexpression downregulates IFN production in human cells infected by seasonal H3N2 and H1N1 human influenza A viruses that activate IRF3.
Fig 4.
LGP2 inhibits STAT1 activation in human cells infected with the H3N2 Wisc05 and H1N1 USSR/79 viruses and has no detectable role in human cells infected with the H1N1 Bris07 virus. HeLa cells were transfected with LGP2 or a control plasmid for 24 h, followed by infection with H3N2 Wisc05 (left panel), H1N1 Bris07 (middle panel), or H1N1 USSR/79 (right panel) virus. The levels of phosphorylated STAT1 and of the NS1 protein at 9 h after infection were assayed by immunoblots.
The absence of LGP2 in mouse cells increases IFN production induced by an H3N2 virus that activates IRF3.
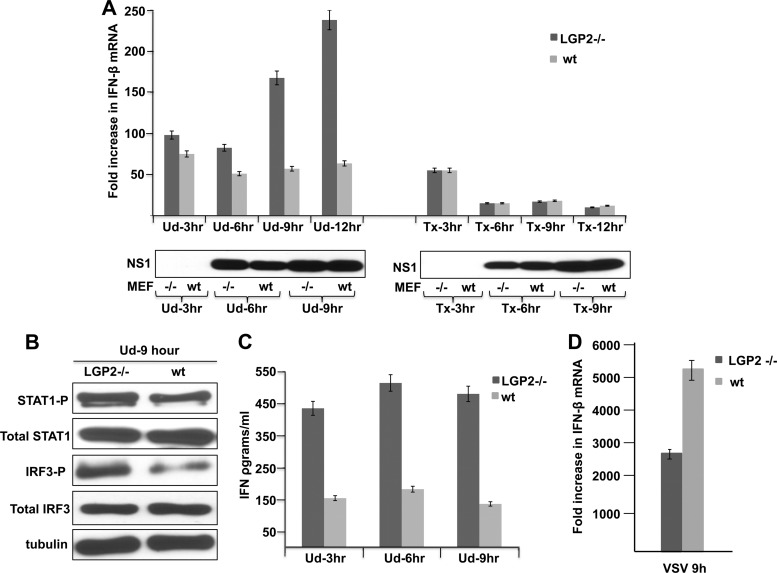
Because siRNA knockdown did not deplete LGP2 from influenza A virus-infected human cells, we evaluated the effect of LGP2 depletion by using LGP2−/− mouse embryo cells (MEFs). These MEFs were from LGP2−/− mice that were generated on a pure C57BL/6 background (17). We infected wild-type (LGP2+/+) and LGP2−/− MEFs with either H3N2 Ud or H1N1 Tx91 virus and measured the amount of IFN-β mRNA produced at various times after infection using quantitative RT-PCR (Fig. 5A). The amount of IFN-β mRNA produced in H3N2 Ud virus-infected LGP2−/− MEFs was greater than that produced in wt MEFs, and the magnitude of this difference increased with time postinfection, from 20% higher at 3 h to 4- to 5-fold higher at 12 h after infection. The IFN-β mRNAs produced in Ud virus-infected MEFs are derived from IFN-β pre-mRNAs that have escaped the inhibition of their 3′-end processing resulting from the binding of the NS1 protein to CPSF30 (9). Because little or no NS1 protein was detected at 3 h postinfection (Fig. 5A), minimal inhibition of the 3′-end processing of IFN-β pre-mRNA is expected to occur, accounting for the production of IFN-β mRNA in both wt and LGP2−/− MEFs at 3 h postinfection. At later times after infection, the amount of IFN-β mRNA did not increase in wt infected MEFs but did increase in infected LGP2−/− MEFs, resulting in the 4- to 5-fold higher level of IFN-β mRNA in infected LGP2−/− MEFs at 12 h after infection. This increase is attributable to increased transcription of the IFN-β gene, due at least in part to the increased activation of IRF3 that was observed (Fig. 5B). The increase in IFN-β mRNA resulted in a 3- to 4-fold increase in secreted IFN-β detected by an enzyme-linked immunosorbent assay (ELISA) (Fig. 5C) and an increase in the activation of the STAT1 transcription factor that is induced by IFN-β (Fig. 5B). These results demonstrate that LGP2 substantially downregulates the IRF3 activation pathway and, hence, the production of IFN-β mRNA in MEFs infected with the H3N2 Ud virus.
Fig 5.
LGP2 downregulates IFN production in MEFs infected with the H3N2 Ud virus but has no effect on IFN production in MEFs infected with the H1N1 Tx91 virus. (A) Increase in IFN-β mRNA at the indicated times after infection of wt and LGP2−/− MEFs with either the H3N2 Ud virus or the H1N1 Tx91 virus. (B) Levels of activated (phosphorylated) STAT1 and IRF3 at 9 h after infection of wt and LGP2−/− MEFs with Ud virus, as assayed by immunoblots. (C) Amount of IFN produced during infection of wt and LGP2−/− MEFs with Ud virus. (D) LGP2 upregulates IFN-β mRNA in MEFs infected with the VSV Indiana strain. IFN-β mRNA levels were measured by quantitative RT-PCR carried out in triplicate.
In contrast, the same small amount of IFN-β mRNA was produced in H1N1 Tx91 virus-infected wt and LGP2−/− MEFs (Fig. 5A). The production of some IFN-β mRNA in both wt and LGP2−/− MEFs at 3 h postinfection presumably occurred because little or no NS1 protein was produced by 3 h after infection. No increase in IFN-β mRNA was detected at later times in either wt or LGP2−/− MEFs (Fig. 5A), and little or no IFN-β protein was detected by an ELISA (data not shown). Consequently, we did not detect any role for LGP2 in MEFs infected with the H1N1 Tx91 virus.
Using MEFs, dendritic cells, or macrophages from these LGP2−/− mice, it has been shown that LPG2 upregulated rather than downregulated IFN production after infection with three other viruses, Sendai virus, dengue virus type 2, and West Nile virus (17). Here we tested another virus, vesicular stomatitis virus (VSV), in these LGP2−/− MEFs (Fig. 5D). The amount of IFN-β mRNA in LGP2−/− MEFs was lower than that in LGP2+/+ MEFs during VSV infection, showing that LGP2 upregulates IFN production during infection by this virus.
DISCUSSION
Here we establish that LGP2 downregulates the production of IFN in cells infected by wild-type seasonal influenza A viruses whose NS1 proteins do not inhibit the activation of IRF3 and IFN-β transcription. The strongest evidence for this conclusion was provided by our experiments using LGP2−/− MEFs. Infection of these LGP2-negative MEFs with a seasonal H3N2 virus that activates IRF3 resulted in a 3- to 5-fold increase in IFN production relative to the production in wt MEFs expressing LGP2 (LGP2+/+ MEFs). In contrast, infection of these LGP2−/− MEFs with VSV (present study) or with other viruses (17) resulted in a decrease in IFN production relative to that in infection of the wt LGP2+/+ MEFs. In addition, we did not detect any role for LGP2 in IFN production in cells infected with a seasonal influenza A virus that does not activate IRF3 and IFN transcription. Thus, downregulation of IFN production by LGP2 has so far been observed only during infection by seasonal influenza A viruses that activate IRF3 and IFN transcription.
Our experiments using human cells employed LGP2 overexpression because siRNA knockdowns failed to deplete the LGP2 protein that was synthesized during influenza A virus infection. We showed that in human cells infected with seasonal influenza A viruses that activate IRF3 and IFN transcription, overexpression of LGP2 or its RD decreased STAT1 activation and IFN-β transcription approximately 10-fold. Consequently, these results, like the MEF results, show that LGP2 negatively affects the RIG-I-mediated activation of IFN transcription in cells infected with these seasonal influenza A viruses. We conclude that one or more of the mechanisms that have been reported for the LGP2-mediated inhibition of RIG-I signaling operate in cells infected by H3N2 and H1N1 influenza A viruses that activate IRF3 (8, 14, 22).
Currently circulating H1N1 viruses effectively inhibit the RIG-I-mediated activation of IRF3 and the production of IFN via their NS1 proteins (2, 9, 18). It is therefore not surprising that further downregulation of this RIG-I pathway by LGP2 cannot be detected in human or mouse cells.
Like the attenuated H1N1 PR8 virus that does not encode an NS1 protein (1), IFN transcription and the production of IFN are induced by seasonal H3N2 and some seasonal H1N1 viruses (9). However, the effects of LGP2 on IFN production induced by the seasonal viruses and the attenuated PR8 virus are different. As shown here, LGP2 downregulates IFN production induced by these seasonal viruses, whereas LGP2 has no effect on the production of IFN induced by the attenuated PR8 virus (15). These results indicate that the signaling pathway for the activation of IFN transcription that is induced by wt seasonal viruses differs from the pathway that is induced by the attenuated PR8 virus that does not encode an NS1 protein.
In cells infected by the seasonal influenza A viruses that activate IRF3 and IFN transcription, the production of mature IFN mRNA is inhibited because the viral NS1 protein binds CPSF30, thereby inhibiting the 3′-end processing of IFN pre-mRNA (7, 12, 19). However, as further documented here, significant levels of IFN mRNA and IFN are produced at early times after infection (3 to 6 h postinfection), prior to the time at which the NS1 protein is synthesized in sufficient amounts to efficiently block the 3′-end processing of IFN pre-mRNA. During these early times, the production of IFN is downregulated only by LGP2 (Fig. 5A). At later times, IFN production is downregulated by both LGP2 and the NS1 protein-mediated inhibition of IFN pre-mRNA processing. It is intriguing that LGP2, a host protein induced during influenza A virus infection, actually downregulates the host antiviral IFN response. Further, it will be important to determine the mechanism by which LGP2 transcription is activated during influenza A virus infection, particularly as it apparently does not require activated IRF3 or IFN.
LGP2 has recently been shown to have another role during West Nile virus infection of mice, specifically regulation of CD8+ T cell survival and fitness (17). It will be of great interest to determine whether LGP2 has a similar role during influenza A virus infection.
ACKNOWLEDGMENT
This investigation was supported by NIH grant AI-011772 to R.M.K.
Footnotes
Published ahead of print 25 July 2012
REFERENCES
- 1. Garcia-Sastre A, et al. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324–330 [DOI] [PubMed] [Google Scholar]
- 2. Hale BG, Randall RE, Ortin J, Jackson D. 2008. The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol. 89:2359–2376 [DOI] [PubMed] [Google Scholar]
- 3. Haller O, Kochs G, Weber F. 2006. The interferon response circuit: induction and suppression by pathogenic viruses. Virology 344:119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kato H, et al. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101–105 [DOI] [PubMed] [Google Scholar]
- 5. Katze MG, He Y, Gale M., Jr 2002. Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2:675–687 [DOI] [PubMed] [Google Scholar]
- 6. Kawai T, et al. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981–988 [DOI] [PubMed] [Google Scholar]
- 7. Kochs G, Garcia-Sastre A, Martinez-Sobrido L. 2007. Multiple anti-interferon actions of the influenza A virus NS1 protein. J. Virol. 81:7011–7021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Komuro A, Horvath CM. 2006. RNA- and virus-independent inhibition of antiviral signaling by RNA helicase LGP2. J. Virol. 80:12332–12342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kuo RL, Zhao C, Malur M, Krug RM. 2010. Influenza A virus strains that circulate in humans differ in the ability of their NS1 proteins to block the activation of IRF3 and interferon-beta transcription. Virology 408:146–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Loo YM, et al. 2008. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 82:335–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Meylan E, et al. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167–1172 [DOI] [PubMed] [Google Scholar]
- 12. Nemeroff ME, Barabino SM, Li Y, Keller W, Krug RM. 1998. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′ end formation of cellular pre-mRNAs. Mol. Cell 1:991–1000 [DOI] [PubMed] [Google Scholar]
- 13. Rothenfusser S, et al. 2005. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J. Immunol. 175:5260–5268 [DOI] [PubMed] [Google Scholar]
- 14. Saito T, et al. 2007. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc. Natl. Acad. Sci. U. S. A. 104:582–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Satoh T, et al. 2010. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc. Natl. Acad. Sci. U. S. A. 107:1512–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Seth RB, Sun L, Ea CK, Chen ZJ. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF 3. Cell 122:669–682 [DOI] [PubMed] [Google Scholar]
- 17. Suthar MS, et al. 26 July 2012. The RIG-I-like receptor LGP2 controls CD8+ T cell survival and fitness. Immunity. doi:10.1016/j.immuni.2012.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Talon J, et al. 2000. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 74:7989–7996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Twu KY, Noah DL, Rao P, Kuo R-L, Krug RM. 2006. The CPSF30 binding site on the NS1A protein of influenza A virus is a potential antiviral target. J. Virol. 80:3957–3965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Venkataraman T, et al. 2007. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J. Immunol. 178:6444–6455 [DOI] [PubMed] [Google Scholar]
- 21. Xu LG, et al. 2005. VISA is an adapter protein required for virus-triggered IFN-β signaling. Mol. Cell 19:727–740 [DOI] [PubMed] [Google Scholar]
- 22. Yoneyama M, et al. 2005. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 175:2851–2858 [DOI] [PubMed] [Google Scholar]
- 23. Yoneyama M, et al. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730–737 [DOI] [PubMed] [Google Scholar]