Abstract
Activation of oncogenes or inhibition of WEE1 kinase deregulates cyclin-dependent kinase (CDK) activity and leads to replication stress; however, the underlying mechanism is not understood. We now show that elevation of CDK activity by inhibition of WEE1 kinase rapidly increases initiation of replication. This leads to nucleotide shortage and reduces replication fork speed, which is followed by SLX4/MUS81-mediated DNA double-strand breakage. Fork speed is normalized and DNA double-strand break (DSB) formation is suppressed when CDT1, a key factor for replication initiation, is depleted. Furthermore, addition of nucleosides counteracts the effects of unscheduled CDK activity on fork speed and DNA DSB formation. Finally, we show that WEE1 regulates the ionizing radiation (IR)-induced S-phase checkpoint, consistent with its role in control of replication initiation. In conclusion, these results suggest that deregulated CDK activity, such as that occurring following inhibition of WEE1 kinase or activation of oncogenes, induces replication stress and loss of genomic integrity through increased firing of replication origins and subsequent nucleotide shortage.
INTRODUCTION
DNA replication is tightly monitored to ensure that the genome is replicated precisely once per cell cycle and that DNA replication is complete before mitosis begins. Conditions for DNA synthesis are rarely ideal, and a number of obstacles must often be dealt with, such as a damaged DNA template and shortage of deoxynucleoside triphosphates (dNTPs), to allow replication fork progression. Stalling replication forks pose serious threats to genome integrity because they can collapse through disassembly of the replication complex and break (6, 11, 24). Such damaged forks may subsequently undergo incorrect repair, leading to genetic changes like chromosomal rearrangements (6, 24). Recent data have also revealed that activated oncogenes can induce DNA replication stress, defined here as replication-associated DNA damage (2, 3, 10). Oncogene-induced replication stress can lead to additional tumor-promoting genetic changes, but it may also serve as a tumor barrier by activation of cell cycle arrest, apoptosis, and/or senescence during early tumor development (32).
WEE1 and CHK1 kinases have major roles in suppressing DNA replication stress (4, 23, 27, 42), and attenuation of their function can contribute to carcinogenesis and cause cell death (40). The massive amount of DNA breakage is likely mediated by DNA endonuclease activity, and recent studies suggest that this is mediated by the endonuclease MUS81 (12, 14, 15). Notably, the mechanisms by which oncogenes or inhibition of checkpoint kinases can lead to endonuclease-mediated DNA breakage are poorly understood. It is also not fully understood if these breaks also play a role in inducing fork stalling or if they are temporally delayed events secondary to the fork stalling.
As both oncogenes and checkpoint kinases are regulators of cyclin-dependent kinase (CDK) activity, we previously proposed that most of the DNA replication stress arises due to elevated CDK activity that deregulates the DNA replication process at the level of initiation (40). However, this hypothesis has yet to be substantiated experimentally. It is not clearly understood how enhanced initiation can lead to DNA breakage. It also remains to be investigated whether the mitotic kinase WEE1 regulates replication initiation. In fact, a recent study found that inhibition of WEE1 causes replication fork stalling without evidence of initiation defects (12).
Notably, a recent study suggested that oncogenes lead to DNA replication stress due to nucleotide shortage (5). It was proposed that oncogene-induced forced S-phase entry generates a situation with insufficient nucleotide pools for normal replication, leading to replication fork collapse and DNA breakage. The low nucleotide pool was suggested to be a result of an unbalanced activation of nucleotide biosynthesis genes (5).
Based on these studies, we hypothesized that increased replication initiation might lead to nucleotide shortage and that this may be a major underlying mechanism causing DNA replication stress when CDK activity is elevated due to inhibition of WEE1 or oncogene activation. To investigate this hypothesis, we explored the effects of inhibiting WEE1 kinase. We found that acute activation of CDK activity by inhibition of WEE1 kinase stimulates replication through firing of additional origins, leading to nucleotide insufficiency. This reduces replication fork speed and leads to fork stalling followed by increased binding of SLX4-MUS81 nuclease to regions of stalled replication forks, ultimately leading to DNA double-strand breakage. Thus, our results demonstrate for the first time that WEE1 is a major regulator of replication initiation and that increased initiation followed by increased nucleotide consumption mediates the DNA replication stress response.
MATERIALS AND METHODS
Cell culture and chemicals.
The human osteosarcoma cell line U2OS and the telomerized human normal fetal lung fibroblast cell line TIG-3 were grown in Dulbecco's modified Eagle medium (GIBCO) with 10% fetal bovine serum (FBS). For small interfering RNA (siRNA)-mediated knockdown, U2OS cells were transfected using Lipofectamine RNAiMAX (Invitrogen) or Oligofectamine (Invitrogen) according to the manufacturer's protocols. The following oligonucleotide sequences were ordered from Sigma-Aldrich: UNC (universal negative control); WEE1-1, 5′-GGAAAAAGGGAAUUUGAUG(dT)(dT)-3′; WEE1-2, 5′-GGGAAUUUGAUGUGCGACA(dT)(dT)-3′; WEE1-3, 5′-GGUAUAUUCAUUCAAUGUC(dT)(dT)-3′; CDK1, 5′-GAUGUAGCUUUCUGACAAAAA(dT)(dT)-3′; CDK2, 5′-GUUUCAGUAUUAGAUGCAC(dT)(dT)-3′; CDT1, 5′-GCGCAAUGUUGGCCAGAUC(dT)(dT)-3′; MUS81-1, 5′-CAGCCCUGGUGGAUCGAUA(dT)(dT)-3′; MUS81-2, 5′-GGGUAUACCUGGUGGAAGA(dT)(dT)-3′; MUS81-3, 5′-CAGGAGCCAUCAAGAAUAA(dT)(dT)-3′; SLX4-1, 5′-AAACGUGAAUGAAGCAGAA(dT)(dT)-3′; and SLX4-2, 5′-GAACGAAGUCGCACAGAAG[dT][dT]-3′.
Experiments with siRNA-transfected cells were performed as indicated in the figure legends. Inhibition of WEE1 kinase activity was achieved by the addition of MK-1775 at a final concentrations of 100 nM, 300 nM, 1 μM, or 2 μM. CDK activity was inhibited by addition of roscovitine (Calbiochem) to a final concentration of 25 μM, and CDK1 activity was inhibited by addition of RO-3306 (Calbiochem) to a final concentration of 10 μM. Nucleosides were dissolved in phosphate-buffered saline (PBS) and added to the growth medium at a final concentration of 5 μM. When stated in the figure legends, Embryomax ES nucleosides (Millipore; cytidine, 0.73 g/liter; guanosine, 0.85 g/liter; uridine, 0.73 g/liter; adenosine, 0.73 g/liter; thymidine, 0.24 g/liter) were used at 1:50 or 1:25.
DNA fiber assay and DNA combing.
Exponentially growing cells were pulse-labeled with 25 μM 5′-chlorodeoxyuridine (CldU) followed by 250 μM 5′-iododeoxyuridine (IdU) for 20 min each. Cells were ruptured on microscope slides by incubation in a drop of lysis buffer (0.5% sodium dodecyl sulfate [SDS], 200 mM Tris [pH 7.4], 50 mM EDTA) for ∼2 min, and DNA fibers were spread by slanting the slide. After the cell lysate had dried, the DNA was fixed in 3:1 methanol-acetic acid for 10 min. Fibers were denatured in 2.5 M HCl for 60 min at 4°C. For immunodetection of CldU- and IdU-labeled tracks, fiber spreads were incubated with rat monoclonal antibody against bromodeoxyuridine (BrdU; 1:1,000; AbD Serotec) and mouse monoclonal anti-BrdU antibody (1:200; Becton, Dickinson), respectively, overnight at 4°C. Slides were then incubated with Alexa Fluor 488-conjugated goat anti-mouse IgG (1:500; Invitrogen) and Alexa Fluor 555-conjugated goat anti-rat IgG (1:500; Invitrogen) for 1 h at room temperature. Fibers were examined using an Axiovert 200 M LSM 520 (Carl Zeiss, Inc.) confocal microscope using a 63× c-Apochromat objective with a numerical aperture (NA) of 1.2. The lengths of CldU (Alexa Fluor 555; red)- and IdU (Alexa Fluor 488; green)-labeled patches were measured using ImageJ software (http://rsb.info.nih.gov/ij/). For DNA combing, asynchronous exponentially growing U2OS cells were pulse-labeled with BrdU (25 μM) for 45 min. Single-molecule analysis of DNA replication by molecular combing was carried out as described previously (44).
Immunoblotting and antibodies.
Cells were lysed on ice in radioimmunoprecipitation assay (RIPA) buffer and processed as previously described (18). Phospho-CHK1 antibody (CHK1-pSer317), phospho-CDC2 (Tyr15), CDC2, phospho-Ser CDK substrates (2324), and phospho-p53 (pSer15) antibodies were purchased from Cell Signaling Technology. Antibodies against MCM7 (DCS-141) and CHK1 (DCS-310) were described previously (39). Anti-CDC45 (sc-55569), anti-p-CDC2 p34 (Thr14/Tyr15), and anti-CDK2 (D-12) were purchased from Santa Cruz Biotechnology, Inc. Anti-phosphorylated H2AX (JBW301) and anti-phosphorylated histone H3 Ser10 (06-570) were purchased from Millipore. Anti-MUS81 (MTA30 2G10/3) and anti-PCNA (PC10) were purchased from Abcam, and anti-RPA (Ab-3) and anti-CDC2/CDK1 (Ab-1) were purchased from EMD. Antiactin (MAB1501) was purchased from Chemicon International. Anti-SLX4 was a gift from John Rouse.
Flow cytometry.
Cells were prepared for flow cytometry as previously described (18). The cells were fixed in 70% ethanol, permeabilized in 0.2% Triton X-100 (Fluka), stained with primary antibodies diluted in PBS–1% FBS or in flow buffer (0.1% Igepal CA-630, 6.5 mM Na2HPO4, 1.5 mM KH2PO4, 2.7 mM KCl, 137 mM NaCl, 0.5 mM EDTA [pH 7.5]) containing 4% nonfat milk and incubated with secondary antibodies (Alexa Fluor 488, 568, and 647 anti-rabbit and anti-mouse antibodies [Invitrogen] at 1:1,000) diluted in PBS–1% FBS or in flow buffer. Where indicated in the figure legends, cells were preextracted in detergent buffer (20 mM HEPES, pH 7.4; 50 mM NaCl; 3 mM MgCl2; 300 mM sucrose; 0.5% Triton X-100) for 5 min on ice prior to fixation. For EdU detection, a Click-iT 5-ethynyl-2′-deoxyuridine (EdU) cell proliferation assay kit (Invitrogen) was used according to the manufacturer's protocol. DNA was stained with 10 μg/ml propidium iodide (Sigma) and 20 μg/ml RNase (Sigma), with Hoechst 33258 (1.5 μg/ml), or with Cell Cycle 633 (Invitrogen). The samples were analyzed on a FACSCalibur flow cytometer (BD Biosciences) using CellQuest software (BD Biosciences) or on an LSRII flow cytometer (BD Biosciences) using Diva software. The bar-coding flow cytometry technique (21) (see Fig. S1G in the supplemental material) was used to eliminate variation in antibody staining or variation in the EdU Click-iT reaction between individual samples. Four fixed samples were stained with four different concentrations (0.125, 0.031, 0.0062, and 0.00078 ng/μl) of Pacific Blue (Invitrogen) for 30 min in the dark at room temperature and subsequently mixed in one tube. The mixed cells were then stained with antibody to CDC45 (1:100, sc-55569; Santa Cruz) or RPA (MS-691P1; Neomarkers) or labeled via the EdU Click-iT reaction. Flow cytometry analysis was performed on a LSRII flow cytometer (BD Biosciences) using Diva software. For analysis, the four samples were gated on the Pacific Blue signal before analysis of CDC45 or EdU median values in each sample.
Microscopy and immunofluorescence.
Microscopy and immunofluorescence analysis were performed as previously described (18). Where indicated in the figure legends, cells were preextracted in detergent buffer (20 mM HEPES, pH 7.4; 50 mM NaCl; 3 mM MgCl2; 300 mM sucrose; 0.5% Triton X-100) for 5 min on ice prior to fixation. RPA, 53BP1, cyclin B, CDC45L, and γH2AX were stained with the following antibodies: anti-phosphorylated H2AX (2577) from Cell Signaling, cyclin B (sc-245), 53BP1 (H-300), and CDC45L (sc-55569) from Santa Cruz, and anti-RPA (Ab-3) from Calbiochem. Cells were visualized using a Leica Leitz DMRXE or Zeiss Axiovert microscope, and pictures were obtained using a 63× objective and a DFC340 FX digital camera system. For automated counting of 53BP1 foci, pictures were obtained with an Olympus ScanR̂ system. At least 580 cells were analyzed using CellProfiler software (7).
Pulsed-field gel electrophoresis.
After treatment, cells were harvested, cast in 1% agarose plugs (InCert agarose; Lonza) (106 cells/plug), and incubated in 0.5% EDTA, 1% N-laurylsarcosyl, 1 mg/ml proteinase K at 50°C overnight. Plugs were washed four times in Tris-EDTA (TE) buffer before they were loaded onto a 1% agarose (chromosomal grade) gel and separated by pulsed-field gel electrophoresis for 20 h (CHEF-DR II system; 120° angle, 60- to 240-s switch time, 4 V/cm; Bio-Rad). DNA was visualized on the gel by ethidium bromide (EtBr) staining.
iPOND.
iPOND was essentially performed as previously described (38). After treatments, cells were cross-linked in 1% formaldehyde in PBS for 20 min at room temperature and afterwards quenched using glycine (0.125 M final concentration) for 5 min. The cells were washed three times in PBS, collected by cell scraping, and permeabilized in 0.25% Triton X in PBS for 10 min on ice. Subsequently, the cells were incubated with the Click-iT reaction mixture (Invitrogen) according to the manufacturer's protocol, using 10 μM biotin-azide diluted in dimethyl sulfoxide (DMSO) for 1 to 2 h. DMSO alone was used in the non-Click-iT reaction control. The cells were lysed using 1% SDS, 50 mM Tris (pH 8.0), 2.5 μg/ml leupeptin, 2.5 μg/ml aprotinin and then sonicated for 6 min at 5°C (Bioruptor). Cell lysates were cleared by fast centrifugation for 10 min at 4°C. Protein concentration was determined, and 5% of each sample was used as input (1% was normally loaded). The remaining 95% of each sample was diluted 1:1 (vol/vol) with PBS containing 2.5 μg/ml leupeptin and 2.5 μg/ml aprotinin, and this mixture was incubated with washed streptavidin-agarose beads overnight at 4°C in a rotator. Afterwards, the beads were washed once in the lysis buffer, once in 1 M NaCl, and then twice in lysis buffer. The beads were added to 2× loading sample buffer (LSB; 1× LSB is 50 mM Tris [pH 6.8], 100 mM dithiothreitol [DTT], 2% bromophenol blue, 10% glycerol) for 25 min at 95°C to elute the captured proteins and reverse the cross-link. Finally, the proteins were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) and detected by immunoblotting.
dATP/ATP ratio measurement.
To assess deoxynucleotide availability, we made small-molecule extracts of ∼2 × 107 exponentially growing cells by lysing the cells in 500 μl ice-cold 10% trichloroacetic acid (TCA)–15 mM MgCl2. After centrifugation, the supernatant was ether-extracted seven times to remove TCA and then used for ATP and dATP determination. dATP was determined by a primer extension assay on a dA-specific template essentially as described previously (37). The amounts of extended products were quantified on a Storm phospho-imager. ATP was measured indirectly using a luciferase-based ATP determination kit from Biaffin GmbH & Co KG according to the instructions. Statistical analysis of dATP/ATP ratios resulting from different treatments was performed using a column statistics test (GraphPad Prism software; GraphPad, La Jolla, CA).
RESULTS
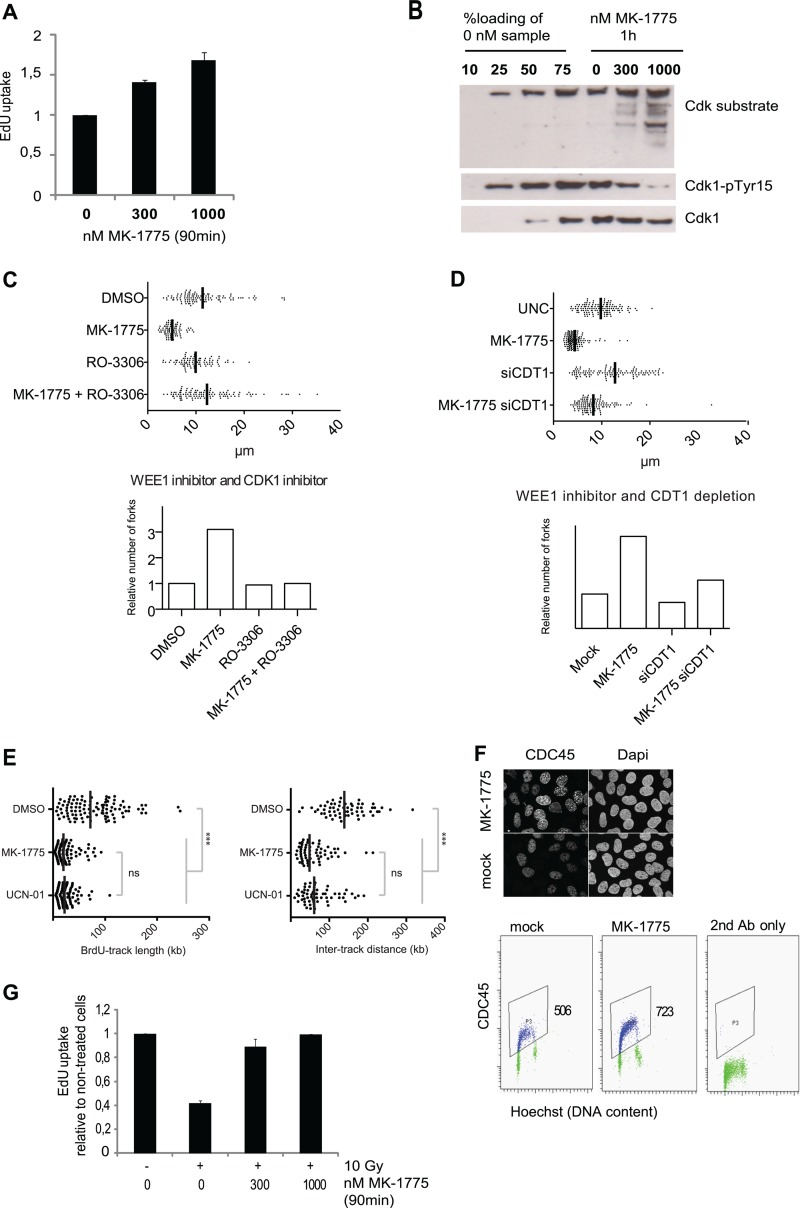
We recently identified a critical role for CDK regulation in genome maintenance during S phase (4). To understand the underlying mechanisms, we took advantage of MK-1775, a small molecule inhibitor of WEE1 kinase (17). We found that inhibition of WEE1 kinase by 300 or 1,000 nM MK-1775 led to increased uptake of the nucleoside analog EdU during the 30 to 90 min after MK-1775 addition (Fig. 1A). This indicates that WEE1 inhibition stimulates DNA replication. As expected, MK-1775 also led to decreased tyrosine 15 inhibitory phosphorylation on CDK1 and increased phosphorylation of CDK substrates (Fig. 1B), consistent with a rapid increase in CDK activity. The induction of EdU uptake by MK-1775 was reduced in cells partially depleted of CDK1 or CDK2 by siRNA transfection, as well as in cells cotreated with the CDK inhibitor roscovitine or with the CDK1-specific inhibitor RO-3306 (see Fig. S1 in the supplemental material). These results suggest that WEE1 inhibition causes a rapid increase in DNA replication due to elevated activities of CDK1 and CDK2.
Fig 1.
WEE1 suppresses CDK activity to control initiation of DNA replication. (A) Total cellular EdU uptake following treatment with the WEE1 inhibitor MK-1775. MK-1775 (300 or 1,000 nM) was added to U2OS cells, and after 30 min, cells were labeled with 1 μM EdU for 1 h. Cells were fixed and analyzed by the bar-coding flow cytometry method. Average EdU median values are shown relative to untreated cells. Error bars show standard errors of the means (SEM) (300 nM, n = 4; 1,000 nM, n = 3). (B) U2OS cells were treated with MK-1775 (0, 300, or 1,000 nM), and cell lysates were collected after 1 h and processed for Western blotting. The left four lanes were loaded with different amounts of the untreated sample (0 nM). (C) (Top) Replication track length analyzed by DNA fiber spreading. The WEE1 inhibitor MK-1775 (1 μM) and the CDK1 inhibitor RO-3306 (10 μM) were added for 30 min before pulses with CldU and subsequently with IdU of 20 min each. The total length of CldU and IdU was measured. (Bottom) Relative fork number calculated by dividing cellular EdU incorporation from panel B by the corresponding replication fork track length. (D) (Top) Replication track length analyzed by DNA fiber spreading. Cells were transfected with siRNA, and 48 h later the WEE1 inhibitor MK-1775 was added to a final concentration of 1 μM for 30 min followed by pulses with CldU and IdU. Total length of CldU and IdU was measured. (Bottom) Relative fork number calculated by dividing cellular EdU incorporation from panel B by the corresponding replication fork track length. (E) Single-molecule analysis of DNA replication by molecular combing. DMSO (0.1%), MK-1775 (1 μM), and UCN-01 (300 nM) were added 45 min before cells were pulse-labeled with BrdU (25 μM, 45 min). (Left) Distribution of BrdU track lengths. ***, P < 10−4; ns, nonsignificant (P = 0.156) (Mann-Whitney; n > 100). (Right) Distribution of intertrack distances. ***, P < 10−4; ns, P = 0.157 (Mann-Whitney; n = 60). (F) Measurement of CDC45 chromatin loading after preextraction of cells with detergent. U2OS cells were treated with 1 μM MK-1775, preextracted for 5 min in detergent buffer, fixed, and stained for CDC45L for immunofluorescence (above) and flow cytometry (below), respectively. Values are medians for the gated cell populations. (G) Abrogation of the IR-induced S-phase checkpoint by MK-1775. U2OS cells were treated with MK-1775 (0, 300, and 1,000 nM) at 15 min before IR (10 Gy). After 15 min, cells were labeled with 1 μM EdU for 1 h, fixed, and analyzed with the bar-coding method for flow cytometry. Median values relative to untreated cells are shown. Error bars show SEM (for 300 nM, n = 4; for 1,000 nM, n = 2).
The increased EdU uptake could in theory be due to increased initiation or increased fork rate, or both. To explore this issue, we analyzed replication fork rates by sequential labeling with the halogenated thymidine analogues CldU and IdU. Cells were lysed, and the DNA fibers were spread on microscope slides and stained with specific antibodies against CldU and IdU. We found that inhibition of WEE1 markedly reduced the replication fork rates (Fig. 1C), despite the total EdU uptake being increased (Fig. 1A). Addition of the CDK1 inhibitor RO-3306 (45) could partially normalize fork speed following WEE1 inhibition (Fig. 1C). Taken together, these observations suggest that replication origin firing is increased following WEE1 ablation due to unscheduled CDK activity. Similarly, depletion of CDK1 kinase also rescued the fork rates following MK-1775 treatment (see Fig. S3 in the supplemental material). WEE1 depletion also slows the fork rate (see Fig. S4 in the supplemental material). We then calculated a relative number of active replication forks based on our measurements of the total EdU uptake and fork rates (Fig. 1C; also, see Fig. S3 and S4 [bottom histogram blot] in the supplemental material). The results of these calculations support the theory that in response to inhibition or depletion of WEE1, replication initiation is increased due to unscheduled CDK activity.
To investigate whether the reduced fork rate may be a downstream consequence of the increased initiation, we measured fork speed following partial codepletion of the replication initiation factor CDT1 (36). Indeed, depletion of CDT1 could normalize fork speed in response to WEE1 inhibition by MK-1775 (Fig. 1D; also, see Fig. S2 in the supplemental material), as well as in response to WEE1 depletion (see Fig. S5 in the supplemental material). These results strongly suggest that unscheduled CDK activity due to inhibition of WEE1 activity leads to increased initiation followed by a decrease in fork rate. Consistent with increased replication initiation, we also observed decreased interorigin distance by DNA combing following treatment with MK-1775, to an extent similar to that achieved with treatment with the CHK1 inhibitor UCN-01 (Fig. 1E).
Furthermore, we also measured the extent of chromatin loading of the replication factor CDC45 in response to WEE1 inhibition, following preextraction of cells with detergent before fixation. The loading of CDC45 was markedly increased in S phase 1 h after the addition of MK-1775 (Fig. 1F), confirming that replication initiation factors accumulate when stimulated by the increased CDK activity. The radiation-induced S-phase checkpoint is thought to mainly suppress replication initiation (as opposed to replication elongation) (22), and based on the results so far, we reasoned that WEE1 would be required for the S-phase checkpoint. We treated cells with MK-1775 prior to ionizing radiation, and this partly reconstituted the irradiated cells' ability to incorporate EdU compared to nonirradiated cells, thus indicating that MK-1775 treatment abrogated the S-phase checkpoint (Fig. 1G; also, see Fig. S6 in the supplemental material). This result lends further support for a role of WEE1 activity in control of replication initiation.
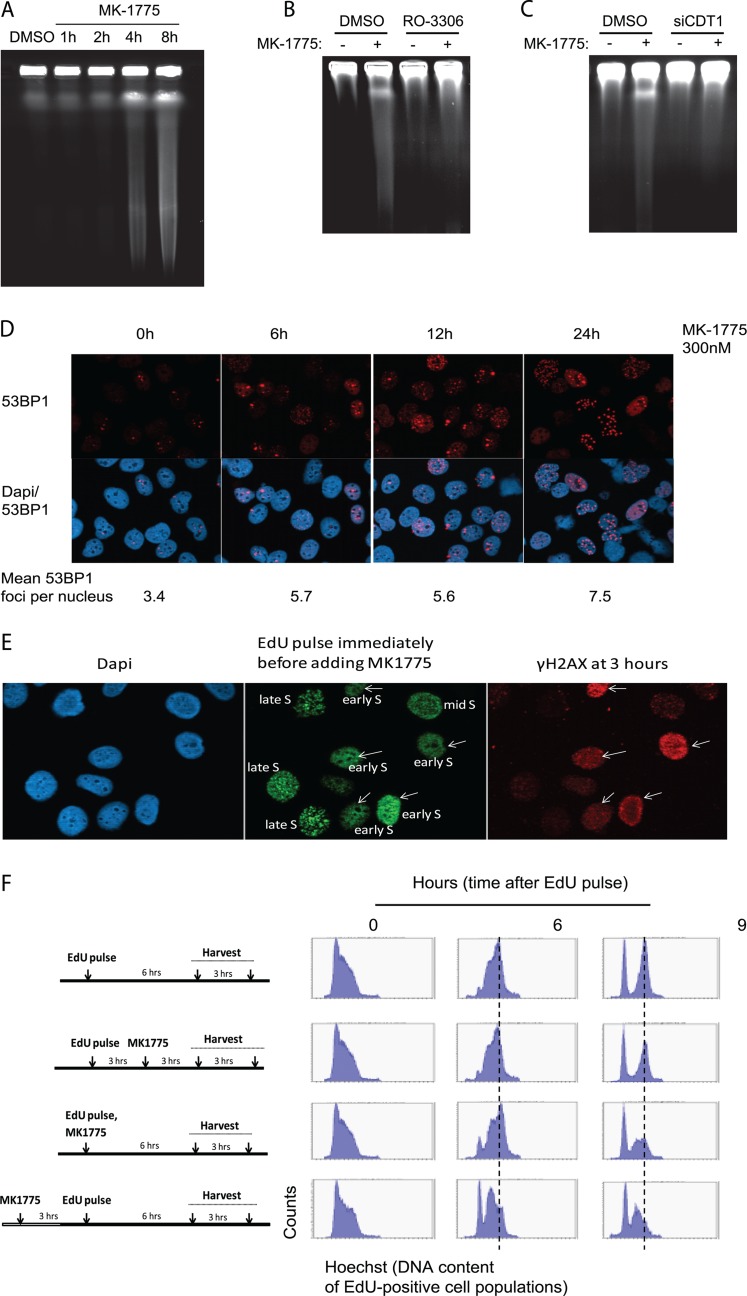
WEE1 depletion leads to marked accumulation of replication-associated DNA double-strand breaks (4). Analysis of DNA breaks by pulsed-field gel electrophoresis (PFGE) showed that as little as 4 h of WEE1 inhibition induced DSBs (Fig. 2A). This induction of DSBs was abrogated by addition of the CDK1 inhibitor RO-3306 (Fig. 2B) or by partial depletion of CDT1 (Fig. 2C). Together, these results suggest that DNA breaks are induced in response to WEE1 inhibition as a consequence of increased initiation mediated by unscheduled CDK activity. Another sign of replication stress is the occurrence of 53BP1 foci in the subsequent G1 phase following cell division (16, 28). In response to MK-1775 we observed an induction of 53BP1 foci at 6 to 24 h (Fig. 2D). To confirm that these 53BP1 foci were induced in G1 phase, we simultaneously stained cells with 53BP1, cyclin B, and EdU at various times following MK-1775 addition, when a short pulse of EdU was added to label S-phase cells before fixation (see Fig. S7 in the supplemental material). 53BP1 foci increased at 6 to 24 h after WEE1 inhibition predominantly in cells negative for EdU and cyclin B. Thus, addition of MK-1775 causes 53BP1 foci in the subsequent G1 phase, consistent with these cells having experienced replication stress in the previous S phase. Some of the 53BP1 foci could potentially also be related to effects of MK-1775 on the G2/M transition.
Fig 2.
DNA damage occurs rapidly after WEE1 inhibition as a consequence of the increased initiation. (A) Pulsed-field gel electrophoresis. U2OS cells were treated with the WEE1 inhibitor MK-1775 (2 μM) for the indicated duration. (B) Pulsed-field gel electrophoresis as done for panel A with the WEE1 inhibitor MK-1775 (2 μM) and the CDK1 inhibitor RO-3306 (10 μM) for 4 h. (C) As described for panel A with siRNA against CDT1 for 48 h and MK-1775 (2 μM) for 4 h. (D) Immunofluorescence images of 53BP1 foci after 300 nM MK-1775 treatment. U2OS cells were preextracted with detergent before fixation at the indicated times and stained for 53BP1 and for DNA with DAPI. Numbers are average 53BP1 foci per nuclei (at least 580 cells were counted per treatment). (E) Strong γH2AX staining occurs preferentially in cells that were in early S phase at the time of MK-1775 addition. U2OS cells were pulsed with EdU (10 μM, 10 min) immediately before MK-1775 addition (1 μM), and cells were harvested and stained for γH2AX and EdU at 3 h later. The pattern of EdU foci was used to distinguish between early-, mid-, and late-S-phase cells. (F) S-phase progression is delayed in cells that were in late G1/early S phase at the time of MK-1775 addition (bottom two panels), while cells in late S phase are less affected (second panel from top). EdU pulse-labeling (10 μM, 10 min) and MK-1775 treatment (1 μM) were done at the indicated times. MK-1775 was present in the cell culture medium until cell harvest. During analysis, the EdU-positive cells were gated in each sample, and DNA profiles (cell cycle profiles) are shown for the gated EdU-positive cell populations.
To further examine the role of WEE1 inhibition on DNA replication, we analyzed DNA damage in S-phase cells by combining EdU and H2AX staining. We noted that cells with strong γH2AX staining that appeared 3 h after addition of MK-1775 were almost exclusively cells that had been in early S phase at the time of MK-1775 addition (Fig. 2E and data not shown). Consistent with these findings, time-lapse imaging of GFP-PCNA expressing U2OS cells showed that cells in late S phase completed S phase normally, while cells in early S phase were often impaired in S-phase progression after MK-1775 (data not shown). Furthermore, we analyzed cell cycle progression following EdU pulse-labeling to distinguish between cells that were in early versus late S phase at the time of addition of MK-1775 (Fig. 2F). By gating for the EdU positive populations, we could follow the cell cycle progression of only the pulse-labeled cells. The results confirmed that cells that were in late S phase at the time of addition of MK-1775 (Fig. 2F, second from top) completed S phase as untreated cells (Fig. 2F, top). However, a proportion of cells that were in early S phase, or of cells entering S phase at the time of addition of MK-1775, arrested during S-phase progression (Fig. 2F, 9 h, bottom panels). Thus, early-S-phase cells, which would be expected to rely most heavily on proper regulation of replication initiation, are most affected by WEE1 inhibition both in terms of replication progression and induction of DNA damage. These results are consistent with deregulated CDK activity causing deficient replication almost immediately in early S phase, whereas DNA breaks appear at later time points.
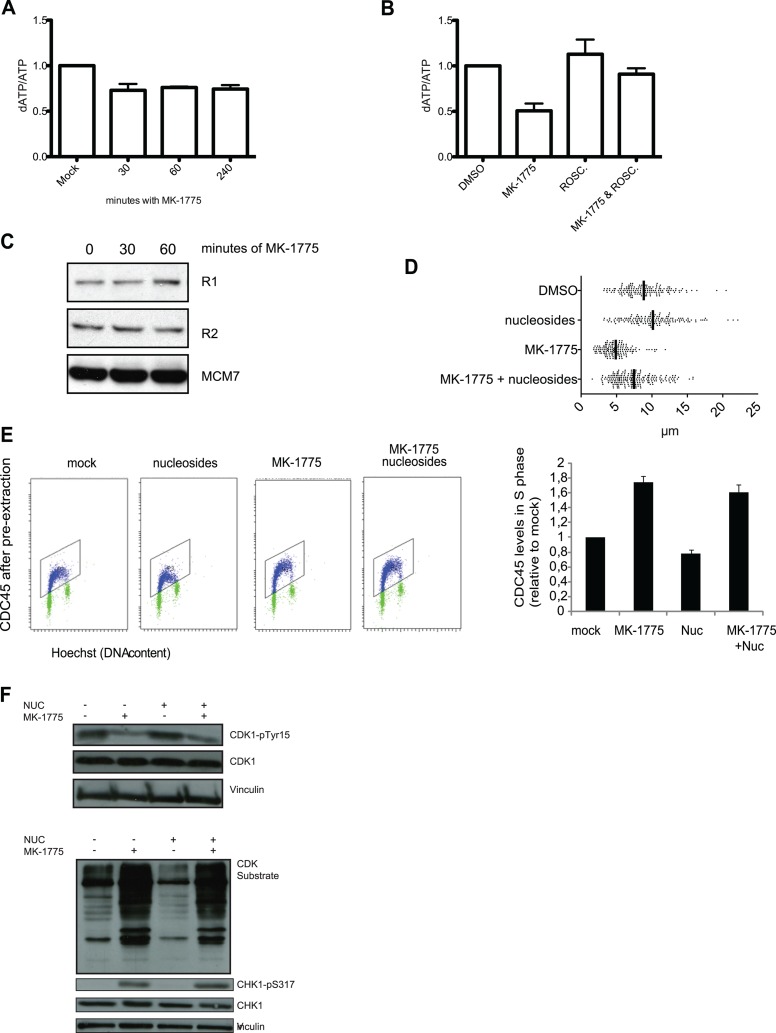
A key unresolved issue was how enhanced initiation can lead to DNA replication stress. We hypothesized that replication deficiency occurs due to insufficient amounts of required replication factors. The cellular availability of required replication factors may not be sufficient to carry out replication under conditions where a massive number of origins are fired simultaneously. Imbalance in dNTP pools are known to cause dysfunctional replication (35, 43). To address whether deoxynucleotide pools were affected, we measured intracellular dATP levels after inhibiting WEE1 with MK-1775 (37). Indeed, the amount of dATP was decreased following MK-1775 treatment compared to mock-treated cells (Fig. 3A), suggesting that increased origin firing due to unscheduled CDK activity leads to increased deoxynucleotide consumption. Given that WEE1 inhibition leads to a reduction in intracellular dATP levels, concomitant with increased CDK activity and increased origin firing, we hypothesized that inhibition of CDK activity by exposure to the CDK inhibitor roscovitine would restore intracellular dATP to control levels. To this end, we quantified dATP levels in small-molecule extracts prepared following MK-1775, roscovitine, or roscovitine-plus-MK-1775 exposure. Indeed, dATP levels were similar to those in DMSO-treated control cells following 30 min exposure to roscovitine (Fig. 3B), indicating that the reduction in dATP levels observed upon WEE1 inhibition is dependent upon elevated CDK activity. Importantly, the protein levels of ribonucleotide reductase were not altered (Fig. 3C), and addition of nucleosides to the cell culture medium partially normalized replication fork rates in MK-1775-treated cells (Fig. 3D). Next, we investigated if nucleoside addition could suppress additional replication complications after deregulated CDK activity by MK-1775 treatment. We found that the increased chromatin binding of the initiation factor CDC45 after MK-1775 treatment persisted in the presence of nucleosides in the growth medium (Fig. 3E). Furthermore, nucleoside addition had little impact on CDK activation and ATR-mediated CHK1 phosphorylation (Fig. 3F). This indicates that WEE1 inhibition still leads to unscheduled CDK activity and some replication complications in the presence of exogenously added nucleosides, which is possibly due to enhanced initiation depriving cells of a number of additional key replication factors.
Fig 3.
WEE1 inhibition leads to enhanced nucleotide consumption, and nucleoside addition normalizes replication fork speed. (A) U2OS cells were exposed to MK-1775 (1 μM) for the indicated times or mock exposed to DMSO only. Small-molecule extracts were tested for dATP and ATP content, and the results are presented as dATP/ATP ratios. Data are means ± SEM for three biological replicates. (B) U2OS cells were exposed to MK-1775 (1 μM), roscovitine (25 μM), or MK-1775 plus roscovitine for 30 min or DMSO only, and dATP was quantified as for panel A. Data are mean dATP/ATP ratios ± SEM for three (DMSO, MK-1775, and roscovitine) or four (MK-1775 plus roscovitine) biological replicates. *, P < 0.05. (C) U2OS cells were treated with MK-1775 (1 μM) for the indicated periods. The cells were harvested and processed for immunoblotting. Cells were stained with antibodies against the R1 and R2 subunits of ribonucleotide reductase and MCM7 as a loading control. (D) Replication track length analyzed by DNA fiber spreading. Cells were transfected with siRNA and 48 h later pulsed with CldU and subsequently with IdU for 20 min each. Total length of CldU and IdU was measured. The WEE1 inhibitor MK-1775 was added to a final concentration of 1 μM for 30 min before the pulses with CldU and IdU. Nucleosides (5 μM) were added to growth medium simultaneously with MK-1775. (E) U2OS cell were treated with MK-1775 (0 or 1000 nM) in the presence or absence of extra nucleosides (Embryomax ES nucleosides at 1:50) for 1 h. Cells were treated and processed as for Fig. 1F and analyzed by the flow cytometry bar-coding method. The graph shows quantification of CDC45 levels in S phase. Background levels in S phase after staining with the second antibody alone were subtracted. Data are means and SEM from three independent experiments. (F) U2OS cells were treated with MK-1775 (2 μM), with or without nucleosides (5 μM) for 30 min. The cells were harvested and processed for immunoblotting. Cells were stained with antibodies against CDK1-pY15, CDK1, CHK1-p317, CHK1, and CDK substrates and vinculin as a loading control.
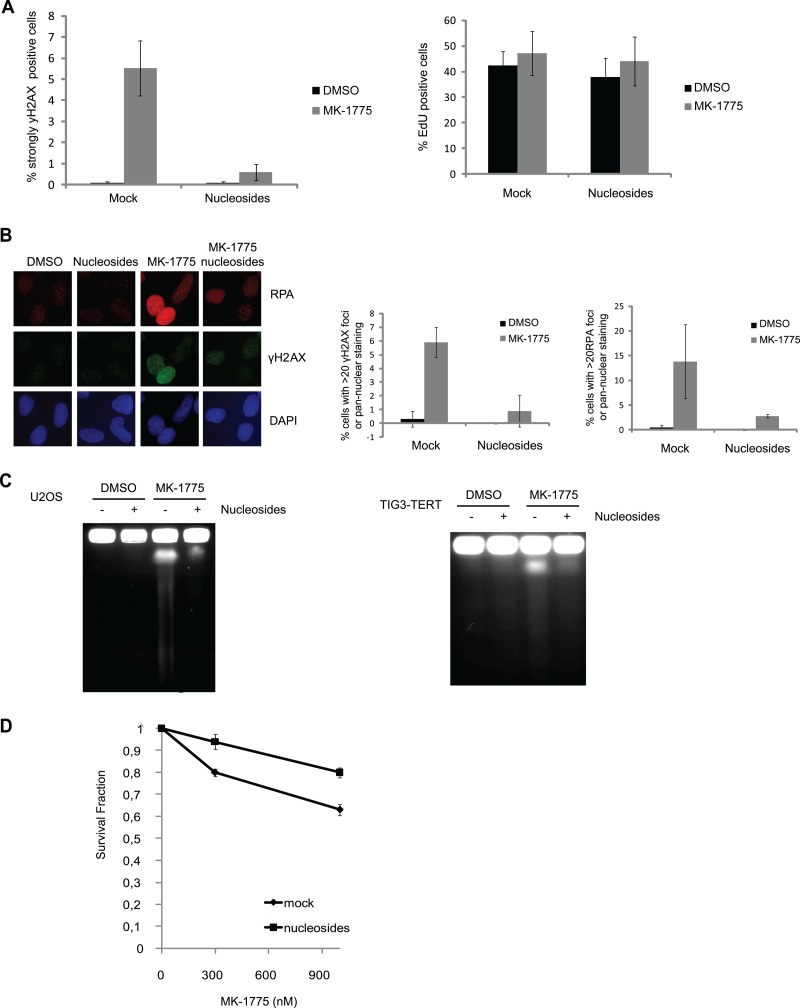
Given that nucleotide metabolism had an impact on fork progression, we asked if actual DNA damage following WEE1 inhibition was affected by nucleotide levels. We added nucleosides to MK-1775-treated cells, and this markedly reduced the number of cells containing the highest levels of γH2AX staining detected by fluorescence-activated cell sorting (FACS) analysis (Fig. 4A; also, see Fig. S8 in the supplemental material) and immunofluorescence (Fig. 4B). However, many S-phase cells showed moderately increased RPA and γH2AX staining in response to MK-1775, regardless of nucleoside addition (Fig. 4B; see Fig. S8 and S9 in the supplemental material; also data not shown). This indicates that nucleoside addition cannot fully normalize S-phase progression. To confirm that addition of nucleosides suppressed the induction of DNA breaks after WEE1 inhibition, we also measured DNA double-strand-break formation by PFGE. Importantly, addition of nucleosides inhibited MK-1775-induced DNA double-strand breaks both in U2OS cells and in Tig3-TERT immortalized human fibroblasts (Fig. 4C). These effects cannot be explained by altered number of S-phase cells or by decreased effects on CDK activity following addition of nucleosides. Addition of nucleosides did not alter the number of S-phase cells (Fig. 4A, right) or markedly change cell cycle progression after 2, 4, or 8 h of MK-1775 treatment (see Fig. S10 in the supplemental material). Addition of nucleosides could also partially restore the reduction in clonogenic survival following MK-1775 treatment (Fig. 4D). Taken together, our data show that unscheduled CDK activity results in increased origin firing and nucleotide shortage. Nucleotide shortage may not be the sole reason for the replication deficiencies, but this shortage has a marked impact on fork progression, and these forks are then subsequently encountering DNA breakage.
Fig 4.
Nucleoside addition suppresses DNA damage induced by deregulation of the WEE1-CDK pathway. (A) U2OS cells were treated with MK-1775 (2 μM), with or without nucleosides (5 μM) for 4 h. EdU (10 μM) was added during the last 30 min. The cells were harvested and processed for FACS. Cells were stained with propidium iodide (PI) and antibody against γH2AX, and EdU was detected through the Click-iT reaction. Data are means ± SEM from five experiments (left) and three experiments (right). (B) Cells were seeded on coverslips and analyzed as described for panel A. The cells were fixed and stained with DAPI and for antibodies against RPA and γH2AX. Data are means ± SEM from three experiments. (C) Pulsed-field gel electrophoresis. U2OS (left) and Tig3-tert (right) cells were treated with the WEE1 inhibitor MK-1775 (2 μM) and nucleosides (5 μM) for 4 h. (D) Clonogenic survival after 6 h treatment with MK-1775 (0, 300, and 1,000 nM) in the presence (nucleosides) and absence (mock) of Embryomax ES nucleosides at 1:25. Survival is shown as a fraction of the cells treated with 0 nM MK-1775. Data are means ± SEM from three independent experiments. The cloning efficiency of cells treated with nucleosides alone relative to untreated cells was 0.94 ± 0.04.
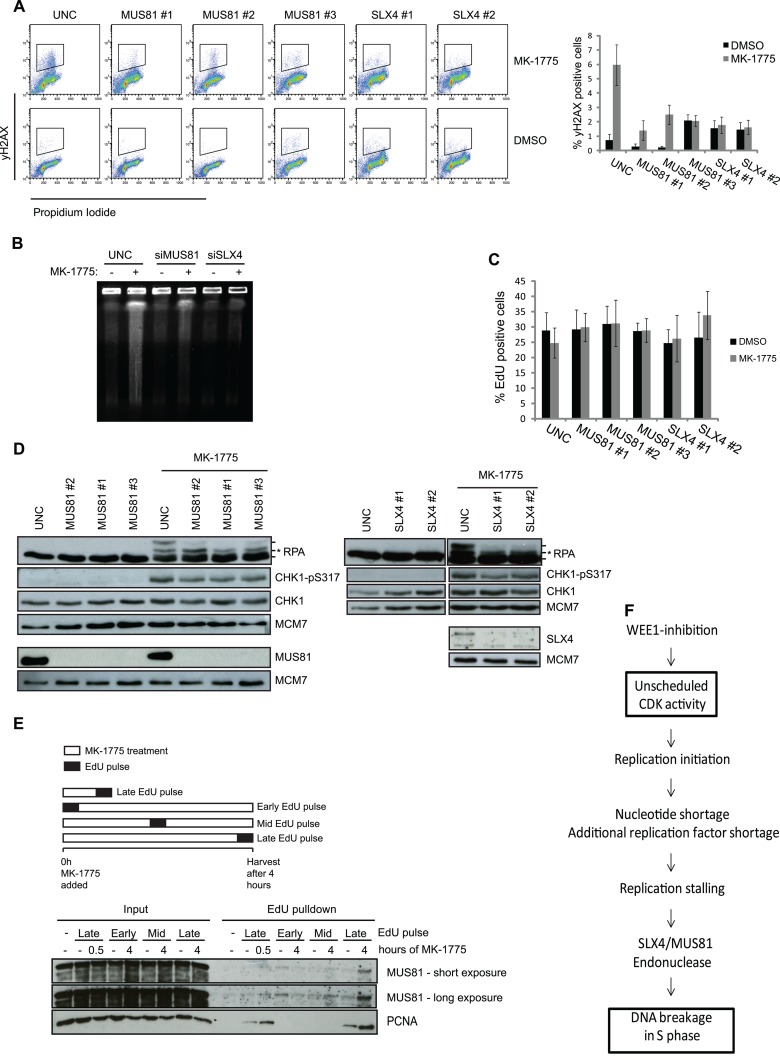
As our results indicate that induction of DNA breakage following inhibition of WEE1 is tightly coupled to replication stalling, the DNA breaks are likely caused by endonucleases with a role in processing replication fork lesions. Stalled replication forks provide substrates for endonucleases such as MUS81-EME1 (15, 24). To better understand the involvement of MUS81 in this process, we tested the effects of depleting MUS81 and its regulator SLX4 (13, 31, 41). We found that depletion of either MUS81 or SLX4 could abrogate the majority of MK-1775-induced γH2AX (Fig. 5A) and DNA double-strand breaks measured by PFGE (Fig. 5B). Depletion of MUS81 or SLX4 did not alter DNA replication (Fig. 5C). We next investigated if MUS81 or SLX4 had an impact on ATR checkpoint signaling after WEE1 inhibition. We found that depletion of MUS81 or SLX4 did not reduce ATR-mediated phosphorylation of CHK1 or RPA upon addition of MK-1775 (Fig. 5D), indicating that replication complications persisted when the SLX4-MUS81 endonuclease complex was depleted. Under these conditions, the SLX4-MUS81 endonuclease appears to act downstream of replication fork stalling to induce DNA breakage. To further confirm that the SLX4-MUS81 complex functions downstream of fork stalling, we performed iPOND on cells treated with MK-1775 for 30 min or 4 h. We added EdU at various points during WEE1 inhibition to mark replicated DNA, and we then purified EdU containing chromatin fragments and prepared them for immunoblotting (Fig. 5E) (38). After 4 h of MK-1775 treatment there was a significant increase of MUS81 binding on chromatin compared to 0 and 30 min treatment (Fig. 5E), consistent with the appearance of DNA DSBs at 4 h (Fig. 2A). This suggests that MUS81 acts on stalled replication forks after DNA replication has already been affected by the deregulated CDK activity. As expected, PCNA is present on chromatin in the samples where the EdU pulse was given just prior to harvesting. In addition, we detected increased PCNA binding after MK-1775 treatment (Fig. 5E), which further supports the notion that the elevated CDK activity stimulates DNA replication (Fig. 1A; also, see Fig. S1 and S3 in the supplemental material). Together, these results suggest that replication deficiency enables the SLX4-MUS81 endonuclease complex to cleave stalled forks after prolonged WEE1 inhibition.
Fig 5.
DNA DSBs formation but not ATR signaling is mediated by SLX4-MUS81 after WEE1 inhibition. (A) U2OS cells were transfected with siRNAs targeting MUS81 and SLX4. Transfection with UNC was included as a negative control. The cells were treated with EdU (10 μM) for 30 min prior to harvest, and at 48 h posttransfection the cells were harvested and processed for FACS. Cells were stained with PI and antibody against γH2AX, and EdU was detected through the Click-iT reaction. FACS analysis was carried out with cells stained for γH2AX and PI, and data are means ± SEM from a minimum of five experiments. (B) U2OS cells were reverse transfected with siRNA targeting MUS81 and SLX4 for 48 h, treated with the WEE1 inhibitor MK-1775 (2 μM) for 4 h, and processed for PFGE. (C) EdU graphs from the experiment whose results are shown in panel A. (D) Cells were treated as for panel A and then harvested and processed for immunoblotting. Cell lysates were stained with antibodies against RPA, CHK1-p317, CHK1, MUS81, SLX4, and MCM7 as a loading control. In the RPA panels, the asterisk shows the ATR-dependent RPA phosphorylation, whereas the upper RPA band is the DNA-PK mediated phosphorylation of S4 and S8. (E) Cells were pulsed with 12 min of EdU at the start, middle, or end of MK-1775 treatment. The cells were harvested at the indicated time points and processed for iPOND to pull down nascent proteins. Lysates were processed for immunoblotting using antibodies against MUS81 and PCNA. A minus sign denotes a non-Click-iT control, where DMSO was used instead of biotin-azide to detect incorporated EdU. (F) Diagram depicting how increased CDK activity following WEE1 inhibition can lead to DNA breakage in S-phase cells via increased origin firing and ensuing enhanced nucleotide consumption.
DISCUSSION
Here, we show that WEE1 is part of a cell-autonomous mechanism that limits the activity of CDKs in order to suppress initiation of replication. Failure of this control mechanism results in increased DNA synthesis and nucleotide insufficiency that reduces replication fork speed and leads to fork stalling and DNA DSBs (see the model in Fig. 5F). Hence, our results may help explain how deregulated CDK activity, such as that occurring after the activation of oncogenes or suppression of checkpoint kinases, can induce replication stress and loss of genomic integrity.
WEE1-CDK regulation of origin firing.
Our data show for the first time that the mitotic kinase WEE1 regulates replication initiation. We previously demonstrated that WEE1 suppresses formation of DNA breakage during normal S-phase progression in a process dependent upon CDK activity and replication initiation factors (4). However, it has not previously been shown that WEE1 controls origin firing. In another recent report, it was proposed instead that WEE1 controls DNA replication via direct interaction with MUS81 endonuclease (12). Our data are in agreement with a role for WEE1 in regulation of origin firing due to the direct WEE1 phosphorylation of the tyrosine 15-inhibitory phosphorylation on CDK1 and CDK2 (33, 46). In the case of reduced WEE1 activity, the inhibitory phosphorylation on CDK1 and CDK2 is decreased, leading to hyperactivation of CDK1 and CDK2 and thereby increased origin firing. Similarly, reduced CHK1 kinase activity, which leads to decreased inhibitory phosphorylation on CDK1 and CDK2 through activation of the CDC25A phosphatase, also causes increased origin firing (19, 34).
We observed that replication fork speeds were decreased following WEE1 inhibition; however, these effects were rescued by CDK inhibition or depletion of CDT1. Thus, the stalled replication in response to WEE1 inhibition occurred as a downstream consequence of the increased initiation. Interestingly, codepletion of the replication factor CDC7 or roscovitine treatment partly rescued the reduced fork speed that was observed in response to CHK1 inhibition (34). However, following CHK1 inhibition, fork speeds are not completely rescued by roscovitine treatment or by codepletion of the replication factor CDC7 (34). A difference between WEE1 and CHK1 may be that CHK1 has multiple additional targets that may be important for fork progression. In agreement with this, we found that DSB formation following CHK1 inhibition could not be significantly rescued by supplying cells with nucleosides in the growth medium (data not shown). Taken together, this argues that whereas both WEE1 and CHK1 contribute to suppression of CDK activity to control replication initiation, CHK1 has additional targets that further promote fork progression and stability.
Nucleoside effects on replication stress.
Earlier work established the importance of nucleotide pools for normal DNA replication (1, 9, 35). Hydroxyurea inhibits ribonucleotide reductase, thereby lowering the dNTP pools, and this leads to low DNA replication rates, which is compensated for by the activation of dormant origins. We show that the nucleotide pools, as measured by dATP levels, are reduced by WEE1 inhibition, which is likely due to deregulated origin firing and an overall increase in DNA synthesis. We find it plausible that the decreased pools would further exacerbate the stressful situation by supporting the activation of additional dormant origins. In line with this, an increase of nucleoside supply had a significant impact on the perturbed replication and the DNA damage occurring after acute CDK activation. Interestingly, recent data from budding yeast suggested that increased origin firing in early S phase causes nucleotide shortage and subsequent fork stalling (29). Intriguingly, we found that exogenously supplied nucleosides had little effect on activation of ATR-dependent CHK1 phosphorylation. We also noticed that nucleoside addition could not fully rescue the fork rate. This suggests that a number of replication forks still stall and thus activate ATR under these conditions (8, 26). Hence, activation of the ATR pathway is likely not a symptom of nucleotide pool depletion alone but may be due to fork stalling following the enhanced initiation, depriving cells of a larger number of replication factors.
The observation that pannuclear γH2AX and massive DNA cleavage takes place with a 3- to 4-h delay implies that the replication forks are remodeled to reveal a substrate at this point. In addition, the nuclease activity may be stimulated by elevated CDK activity, as recently suggested (30). The 3- to 4-h delay coincides with the observation that there is impaired S-phase progression of a subset of cells that were inhibited from early S phase to late S phase, as well as the observation that these cells were almost exclusively the ones with high pannuclear γH2AX staining. This suggests that the DNA cleavage is delayed and that WEE1 inhibition early in S phase primes the cells for catastrophic widespread fork collapse and subsequent cleavage. The onset of the DSB/pannuclear γH2AX phenotype later in S phase may be related to rising levels of cyclins (or other cell cycle-dependent factors), allowing the CDK activity to surpass a threshold that activates the nuclease complex.
Potential relevance for tumorigenesis.
Genomic instability is a hallmark of cancer that contributes to the genetic changes underlying carcinogenesis. This instability can for example be caused by defective DNA damage repair and reactive oxygen species. DNA replication stress induced by oncogenes has recently been suggested as another model to explain the genomic instability of cancer (32). However, the mechanisms underlying oncogene-induced replication-associated DNA damage are largely unknown. Our results now support and further extend the recent work by Bester et al., who showed that oncogenes driving uncoordinated S-phase entry generate a situation with insufficient nucleotide pools for normal replication (5). Furthermore, the deregulation of CDK activity has been suggested to be an important event in the tumorigenesis process (25). In this respect, it is intriguing that prostate epithelium, which expresses low WEE1 levels, is prone to develop tumors (20). Altogether, we propose that enhanced CDK activity in early stages of cancer development forces cell proliferation and DNA replication, which can result in low nucleotide pools and DNA damage.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Danish Cancer Society, the Novo Nordisk Foundation, the Lundbeck Foundation, the Danish Medical Research Council, the Norwegian Cancer Society, the Norwegian Radium Hospital Research Foundation, the Research Council of Norway, and the South-Eastern Norway Regional Health Authority for their generous support of this work.
We thank Kay Schink for technical advice regarding microscopy.
Footnotes
Published ahead of print 20 August 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Anglana M, Apiou F, Bensimon A, Debatisse M. 2003. Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell 114:385–394 [DOI] [PubMed] [Google Scholar]
- 2. Bartkova J, et al. 2005. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434:864–870 [DOI] [PubMed] [Google Scholar]
- 3. Bartkova J, et al. 2006. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 444:633–637 [DOI] [PubMed] [Google Scholar]
- 4. Beck H, et al. 2010. Regulators of cyclin-dependent kinases are crucial for maintaining genome integrity in S phase. J. Cell Biol. 188:629–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bester AC, et al. 2011. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell 145:435–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Branzei D, Foiani M. 2010. Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell Biol. 11:208–219 [DOI] [PubMed] [Google Scholar]
- 7. Carpenter AE, et al. 2006. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7:R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cimprich KA, Cortez D. 2008. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 9:616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Courbet S, et al. 2008. Replication fork movement sets chromatin loop size and origin choice in mammalian cells. Nature 455:557–560 [DOI] [PubMed] [Google Scholar]
- 10. Di Micco R, et al. 2006. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 444:638–642 [DOI] [PubMed] [Google Scholar]
- 11. Dimitrova DS, Gilbert DM. 2000. Temporally coordinated assembly and disassembly of replication factories in the absence of DNA synthesis. Nat. Cell Biol. 2:686–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dominguez-Kelly R, et al. 2011. Wee1 controls genomic stability during replication by regulating the Mus81-Eme1 endonuclease. J. Cell Biol. 194:567–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fekairi S, et al. 2009. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell 138:78–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Forment JV, Blasius M, Guerini I, Jackson SP. 2011. Structure-specific DNA endonuclease Mus81/Eme1 generates DNA damage caused by Chk1 inactivation. PLoS One 6:e23517 doi:10.1371/journal.pone.0023517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hanada K, et al. 2007. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat. Struct. Mol. Biol. 14:1096–1104 [DOI] [PubMed] [Google Scholar]
- 16. Harrigan JA, et al. 2011. Replication stress induces 53BP1-containing OPT domains in G1 cells. J. Cell Biol. 193:97–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hirai H, et al. 2009. Small-molecule inhibition of Wee1 kinase by MK-1775 selectively sensitizes p53-deficient tumor cells to DNA-damaging agents. Mol. Cancer Ther. 8:2992–3000 [DOI] [PubMed] [Google Scholar]
- 18. Jørgensen S, et al. 2007. The histone methyltransferase SET8 is required for S-phase progression. J. Cell Biol. 179:1337–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Katsuno Y, et al. 2009. Cyclin A-Cdk1 regulates the origin firing program in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 106:3184–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kiviharju-af Hallstrom TM, et al. 2007. Human prostate epithelium lacks Wee1A-mediated DNA damage-induced checkpoint enforcement. Proc. Natl. Acad. Sci. U. S. A. 104:7211–7216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Krutzik PO, Nolan GP. 2006. Fluorescent cell barcoding in flow cytometry allows high-throughput drug screening and signaling profiling. Nature Methods 3:361–368 [DOI] [PubMed] [Google Scholar]
- 22. Labib K, De Piccoli G. 2011. Surviving chromosome replication: the many roles of the S-phase checkpoint pathway. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366:3554–3561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lam MH, Liu Q, Elledge SJ, Rosen JM. 2004. Chk1 is haploinsufficient for multiple functions critical to tumor suppression. Cancer Cell 6:45–59 [DOI] [PubMed] [Google Scholar]
- 24. Lambert S, Froget B, Carr AM. 2007. Arrested replication fork processing: interplay between checkpoints and recombination. DNA Repair 6:1042–1061 [DOI] [PubMed] [Google Scholar]
- 25. Lloyd AC, et al. 1997. Cooperating oncogenes converge to regulate cyclin/cdk complexes. Genes Dev. 11:663–677 [DOI] [PubMed] [Google Scholar]
- 26. Lopez-Contreras AJ, Fernandez-Capetillo O. 2010. The ATR barrier to replication-born DNA damage. DNA Repair 9:1249–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lopez-Contreras AJ, Gutierrez-Martinez P, Specks J, Rodrigo-Perez S, Fernandez-Capetillo O. 2012. An extra allele of Chk1 limits oncogene-induced replicative stress and promotes transformation. J. Exp. Med. 209:455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lukas C, et al. 2011. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat. Cell Biol. 13:243–253 [DOI] [PubMed] [Google Scholar]
- 29. Mantiero D, Mackenzie A, Donaldson A, Zegerman P. 2011. Limiting replication initiation factors execute the temporal programme of origin firing in budding yeast. EMBO J. 30:4805–4814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Matos J, Blanco MG, Maslen S, Skehel JM, West SC. 2011. Regulatory control of the resolution of DNA recombination intermediates during meiosis and mitosis. Cell 147:158–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Munoz IM, et al. 2009. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol. Cell 35:116–127 [DOI] [PubMed] [Google Scholar]
- 32. Negrini S, Gorgoulis VG, Halazonetis TD. 2010. Genomic instability–an evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 11:220–228 [DOI] [PubMed] [Google Scholar]
- 33. Parker LL, Piwnica-Worms H. 1992. Inactivation of the p34cdc2-cyclin B complex by the human WEE1 tyrosine kinase. Science 257:1955–1957 [DOI] [PubMed] [Google Scholar]
- 34. Petermann E, Woodcock M, Helleday T. 2010. Chk1 promotes replication fork progression by controlling replication initiation. Proc. Natl. Acad. Sci. U. S. A. 107:16090–16095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Poli J, et al. 2012. dNTP pools determine fork progression and origin usage under replication stress. EMBO J. 31:883–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Remus D, Diffley JF. 2009. Eukaryotic DNA replication control: lock and load, then fire. Curr. Opin. Cell Biol. 21:771–777 [DOI] [PubMed] [Google Scholar]
- 37. Roy B, et al. 1999. Simultaneous determination of pyrimidine or purine deoxyribonucleoside triphosphates using a polymerase assay. Anal. Biochem. 269:403–409 [DOI] [PubMed] [Google Scholar]
- 38. Sirbu BM, et al. 2011. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev. 25:1320–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sørensen CS, et al. 2005. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat. Cell Biol. 7:195–201 [DOI] [PubMed] [Google Scholar]
- 40. Sørensen CS, Syljuåsen RG. 2012. Safeguarding genome integrity: the checkpoint kinases ATR, CHK1 and WEE1 restrain CDK activity during normal DNA replication. Nucleic Acids Res. 40:477–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Svendsen JM, et al. 2009. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell 138:63–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Syljuåsen RG, et al. 2005. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol. Cell. Biol. 25:3553–3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tourriere H, Pasero P. 2007. Maintenance of fork integrity at damaged DNA and natural pause sites. DNA Repair (Amst.) 6:900–913 [DOI] [PubMed] [Google Scholar]
- 44. Tuduri S, et al. 2009. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nature Cell Biol. 11:1315–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vassilev LT, et al. 2006. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc. Natl. Acad. Sci. U. S. A. 103:10660–10665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wu CL, et al. 2001. Cables enhances cdk2 tyrosine 15 phosphorylation by Wee1, inhibits cell growth, and is lost in many human colon and squamous cancers. Cancer Res. 61:7325–7332 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.