Abstract
Selenium (Se) is an essential trace element used for biosynthesis of selenoproteins and is acquired either through diet or cellular recycling mechanisms. Selenocysteine lyase (Scly) is the enzyme that supplies Se for selenoprotein biosynthesis via decomposition of the amino acid selenocysteine (Sec). Knockout (KO) of Scly in a mouse affected hepatic glucose and lipid homeostasis. Mice lacking Scly and raised on an Se-adequate diet exhibit hyperinsulinemia, hyperleptinemia, glucose intolerance, and hepatic steatosis, with increased hepatic oxidative stress, but maintain selenoprotein levels and circulating Se status. Insulin challenge of Scly KO mice results in attenuated Akt phosphorylation but does not decrease phosphorylation levels of AMP kinase alpha (AMPKα). Upon dietary Se restriction, Scly KO animals develop several characteristics of metabolic syndrome, such as obesity, fatty liver, and hypercholesterolemia, with aggravated hyperleptinemia, hyperinsulinemia, and glucose intolerance. Hepatic glutathione peroxidase 1 (GPx1) and selenoprotein S (SelS) production and circulating selenoprotein P (Sepp1) levels are significantly diminished. Scly disruption increases the levels of insulin-signaling inhibitor PTP1B. Our results suggest a dependence of glucose and lipid homeostasis on Scly activity. These findings connect Se and energy metabolism and demonstrate for the first time a unique physiological role of Scly in an animal model.
INTRODUCTION
Selenium (Se) is an essential trace element acquired through the diet that has been implicated in brain (53), immune, and thyroid function (49), in fertility (2), and in cancer prevention (43). Dietary Se is found in inorganic or organic forms. Se is mostly utilized for biosynthesis of the unique amino acid selenocysteine (Sec), which is cotranslationally incorporated into selenoproteins (36), functioning primarily in redox reactions. The Sec incorporation mechanism involves de novo synthesis of Sec via selenophosphate (Se∼P), which is synthesized by selenophosphate synthetases (SPS) (60). Se∼P is enzymatically attached to the O-phosphoseryl-tRNA[Ser]Sec, which is then converted to the specific selenocysteyl-tRNA[Ser]Sec used in the selenoprotein translation (54, 61). Se is thought to enter the Se∼P pool for Sec biosynthesis either from diet or via recycling after selenoprotein degradation and release of Sec.
Selenocysteine lyase (Scly) is responsible for cellular Sec decomposition to mobilize Se for utilization in selenoprotein synthesis (10, 41). Scly was first isolated and characterized from pig liver (18) and subsequently shown to break down Sec into alanine and selenide (41). Scly has been the target of several in vitro studies: it was reported to interact with SPS (58), and its crystal structure revealed the mechanism for the enzyme reaction specificity toward Se (10, 46). In vivo, Scly was recently shown to be involved in selenoprotein biosynthesis in HeLa cells (30). However, the physiological role of Scly in cellular Se metabolism and in vertebrate whole-body Se homeostasis remains largely unknown.
To assess the role of Scly-mediated Se recycling in vivo, we characterized a whole-body Scly knockout (KO) mouse and examined the animal's physiology. Surprisingly, the Scly KO mice developed significant metabolic disturbances, such as hepatic steatosis, glucose intolerance, hyperinsulinemia, and hyperleptinemia when maintained on an adequate Se diet. These disturbances worsened when animals were exposed to a low dietary Se intake, and the mice developed obesity and hypercholesterolemia. Our results suggest that the observed detrimental metabolic effects are possibly a consequence of inefficient selenoprotein biosynthesis, leading to activation of de novo lipogenesis and attenuation of insulin signaling. Drawing from previous effects of Scly on selenoprotein biosynthesis (30) and in light of the growing evidence demonstrating the involvement of various selenoproteins in mammalian glucose metabolism (8, 20, 37, 39), including glutathione peroxidase 1 (GPx1) (33), selenoprotein S (SelS) (16), and Se-transporter selenoprotein P (Sepp1) (42), our results provide insight into the connection between metabolic energy regulation and overall Se metabolism through the previously unexplored Scly-mediated Se recycling pathway.
MATERIALS AND METHODS
Materials.
All reagents are from Sigma-Aldrich (St. Louis, MO), unless otherwise noted.
Scly KO mouse development.
C57BL/6J mice with deletion of exon 4 from the Scly gene (Scly KO) were generated by the KnockOut Mouse Project Repository, and detailed development of the mouse was described previously (47). Whole-body Scly KO animals were housed and bred in our facility's vivarium and genotyped by PCR of digested tail DNA prior to experiments. Primer sequences for genotyping are given in Table 1. Mice had their KO status further confirmed by quantitative PCR (qPCR) analysis in liver, lung, testis, and spleen tissues (data not shown).
Table 1.
PCR primer sequences used in this study
| Target | Primer sequence |
|
|---|---|---|
| Forward | Reverse | |
| 18S | CGATTGGATGGTTTAGTGAGG | AGTTCGACCGTCTTCTCAGC |
| ACC1 | AAATGACCCATCTGTAATGC | TCAATCTCAGCATAGCACTG |
| GAPDH | TGACATCAAGAAGGTGGTGAA | CCCTGTTGCTGTAGCCGTATTC |
| GLUT2 | GATCATTGGCACATCCTACT | ATCTTTGGTGACATCCTCAG |
| GLUT4 | CCCCAGATACCTCTACATCA | CATCCTTCAGCTCAGCTAGT |
| GPx1 | ACAGTCCACCGTGTATGCCTTC | CTCTTCATTCTTGCCATTCTCCTG |
| GPx4 | TCTGTGTAAATGGGGACGATGC | TCTCTATCACCTGGGGCTCCTC |
| HSL | TGGAACATCACTGAGATTGA | AGGGAGGTGAGATGGTAACT |
| LPL | AGGGCATGTTGACATTTATC | GGGAGTCAATGAAGAGATGA |
| LXRα | TTCTGTCAGAGCAAAGAGCCTC | ACTCCGTTGCAGAATCAGGAGA |
| PGC1α | AGCAGAAAGCAATTGAAGAG | AGGTGTAACGGTAGGTGATG |
| PPARα | GTGTACGACAAGTGTGATCG | TTTCAGATCTTGGCATTCTT |
| PPARγ | TTGATTTCTCCAGCATTTCT | TGTTGTAGAGCTGGGTCTTT |
| SBP2 | GAAGCAGAGGGAGATACCTAAGGC | GGCTCACAGCACTTTCTTGGAG |
| Scly | TCCACTCTATGGTAAGATGCT | CTGTGCAAGTGATAAAATGG |
| Scly KO genotyping | GAGATGGCGCAACGCAATTAAT | CTGGCTGTCCCTGAACTAGCTTCATA |
| Scly WT genotyping | CACAGGTGCGGCCATGAGGG | CTGGCTGTCCCTGAACTAGCTTCATA |
| SelK | TCCACGAAGAATGGGTAGGA | GCTTCTCAGAGCAGACATTTACCT |
| SelI | CCTGACATACTTCGACCCTGA | CAGTCAGGCACATGCTTATGA |
| SelM | TGACAGTTGAATCGCCTAAAGGAG | AACAGCACGAGTTCGGGGTC |
| SelR | TTCGTCCCTAAAGGCAAAGA | CATTCGCAGTCCATGTCCTA |
| SelS | CAGAAGATTGAAATGTGGGACAGC | CCTTTGGGGATGACAGATGAAGTAG |
| SelT | TGAGGCTCCTGCTGCTTC | GGTGGCGTACTGCATCTTTAAT |
| SelW | GGTGCCTCCCCAGAATCTAC | TGGGGGAATTCAGAGAGAGA |
| Sep15 | GCTGTCAGGAAGAAGCACAA | TTTTCATCCGCAGACTTCAA |
| Sepp1 | CCTTGGTTTGCCTTACTCCTTCC | TTTGTTGTGGTGTTTGTGGTGG |
| SOCS-3 | GCGAGAAGATTCCGCTGGTA | CCGTTGACAGTCTTCCGACAA |
| SPS1 | TGAACTGAAAGGCACAGGCTGC | CGCAAGTATCCATCCCAATGC |
| SPS2 | ACCGACTTCTTTTACCCCTTGG | TCACCTTCTCTCGTTCCTTTTCAC |
| TrxR1 | CCTATGTCGCCTTGGAATGTGC | ATGGTCTCCTCGCTGTTTGTGG |
| TrxR2 | GTTCCCCACATCTATGCCATTG | GGTTGAGGATTTCCCAAAGAGC |
| TrxR3 | CTTTGCAAGATGCCAAGAAA | TCATGGCCTCCCAGTTGT |
Animals.
Age-matched homozygous KO mice and wild-type (WT) homozygous littermates and/or C57BL/6J animals derived from The Jackson Laboratory (Bar Harbor, ME) were bred, born, and raised in our vivarium and used in experiments after weaning in accordance with the Institutional Animal Care and Use Committee of the University of Hawaii. Animals were euthanized by CO2 asphyxiation or anesthetized with tribromoethanol prior to collection of tissues or perfusion for histological procedures, respectively. Body weight (BW) was measured prior to sacrifice, and white adipose tissue (WAT) and liver weights were measured before tissues were frozen in liquid nitrogen. Female mice presented a milder phenotype than males and were excluded from analysis for the purpose of this report. All presented experimental data are from male mice, except for data for liver mRNA (Table 2), which include two females.
Table 2.
Expression of selenoproteins and selenoprotein synthesis factors in the liver, eWAT, and testis of age-matched WT and Scly KO mice fed a diet containing adequate Se
| Protein group and name | Relative expression (mean ± SEM) by tissue typea |
|||||
|---|---|---|---|---|---|---|
| Liver |
eWAT |
Testis |
||||
| WT | Scly KO | WT | Scly KO | WT | Scly KO | |
| Selenoproteins | ||||||
| GPx1 | 1.032 ± 0.4335 | 7.794 ± 0.942 | 0.116 ± 0.055 | 0.114 ± 0.039 | 0.033 ± 0.007 | 0.026 ± 0.006 |
| GPx4 | 0.304 ± 0.017 | 1.517 ± 0.197 | 0.446 ± 0.078 | 0.530 ± 0.077 | 2.961 ± 0.093 | 1.872 ± 0.243 |
| SelI | 0.015 ± 0.0006 | 0.057 ± 0.006 | 0.002 ± 0.001 | 0.002 ± 0.001 | 0.023 ± 0.004 | 0.028 ± 0.003 |
| SelK | 0.153 ± 0.018 | 0.212 ± 0.021 | 0.019 ± 0.003 | 0.026 ± 0.002 | 0.224 ± 0.047 | 0.183 ± 0.028 |
| SelM | 0.003 ± 0.0003 | 0.008 ± 0.0003 | 0.013 ± 0.004 | 0.009 ± 0.001 | 0.019 ± 0.002 | 0.020 ± 0.001 |
| SelR | 0.902 ± 0.081 | 1.716 ± 0.202 | 0.156 ± 0.02 | 0.169 ± 0.05 | 0.013 ± 0.004 | 0.017 ± 0.003 |
| SelS | 0.019 ± 0.0031 | 0.043 ± 0.005 | 0.002 ± 0.0004 | 0.006 ± 0.001 | 0.112 ± 0.017 | 0.133 ± 0.016 |
| SelT | 0.083 ± 0.014 | 0.285 ± 0.035 | 0.055 ± 0.021 | 0.041 ± 0.004 | 0.329 ± 0.046 | 0.285 ± 0.019 |
| SelW | 0.018 ± 0.003 | 0.031 ± 0.001 | 0.040 ± 0.01 | 0.019 ± 0.009 | 0.038 ± 0.006 | 0.033 ± 0.004 |
| Sep15 | 0.261 ± 0.045 | 1.267 ± 0.125 | 0.435 ± 0.068 | 0.313 ± 0.046 | 0.568 ± 0.081 | 0.678 ± 0.095 |
| Sepp1 | 11.15 ± 2.907 | 58.92 ± 4.329 | 1.333 ± 0.207 | 1.414 ± 0.423 | 0.384 ± 0.045 | 0.497 ± 0.039 |
| TrxR1 | 0.081 ± 0.019 | 0.283 ± 0.067 | 0.067 ± 0.013 | 0.046 ± 0.014 | 0.065 ± 0.009 | 0.112 ± 0.016 |
| TrxR2 | 0.005 ± 0.001 | 0.014 ± 0.002 | 0.002 ± 0.001 | 0.001 ± 0.0003 | 0.002 ± 0.001 | 0.002 ± 0.001 |
| TrxR3 | 0.021 ± 0.0033 | 0.037 ± 0.003 | 0.022 ± 0.004 | 0.017 ± 0.004 | 1.078 ± 0.156 | 0.538 ± 0.062 |
| Synthesis factors | ||||||
| SPS1 | 0.015 ± 0.003 | 0.049 ± 0.006 | 0.0052 ± 0.001 | 0.0054 ± 0.001 | 0.014 ± 0.0005 | 0.013 ± 0.001 |
| SPS2b | 0.273 ± 0.044 | 1.999 ± 0.219 | 0.0064 ± 0.001 | 0.0129 ± 0.003 | 0.348 ± 0.043 | 0.349 ± 0.067 |
| SBP2 | 0.005 ± 0.001 | 0.015 ± 0.001 | 0.0021 ± 0.0004 | 0.0021 ± 0.0007 | 0.018 ± 0.002 | 0.018 ± 0.002 |
Liver mRNA expression was normalized to 18S rRNA levels, and eWAT and testis values were normalized to GAPDH mRNA levels. Boldface indicates significant difference (P < 0.05, two-tailed unpaired t test) relative to WT values (n = 4 to 6).
SPS2 is also a selenoprotein.
Diets.
Animals were fed standard lab chow containing 0.25 to 0.3 ppm of Se. Diets formulated for specific Se content (Research Diets, Inc., New Brunswick, NJ) were as previously described (24) and contained 12% kcal of fat and 68% of kcal carbohydrate, plus 0.08 ppm of Se in casein (low) or were supplemented with sodium selenite to contain a total of 0.25 ppm (medium) or 1 ppm of Se (high). Food consumption was measured twice weekly for 2 months by weighing the leftovers of 100 g of chow supplied weekly into cages. Each cage had two to three mice, and consumption was averaged and analyzed per cage. Diet pellets had 3.90 kcal/g, and food intake in calories (kcal) was calculated by multiplying this factor by the total consumed food per week.
Se content.
Livers were prepared according to Exova Standard Operating Procedure 7040 for trace metals, and liver Se content was obtained by inductively coupled plasma mass spectrometry (ICP-MS) through comparison with internal standards at Exova Inc. (Santa Fe Springs, CA).
GPx activity assay.
Total serum or liver lysate GPx activity was measured by the coupled-enzyme procedure. Briefly, 1 μl of serum or 40 ng of liver protein was incubated at 37°C for 5 min in buffer containing 50 mM potassium phosphate, pH 7.4, 5 mM EDTA, 5 mM reduced glutathione, 5 μg/ml glutathione reductase, 266 μM NADPH, and 400 μM tert-butyl hydroperoxide. The reaction was read at 340 nm in a SpectraMax M3 spectrometer (Molecular Devices, Sunnyvale, CA), and results were calculated using SoftMax Pro5 software (Molecular Devices).
RNA analysis.
Total RNA was extracted with TRIzol (Invitrogen Life Technologies, Grand Island, NY) and an RNeasy Clean-Up kit (Qiagen, Valencia, CA) and reverse-transcribed (High Capacity cDNA Reverse Transcription; Life Technologies/Applied Biosystems, Carlsbad, CA), and 10 ng of cDNA was used in qPCR with SYBR green (Invitrogen Life Technologies). Results were calculated using the ΔΔ−CT method (where CT is threshold cycle), normalized to 18S rRNA or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA as housekeeping genes. Mouse selenoprotein PCR primers were described previously (25), and additional primer sequences used in this study are found in Table 1.
GTT.
Animals were fasted for 4 h prior to testing and injected with glucose at 1 mg/g of BW. We used strips and/or a glucometer (OneTouch Ultra; LifeScan, Milpitas, CA) for glucose tolerance test (GTT) measurements at time points 0, 30, 60, and 120 min after injection. The area under the curve (AUC) was calculated for individual mice and averaged.
Serum and liver measurements.
An enzyme-linked immunosorbent assay (ELISA) was used to measure serum insulin (Alpco Immunoassays, Salem, NH) and leptin (Crystal Chem Inc., Downers Grove, IL). Serum cholesterol was assayed by commercial kits (Cayman Chemical Company, Ann Harbor, MI, and Wako Pure Chemical Industries, Osaka, Japan) following the manufacturers' protocols, with the two kits showing comparable results in their controls. Hepatic triglycerides were extracted with 5% NP-40, followed by two rounds of 5-min incubations at 100°C, centrifugation at 15,000 × g for 2 min, and measurement with a triglyceride quantification kit (Abcam, Cambridge, MA). Insulin-degrading enzyme (IDE) activity was detected with 5 μg of liver total protein using an InnoZyme Insulysin/IDE Immunocapture Activity assay kit (Calbiochem, Darmstadt, Germany). Oxidative stress was determined by measuring lipid peroxidation end products using an OxiSelect HNE (4-hydroxynonenal)-His Adduct ELISA kit (Cell Biolabs, Inc., San Diego, CA), following the manufacturer's protocol.
Western blotting.
Liver tissues were pulverized and resuspended in CelLytic MT, sonicated, and centrifuged for 10 min at 12,000 × g, and protein supernatant was collected. WATs were homogenized in radioimmunoprecipitation assay (RIPA) buffer (15 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.1% SDS, 0.25% sodium deoxycholate), sonicated, and centrifuged for 5 min at 16,000 × g, and protein supernatant was collected. Samples consisting of 5 to 40 μg of total protein or 1 μl of serum were separated in 4 to 20% SDS-PAGE (Bio-Rad, Hercules, CA), transferred to Immobilon-FL polyvinylidene difluoride (IPFL) membranes (Millipore, Billerica, MA), and probed for 1.5 h with specific antibodies. Detection and analysis of Western blots were performed via an Odyssey Infrared Imager (Li-Cor Biosciences, Lincoln, NE).
Antibodies.
All commercial antibodies used were diluted according to the manufacturer's protocols and included mouse pACC1 Ser79, ACC1, pAkt Ser473, pAkt Thr308, Akt, AMP-activated protein kinase alpha (AMPKα), AMPKα phosphorylated at Thr172 (pAMPKα), peroxisome proliferator-activated receptor gamma (PPARγ), (Cell Signaling, Beverly, MA), GPx1, GPx3 (R&D Systems), insulin (Millipore, Billerica, MA), TATA-binding protein (TBP), PTP1B, IDE (Abcam), SelW, SPS2 (Rockland Inc., Gilbertsville, PA), α-tubulin, TrxR1 (Novus Biologicals, Littleton, CO), SelS, SelK, and β-actin antibodies (Sigma-Aldrich). Rabbit polyclonal Scly antiserum was a kind gift from Nobuyoshi Esaki, Kyoto University, and its development was previously described (30). The rabbit polyclonal Sepp1 antibody against the N-terminal region from mouse Sepp1 expressed in bacteria was produced by Proteintech Group Inc. (Chicago, IL). The first Sec codon (codon 59) of the N-terminal sequence of Sepp1 cDNA was mutated to a cysteine codon using a QuikChange II XL site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA), allowing bacterial synthesis of the Cys-mutant Sepp1 terminating at the second Sec. Purified protein from transformed bacteria was injected into rabbits, and the resulting antisera were affinity purified with the antigen. Antibody specificity was confirmed by Western blotting (data not shown) using liver total protein from Sepp1 KO and WT mice obtained from our colony and described previously (25).
Insulin challenge.
Phosphorylated proteins were detected by Western blotting after insulin challenge, as described previously (42). Briefly, animals were fasted for 4 h prior to injection with a sublethal dose of 10 mU/g of BW of human insulin (Humulin R; Eli Lilly). After 20 min, mice were sacrificed by CO2 asphyxiation, and their livers were collected. The fresh liver was homogenized in a Polytron homogenizer at 15,000 rpm in buffer containing 20 mM Tris, 5 mM EDTA, 10 mM Na4P2O7, 1% NP-40, 1% protease inhibitor cocktail (Sigma-Aldrich), and 1% phosphatase inhibitor cocktail (Pierce-Thermo Scientific, Rockford, IL). Tissue lysates were solubilized by continuous stirring for 1 h at 4°C and centrifuged for 60 min at 14,000 × g. Supernatants were electrophoresed as described above.
Immunohistochemistry.
Liver and pancreas tissues were paraffin embedded. Samples were deparaffinized for staining with hematoxylin and eosin (H&E), and pancreas sections were unmasked by pressure cooking with 0.01 M citric acid, pH 6, followed by 1 h of incubation with anti-insulin (1:100, Millipore), and labeled with a Mouse on Mouse Basic kit with Vectastain ABC and diaminobenzidine (DAB) peroxidase substrate kits (Vector Labs, Burlingame, CA).
Statistics.
Results were plotted and analyzed using Prism software (GraphPad Software Inc., La Jolla, CA). Applied statistical tests varied depending on experimental design and are indicated in the text or figure legends along with P values. All results are means ± standard errors of the means (SEM).
RESULTS
We here demonstrate the involvement of a putative cellular Se recycling enzyme in energy metabolism in an in vivo model. It has been suggested that Sec decomposition is a secondary path to provide Se for selenoprotein production, with diet being the primary path (48). Based on current knowledge of Scly function (30, 41), we predicted that genetic deletion of Scly would result in phenotypes similar to those associated with low dietary Se intake or disruption of Se transport (19, 23), such as male infertility and/or neuromotor impairments. Surprisingly, genetic disruption of the Scly gene did not impair embryonic survival, fertility, or development as Scly KO mice gave birth to viable offspring, without weight or size differences from birth to the juvenile stage compared to their wild-type (WT) counterparts (data not shown). In addition, Scly KO mice fed standard chow containing adequate Se levels had no major neurobehavioral dysfunction (47).
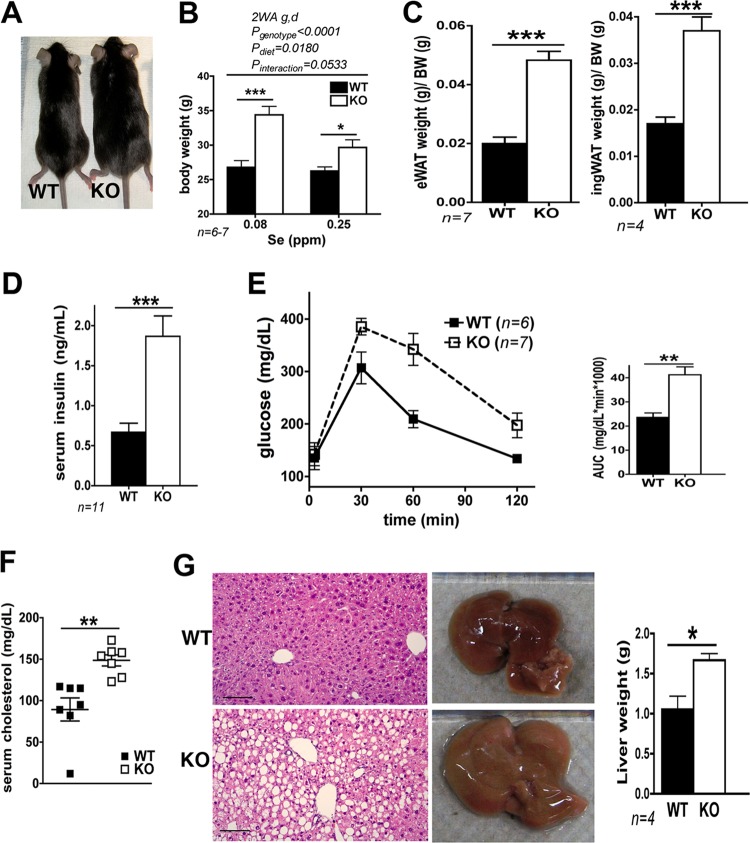
However, after the KO mice reached ∼2 months of age, differences in fat accumulation became apparent. The Scly KO mice had larger epididymal and inguinal white adipose tissues depots (eWAT and ingWAT, respectively) (Fig. 1A) and slightly impaired systemic glucose tolerance (Fig. 1B). Furthermore, histological analysis of Scly KO liver also revealed a subtle hepatic steatosis (Fig. 1C) with twice the amount of triglycerides in the liver tissue (Fig. 1D).
Fig 1.
Male Scly KO mice increase fat accumulation in the WAT and liver and are glucose intolerant and hyperinsulinemic. (A) Scly KO mouse weight of the epididymal (eWAT) and inguinal (ingWAT) fat depots. (B) Glucose tolerance test (GTT). The AUC was calculated for each mouse, averaged, and plotted as a bar graph. (C) Histological sections of the liver stained with H&E. (D) Hepatic triglyceride levels. (E) Serum insulin levels as measured by ELISA. (F) Immunohistochemistry of pancreatic β cells stained with anti-insulin. (G) Insulin-degrading enzyme (IDE) activity. (H) Hepatic levels of IDE visualized by Western blotting. Loading control, TATA-binding protein (TBP). *, P < 0.05; **, P < 0.01; ***, P < 0.001 (by a two-tailed unpaired t test). ns, not significant. Sample size is displayed in graphs. Scale bars, 100 μm (C) and 50 μm (F). Images in these panels represent typical histology from three to four animals/group.
Increased fat weight accompanied by glucose intolerance and hepatic steatosis are the classic characteristics of imbalanced lipid metabolism (9) and are often associated with the early stages of metabolic syndrome, which is additionally characterized by hyperinsulinemia. Therefore, we investigated if insulin function was disturbed in the Scly KO mice. In agreement with the fat accumulation, fasting serum insulin levels were almost 3-fold higher in the KO animals (Fig. 1E). The rise in serum insulin is likely a result of compensatory increased production responding to peripheral insulin resistance as we observed stronger insulin staining in pancreatic β cells of the KO mice (Fig. 1F). The liver is also the major site of insulin degradation, a reaction carried out by insulin-degrading enzyme (IDE). A failure in insulin degradation could generate the hyperinsulinemia phenotype. However, insulin degradation was not affected by disruption of Scly as we did not observe changes in either IDE activity (Fig. 1G) or protein levels in the liver (Fig. 1H). Interestingly, mice maintained normal fasting glucose levels (data not shown) even though hyperinsulinemia and glucose intolerance were present. Taken together, these results indicated that the Scly KO mice were in the early stages of metabolic syndrome.
To further investigate the metabolic events observed in the Scly KO mice, we determined mRNA levels of several transcription factors known to activate lipogenic pathways in the liver, such as peroxisome proliferator-activated receptor gamma and alpha (PPARγ and PPARα), liver X receptor alpha (LXRα), and PPARγ coactivator 1 alpha (PGC1α), as well as major coordinator of hepatic lipogenesis acetyl coenzyme A (acetyl-CoA) carboxylase 1 (ACC1) and hepatic glucose transporter (GLUT2). The transcription factors were upregulated in the liver of Scly KO mice (Fig. 2A), while absence of Scly did not affect the expression of ACC1 or GLUT2 (Fig. 2A). In accord, PPARγ protein levels were higher in the liver of Scly KO mice (Fig. 2B).
Fig 2.
Hepatic expression of genes that regulate energy metabolism and effects on protein levels of Scly KO mice fed an adequate-Se diet (0.25 to 0.3 ppm). (A) qPCR analysis of nuclear receptors and coactivators PPARα, PPARγ, LXRα, and PGC1α, as well as lipogenic enzyme ACC1 and glucose transporter GLUT2. (B) PPARγ protein levels. (C) Se regulation of hepatic Akt phosphorylation in WT mice. (D) Phosphorylation levels of pAkt Ser473, pAkt Thr308, and total Akt after insulin challenge. (E) Protein levels of pAMPKα Thr172 and total AMPKα. (F) Protein levels of pACC1 Ser79 and ACC1. All protein graphs are expressed in arbitrary units (au) and normalized by levels of β-actin or TBP. Protein blots in panels D and E are representative of four animals. ns, not significant. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (by two-tailed unpaired t test, except in panel C, where one-way analysis of variance [1WA] was employed).
Scly KO mice were developing systemic disturbances on glucose and lipid homeostasis. To determine if KO mice were developing impairment of insulin signaling and/or lipid metabolism, we challenged animals fed standard chow with a sublethal dose of insulin and subsequently measured the phosphorylation levels of Akt, the major coordinator of downstream effects of insulin signaling, and AMP kinase alpha (AMPKα), the major coordinator of lipid metabolism in the liver. We determined that Akt phosphorylation was not affected by Se levels in the liver (Fig. 2C) but that Scly KO mice had decreased levels of Akt phosphorylation on serine-473 (pAkt Ser473) and on threonine-308 (pAkt Thr308) in liver (Fig. 2D). These changes are implicated in the attenuated insulin response. Hepatic steatosis and hyperinsulinemia have been linked at the molecular level to increased levels of hepatic ACC1 (27, 59, 66), which is a critical enzyme for de novo lipogenesis. In this process, ACC1 is inactivated by AMPKα phosphorylation. As a result, ACC1 is unable to catalyze malonyl-CoA production, a crucial step for fatty acid biosynthesis in the liver (59). Interestingly, with adequate Se intake, phosphorylation levels of AMPKα on threonine-172 (pAMPKα Thr172) were unaffected as were its levels in the Scly KO mice (Fig. 2E). Additionally, neither ACC1 levels nor phosphorylation on serine-79 (pACC1 Ser79) was increased (Fig. 2F).
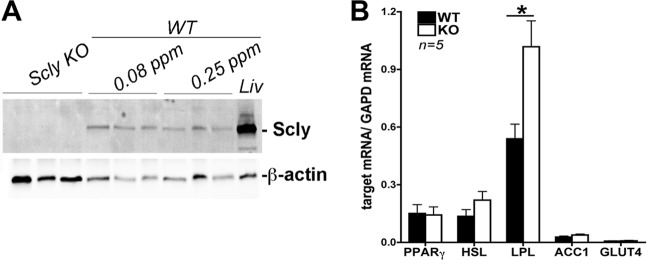
We then determined if disruption of Scly could in fact be interfering with Se and/or the energy metabolism of WAT. Scly had not been previously reported in any murine fat depot, and we here demonstrate its presence for the first time in the eWAT of adult WT mice and show that its expression is not regulated by Se concentration (Fig. 3A). Moreover, its levels in eWAT are much lower than in the liver, where the majority of Scly activity is found (18). Interestingly, qPCR analysis of genes involved in lipid and glucose metabolism regulation revealed that PPARγ, ACC1, hormone-sensitive lipase (HSL), and WAT-glucose transporter GLUT4 levels were unchanged in eWAT. Only lipoprotein lipase (LPL) mRNA levels were upregulated (Fig. 3B), possibly as a mechanism to compensate for the increases in plasma triglycerides (14, 50).
Fig 3.
Epididymal WAT expresses Scly but does not change the expression levels of metabolic genes. (A) Scly levels are not regulated by Se concentration in the eWAT of WT mice. The amount of Se in the diet is indicated (ppm); 40 μg of eWAT total protein was loaded in the gel, while only 10 μg of liver (Liv) total protein was used. (B) mRNA expression levels of metabolic genes PPARγ, hormone-sensitive lipase (HSL), ACC1, and GLUT4 are maintained, while lipoprotein lipase (LPL) expression is increased in the eWAT of Scly KO mice. *, P < 0.05 (two-tailed unpaired Student's t test).
Because Scly is involved in Sec decomposition to salvage Se for selenoprotein production (30), we investigated whether Se levels are altered in the Scly KO mice, thus affecting selenoprotein expression. Plasma glutathione peroxidase 3 (GPx3) and Sepp1 are the main carriers of Se in the bloodstream and are generally acknowledged as biomarkers for Se in the body (11, 22). In the serum of KO animals fed an adequate Se diet (0.25 to 0.3 ppm), GPx3 levels (Fig. 4A), GPx activity (Fig. 4B), and Sepp1 levels (Fig. 4C) did not differ from those of WT animals. Interestingly, we detected a subtle, but significant, ∼10% decrease in Se content in the liver of Scly KO mice even though they were fed the same Se-adequate diet as WT mice (Fig. 4D). These data indicate that the slight reduction in Se in the liver of Scly KO mice does not affect circulating Se or generate whole-body Se deficiency.
Fig 4.
Se levels in the Scly KO mice fed an Se-adequate diet. (A) Western blot of serum GPx3 protein and its relative quantification. (B) Plasma GPx activity assay. (C) Sepp1 levels in the serum measured by Western blotting. Ponceau S staining of blots was used to verify even loading (bottom). (D) Liver Se content measured by ICP-MS. Each square represents one individual animal. The P value was calculated by a two-tailed unpaired t test. ns, not significant.
The fact that the KO mice presented histological changes concomitant to a decrease in Se content in the liver, the main site of Scly expression and function in the body (15), led us to hypothesize that the diminished hepatic Se availability for selenoprotein production could cause a decrease in selenoprotein levels. To test this, we screened liver, eWAT, and testis tissues of Scly KO mice fed a diet containing adequate levels of Se (0.25 to 0.3 ppm of Se) for the expression of several selenoprotein mRNAs, including GPx1, GPx4, thioredoxin reductase 1 (TrxR1), TrxR2, TrxR3, SelI, SelK, SelM, Sepp1, SelR, SelS, SelT, SelW, and the 15-kDa selenoprotein (Sep15). The analysis revealed that most selenoprotein mRNAs were upregulated in the liver but remained unchanged in the testis and eWAT of Scly KO mice (Table 2). Furthermore, selenoprotein synthesis factors SPS1 and SPS2 (58) and SECIS-binding protein (SBP2) (12) mRNAs were also upregulated in the liver (Table 2). Interestingly, SPS2 and SelS were also upregulated in the eWAT, suggesting modulation of their gene expression by Scly activity. Nevertheless, hepatic selenoprotein levels were mostly maintained (Fig. 5A), except for a 10 to 15% decrease in SelS protein levels. Despite normal selenoprotein levels, Scly KO mice presented diminished antioxidant GPx activity (Fig. 5B) with increased hepatic lipid peroxidation (Fig. 5C), indicating that the liver of Scly KO mice had a compromised antioxidant response in comparison to liver from their WT counterparts. The fact that GPx activity was diminished led to the conclusion that, despite normal protein levels, selenoproteins may not be efficiently translated. These results suggest that disruption of Scly may modulate, at least partially, overall efficiency of selenoprotein expression.
Fig 5.

Selenoprotein profile in the liver of Scly KO mice fed an adequate Se diet. (A) Expression of selenoproteins SelS, SelK, TrxR1, SelW, GPx1, and SPS2 in Scly KO mice measured by Western blotting and normalized to the β-actin or α-tubulin level, depending on the blot. (B) Total GPx activity in the liver of Scly KO mice. (C) ELISA results for HNE adduct formation as an indicator of lipid peroxidation and oxidative stress levels. Sample size is displayed in the graphs. *, P < 0.05; **, P < 0.01(two-tailed unpaired t test). au, arbitrary units.
Hyperinsulinemia, decreases in hepatic Akt phosphorylation, and increases in oxidative stress were accompanied by subtle glucose intolerance in these animals. Possibly, the mice were in the early stages of metabolic syndrome, with pancreatic insulin overproduction compensating for the imbalance in glucose metabolism. To test whether the dietary Se levels were masking Scly-dependent disturbances in energy metabolism, we further challenged Se metabolism by feeding Scly KO mice a low-Se diet (0.08 ppm Se) containing equal amounts of fat (12% kcal) and carbohydrate (68% kcal) for 3 months after they were weaned. We hypothesized that, on a low-Se diet, animals would rely more on Scly-mediated Se recycling mechanisms for maintenance of Se homeostasis, and thus the KO would display a stronger phenotype. As expected, under these circumstances and compared to age-matched WT counterparts on the same diet, Scly KO mice developed obesity (Fig. 6A), with increased body weight (Fig. 6B), increased fat depot weights (Fig. 6C), worsening hyperinsulinemia (Fig. 6D) and glucose intolerance (Fig. 6E), increased serum cholesterol (Fig. 6F), and aggravation of hepatic steatosis (Fig. 6G, left panels), with a concomitant increase in liver size (Fig. 6G, right panels and graph). In summary, disruption of Scly accompanied by dietary Se deficiency compounded and worsened the metabolic syndrome phenotype observed in Scly KO mice on an adequate Se diet.
Fig 6.
Scly KO mice fed a diet containing 0.08-ppm of Se for 2.5 months develop obesity, hepatic steatosis, and hypercholesterolemia and worsen their hyperinsulinemia and glucose intolerance. (A) Photograph of age-matched KO mice and WT counterparts. (B) Body weight measurements of Scly KO and WT mice upon sacrifice. 2WA g,d, two-way analysis of variance of genotype and diet. (C) Epididymal (eWAT) and inguinal (ingWAT) fat depot weights. (D) Fasting serum insulin levels. (E) Glucose tolerance test. (F) Fasting serum cholesterol levels. (G) Liver histology following H&E staining of hepatocytes under optical microscopy left) and representative photograph of the liver showing its size (right). Liver weight is displayed as a bar graph. Scale bar, 100 μm. The image is representative of three mice. Sample size is displayed in graphs. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (two-tailed unpaired t test).
The increase in fat deposition in Scly KO animals on a low-Se diet occurred with a subtle (∼15%) alteration in food consumption (Fig. 7A). Interestingly, leptin levels were found to be ∼4-fold higher than in WT mice (Fig. 7B) and ∼5-fold higher in Scly KO mice on an Se-adequate diet than in WT mice. An independent effect of Se deficiency on leptin levels in WT mice was observed (Fig. 7B), which may be due to increased stress accompanying Se deficiency, as found previously in rats administered adrenocorticotropin hormone (6). Despite the >3-fold increase in circulating leptin levels when mice were fed a low-Se diet, these WT mice did not become overweight or obese and did not exhibit signs of metabolic impairment, the primary characteristics correlated with increased leptin levels. Increased circulating leptin leads to leptin resistance in specific areas of the brain where the presence of the leptin receptor controls feeding behavior and satiety. These areas are also known to be important for metabolic regulation, such as the choroid plexus (40) and ventromedial hypothalamus (34). To further corroborate the localized, hepatic effects of Scly disruption, immunostaining of the leptin receptor in these areas of the brain was performed. Scly KO mice fed an Se-adequate diet showed no major difference in leptin receptor staining compared to WT mice (data not shown) even though leptin levels are already increased in circulation in these animals (Fig. 7B). Additional qPCR analysis revealed that the expression of the leptin receptor inhibitor protein, SOCS-3 (1), was also unchanged in the brain but upregulated in the liver of Scly KO mice fed adequate Se (Fig. 7D). Our findings indicate that leptin signaling is activated in liver but not in the brain. The overproduction of leptin could also be a consequence of increased adiposity in WAT, the main site of leptin production, thus contributing toward the unbalanced glucose homeostasis observed in the Scly KO mice.
Fig 7.

Leptin in Scly KO mice. (A) Food consumption measured weekly in animals fed a low-Se diet. The AUC was calculated for individual mice, averaged, and plotted as graph bar (n = 10 animals, housed in four cages). (B) Serum leptin levels in WT and Scly KO mice after being fed on diets containing 0.08 and 0.25 ppm of Se. Pint, P for interaction by two-way analysis of variance. (C) Gene expression of leptin receptor signaling inhibitor SOCS-3 in the brain and in the liver. Sample size is displayed in graphs. RQ, relative quantification. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (two-tailed unpaired t test).
Upon maintenance on a low-Se diet, the levels of the lipid metabolism coordinator AMPKα significantly increased in the liver of Scly KO mice (Fig. 8A). Surprisingly, its phosphorylation levels in fact decreased (Fig. 8A), which explains the sharp upregulation in ACC1 mRNA (Fig. 8B) and the direct increase in phosphorylation of ACC1 (Fig. 8C) we observed in the livers of Scly KO mice. At least under conditions of low Se availability, Scly disruption was affecting ACC1 activation, thus promoting lipogenic pathways in the liver. In addition, in mice fed a low-Se diet, we investigated the levels of insulin signaling inhibitor protein phosphatase 1B (PTP1B). PTP1B dephosphorylates the insulin receptor and the insulin receptor substrate, disrupting the phosphorylation cascade. PTP1B activity and levels are regulated by Se levels (44). Scly KO mice on an adequate-Se diet presented slightly higher levels of PTP1B than WT mice, and these levels increased 3-fold when the mice were fed a low-Se diet (Fig. 8D). Thus, the effect of Scly disruption in insulin signaling may be directly connected to PTP1B.
Fig 8.

Phosphorylation and protein levels of AMPKα, ACC1, and PTP1B in the liver of Scly KO mice fed a low-Se diet (0.08 ppm). (A) Phosphorylation and protein levels of AMPKα in Scly KO mice liver on different Se diets. Although AMPKα levels are increased, phosphorylation is, in fact, slightly decreased. (B) ACC1 mRNA levels are upregulated in Scly KO mice on a low-Se diet. (C) Phosphorylation of ACC1 is mildly increased in the liver of Scly KO mice without changes in ACC1 total levels. (D) PTP1B levels are increased in Scly KO mice on a diet containing 0.25 ppm of Se, and it further increases in a low-Se diet. Phosphorylation was measured in mice challenged with insulin. TBP and β-actin levels were used as loading controls for blots. Sample size is displayed in the graphs, and comparison was performed by two-way analysis of variance (A and D) and by a two-tailed unpaired t test (B and C). *, P < 0.05; **, P < 0.01; ns, not significant.
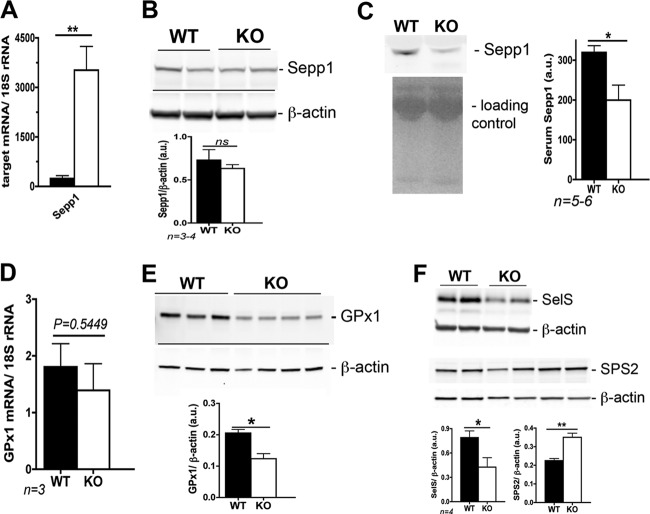
In terms of Se involvement in metabolic syndrome, levels of plasma Sepp1 have been shown to be elevated in human patients with type 2 diabetes (42, 64), a pathology closely related to metabolic syndrome (17). Conversely, Sepp1 KO mice were reported to have improved glucose tolerance and insulin sensitivity in response to a high-fat diet containing similar, adequate Se levels (42). The Scly KO mice had an ∼3-fold upregulation of hepatic Sepp1 mRNA levels even when fed an adequate-Se diet (Table 2), and this increase was even higher (∼10-fold) with low dietary Se intake (Fig. 9A). However, Sepp1 protein levels in the liver did not change (Fig. 9B). At least for the specific case of Sepp1, the mRNA upregulation we detected was not followed by increases in protein expression. Surprisingly, levels of Sepp1 decreased in the serum of KO animals (Fig. 7C).
Fig 9.
Expression of selenoproteins involved in glucose metabolism in Scly KO mice upon a low dietary Se intake (0.08 ppm). (A) Hepatic Sepp1 mRNA levels. (B) Sepp1 protein expression in the liver of Scly KO mice. (C) Serum levels of Sepp1 in Scly KO mice. (D) GPx1 mRNA levels. (E) GPx1 protein levels. (F) SelS and SPS2 protein levels. Protein quantification is shown close to the respective blot, and levels are expressed as arbitrary units (au). *, P < 0.05; **, P < 0.01 (two-tailed unpaired t test).
The selenoprotein GPx1 has also previously been reported to be involved in glucose metabolism when overexpressed in the liver (38). Glutathione from GPx activity has also been shown to regulate PTP1B activity (45). We observed an increase in GPx1 mRNA levels in the liver of Scly KO mice fed an adequate-Se diet (Table 2), but such increase was not accompanied by its protein levels (Fig. 5A). Under low dietary Se intake, GPx1 mRNA was not increased in KO mice (Fig. 9D). Nevertheless, GPx1 protein levels decreased (Fig. 9E), in spite of the metabolic syndrome phenotype. Moreover, SelS protein levels were also decreased in the Scly KO mice upon low dietary Se intake (Fig. 9F) compared to WT mice on the same diet. Surprisingly, SPS2 enzyme, which was demonstrated previously to bind to Scly in vitro (58) and accepts selenide, a reduced selenite, for Se∼P production (57), was increased in the liver of Scly KO mice (Fig. 9F). Such increase suggested a direct role of SPS2 levels on Se mobilized via Scly activity.
DISCUSSION
Diet is the main source of Se, provided Se intake is adequate. When dietary Se levels are insufficient, recycling of Se at the expense of some proteins or tissues to prioritize the Se supply where it is most essential (e.g., brain and testes) may become prominent. Mechanisms for Se prioritization likely evolved in concert with exposure to limited Se availability, allowing organisms to prevent deleterious effects of Se deficiency. We report for the first time the unique physiological role of the putative Se recycling enzyme Scly in vivo in liver metabolism, particularly as a key player for the cross talk between Se and energy metabolic pathways (Fig. 10).
Fig 10.
Schematic view of possible role of Scly in glucose and lipid metabolism in the liver. Scly decomposes Sec (yellow circles) obtained from selenoprotein degradation into alanine (ala) and the Se form selenide. This Se will reenter the selenoprotein synthesis pathway by the action of SPS enzymes. Selenoproteins are mostly involved in cell detoxification reactions, such as breakdown of hydrogen peroxide (H2O2). Oxidative status influences the activity of insulin signaling inhibitor protein PTP1B, a phosphatase that disrupts the insulin signaling cascade by dephosphorylating the insulin receptor (IR) and its substrate (IRS). This results in attenuation of downstream events regulated by Akt, such as gene transcription, glucose uptake, and lipogenesis. Se from Scly activity regulates PTP1B levels (dashed arrow), favoring the negative regulation of insulin signaling.
By disrupting the Scly gene in a mouse, we unveiled a major effect on lipid accumulation in fat depots and liver. As liver is the major site of Scly production and function (18), a hepatic effect was not entirely unexpected. In addition to liver, Scly is expressed at high levels in testis and kidney (30). These tissues also express significant amounts of Sepp1 and/or rely on Sepp1 as an Se source (3). Given the similar expression profiles of Sepp1 and Scly and their roles in Se transport/storage and Sec decomposition, respectively, we first hypothesized that Scly KO mice would display a phenotype similar to that of the Sepp1 KO mice, including reproductive impairment (3) and neurological abnormalities, such as seizures and impaired mobility (5, 52). Se deficiency can also result in developmental and neurological disorders (7), and Sepp1 acts as a carrier to bring Se to brain and other body regions. Nevertheless, we observed neither neurological (47) nor fertility impairment in Scly KO mice on adequate Se intake. In contrast to the metabolic phenotype reported here for Scly KO mice, Sepp1 KO mice are smaller than their WT counterparts and have increased Se content in the liver (52), with improved glucose tolerance and enhanced insulin sensitivity (42). The fact that the Scly KO mouse characteristics are the opposite of those of the Sepp1 KO mice is revealing of the distinct roles and requirements of Scly and Sepp1 in Se metabolism.
Both Scly and Sepp1 are produced primarily in the liver. Sepp1 is largely secreted into the bloodstream, where it transports Se to other organs of the body in the form of Sec residues (4). Sepp1 is a unique protein, containing 10 Sec residues in rodents and humans, most of them clustered in the C-terminal region of the protein. We hypothesized that disruption of Scly might decrease Sepp1 turnover, resulting in elevated levels of this protein, but that was not observed. Nor were the levels of other selenoproteins affected, with the exception of SelS. This suggests that Se derived from Sec decomposition is less utilized for selenoprotein translation when dietary Se is adequate. The lack of changes in these proteins possibly indicates that dietary Se is overcoming the absence of Scly to some extent inside the liver. Since the SPS2 enzyme has been reported to assimilate selenite from external sources (57), thus circumventing the Scly-mediated Se recycling step, the maintenance of SPS2 levels in Scly KO mice could explain the milder phenotype observed under adequate Se intake and the absence of selenoprotein changes. It is also possible that, although levels of selenoproteins are maintained, these proteins may be not incorporating Sec properly, as previously described for TrxR1 in Se deficiency (62). A shift in proper selenoprotein translation toward a less efficient mechanism could lead to elevated oxidative stress. This is observed in the liver of Scly KO mice upon insulin challenge, indicating possibly that, even though levels of selenoproteins are maintained, Sec incorporation may be affected. It could explain why, even without major direct impairments on selenoprotein levels, Scly KO mice displayed a metabolic phenotype when dietary Se levels were adequate. Interestingly, SelS, which has been shown to be regulated by glucose and associated with plasma lipids (20), was found to be decreased in the liver of Scly KO mice, while its mRNA levels were increased, suggesting a major dependence of SelS expression on Se decomposed by the Scly pathway. Scly may also be implicated in tissue-specific regulation of selenoprotein gene transcription since we observed upregulation of virtually all selenoprotein mRNAs in the liver of Scly KO mice, and only a few in eWAT and testis.
Under conditions of low dietary Se, plasma Sepp1 and several hepatic selenoproteins have been shown to be decreased in WT mice (63). To our surprise, low Se intake results in a further decline in selenoprotein translation of plasma Sepp1, hepatic GPx1, and SelS in the Scly KO mice compared to levels in WT mice on the same diet. Interestingly, SPS2 levels were augmented in the livers of these KO mice. Whether SPS2 partially compensates for the low-Se environment remains to be explored.
Previous studies have linked hepatic selenoprotein GPx1 overexpression (32, 38) or increased Sepp1 levels (42) to hyperinsulinemia, hyperglycemia, and insulin resistance. Instead, Scly KO mice showed decreases in hepatic GPx1 and circulating Sepp1 upon low dietary Se intake, when the metabolic phenotype was more prominent. The fact that the Scly KO mice on an Se-limiting diet develop several criteria matching a metabolic syndrome phenotype (e.g., obesity, hyperinsulinemia, hyperleptinemia, glucose intolerance, hypercholesterolemia, and hepatic steatosis), despite decreased levels of Sepp1 in serum, suggests that energy homeostasis in our model may rely on the proper oxidative stress response and on the action of other selenoproteins besides Sepp1. Decreases in GPx activity have also been shown to play a direct role in the regulation of PTP1B, the major inhibitor of the insulin receptor and insulin receptor substrate phosphorylation (45). Binding to glutathione inhibits PTP1B activity, thus allowing the insulin signaling to properly act on its targets. Since we observed attenuation of Akt signaling upon insulin challenge, it is possible that PTP1B is more active in inhibiting the insulin signaling in the liver of Scly KO mice. Because PTP1B expression is regulated by Se levels (44), it is also possible that the hepatic Se decline in Scly KO mice is driving the expression of PTP1B, suggesting that Se derived from Sec decomposition participates in PTP1B regulation. Nevertheless, less GPx activity affects glutathione availability for inhibition of PTP1B activity, favoring the insulin signaling and hepatic lipogenic mechanisms via activation of sterol regulatory element binding protein 1c (SREBP-1c) (45). The observed conflicting effect of Se deficiency and PTP1B levels suggest that additional factors may play a role in the PTP1B dependence on glutathione and Se.
In metabolic terms, it remains to be further investigated if Scly disruption affects primarily pancreatic insulin production and secretion, similar to what is observed upon disruption of the selenoprotein Dio3 gene (39) or upon overexpression of GPx1 (33). We demonstrated that insulin degradation is unchanged in the liver of Scly KO mice (Fig. 1G and H); thus, we ruled out this pathway as a cause for the Scly KO metabolic phenotype, as is the case with other KO mouse models that display hepatic steatosis (21). Alternatively, the presence of the fat droplets in the liver suggests a direct effect on hepatic de novo lipogenesis. Previous studies have shown Se supplementation as a major regulator of AMPKα phosphorylation in colon cancer cells (26) and skeletal muscle of diabetics rats (28). In addition, Sepp1 was pinpointed as the effector protein in the cross talk between Se and lipid metabolism in mice via action upon AMPKα phosphorylation (42). Our data demonstrate that Sepp1 levels depend at least to some extent on the presence of Scly when Se availability is scarce. However, our results of unchanged AMPKα phosphorylation with an adequate-Se diet contrast with the reported role of Sepp1 levels in regulating hepatic lipid biosynthesis. This contrast could be attributed to gender effects on Sepp1 expression (51) and hepatic Se content (65) in rodents. Our study was performed with male mice in contrast to the use of only female mice to establish the involvement of Sepp1 on AMPKα phosphorylation. Under conditions of low dietary Se, however, AMPKα phosphorylation was decreased, leading to increases in ACC1 phosphorylation, an effect that enhances lipogenesis in the liver, and this effect occurred despite the lower circulating Sepp1 levels. The phosphorylation pattern of AMPKα under conditions of Se scarcity could possibly be a direct effect of Scly activity in a lipogenic step that is enhanced by Se deprivation.
Scly KO mice on low-Se intake had high circulating leptin levels and had increased food consumption, indicating that obesity driven by Se deficiency in mice lacking Scly involved leptin-dependent signaling in the liver. However, it should be noted that the elevated food intake in the Scly KO mice is not maintained when the number of calories ingested is normalized to the body weight of individual mice. This discrepancy fosters the possibility that the increase in food intake is a consequence of increases in fat depots and, thus, enhanced leptin production and not a direct effect of the Scly in feeding behavior as no major changes were observed in the brain. Despite these possibilities, leptin overload in livers of Scly KO mice is accompanied by increases in the levels of its signaling inhibitor, the protein SOCS-3, which strengthens the possibility that the rise in leptin is a consequence of adiposity and not a direct effect of Scly disruption.
Interestingly, PPARγ, a member of the nuclear steroid hormone receptor family whose expression is regulated by changes in glucose and lipid homeostasis, is increased in the liver of Scly KO mice (Fig. 2C) but not in the eWAT. Moreover, expression of PGC1α, the coactivator of PPARγ, is also increased in the liver of Scly KO mice under conditions of Se adequacy but not in brown adipose tissue (data not shown), an organ deeply involved in control of energy metabolism and protection from diet-induced obesity (13, 42). PGC1α was previously recognized as a key regulator of Se homeostasis (35, 55), and its localized increase in the liver further corroborates this organ as the origin of the lipid imbalances upon disruption of Scly, with target organs such as eWAT only sensing and responding to the consequences of the primary effect in the liver. An animal transgenic model with disruption of Scly in target tissues could help further corroborate this idea and may reveal additional pathways where Scly is important in our body.
The results presented here could improve our understanding of the controversial influence of Se supplementation on energy metabolism. Until recently, Se was considered a safe supplement that could potentially prevent several diseases in humans. However, the Selenium and Vitamin E Cancer Prevention Trial (SELECT) (29) raised significant questions about Se supplementation. The SELECT revealed that subjects who received high doses of supplemental Se presented a trend, although not significant (P = 0.16), toward increased risk of glucose intolerance. Nevertheless, a 10-year follow-up analysis of SELECT participants revealed both a lack of influence of Se supplementation in the outcome of type 2 diabetes prevalence (31) and lack of effect on prostate cancer protection (35, 56). Even though the glucose intolerance trend was not significant, the result was enough to allow termination of the trial after 7 to 7.5 years. These findings underscore our limited understanding of Se regulation, specifically regarding its influence on energy metabolism. Our findings may shed light on the glucose effects observed in human participants of the SELECT, incorporating the Scly-mediated Se recycling pathway into the overall complex picture of Se regulation of energy metabolism. It is possible that the consequent obesity we observed in our model was not seen in the male subjects of the aforementioned trial because their Se intake was high enough to maintain the Scly-mediated recycling pathway in a slightly downregulated state but not sufficient for avoiding hyperinsulinemia, indicating a complex compartmentalization of Se action in the body. Further studies on the regulation of Scly expression by Se in specific tissues are necessary to test this possibility.
In conclusion, our data revealed that the Scly-mediated Se recycling pathway is involved in lipid and glucose metabolism. We also demonstrated that dietary Se deficiency exacerbates some of the lipid disturbances that Scly disruption causes. When challenged with a low-Se diet and in the absence of functional Sec decomposition, the KO mice developed obesity and metabolic syndrome, with increased hepatic lipogenesis and attenuation of insulin signaling. These data imply that Se metabolism has a significant role in the pathogenesis of obesity. They also revealed new insights into how the Scly-mediated Se recycling pathway could be nonessential for Se functions such as fertility and neurological function but necessary for balanced energy metabolism. We also provide in vivo evidence that Scly is an enzyme involved in the cross talk between Se and energy metabolic pathways, adding a new nutritional and molecular perspective for understanding the pathogenesis of metabolic syndrome.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants R01-DK47320, G12-MD007601, and G12-RR003061.
We thank Miyoko Bellinger from the Histology Core Facility; Jun Panee and Robert Nichols from University of Hawaii, Ann-Marie Zavacki from Brigham and Women's Hospital/Harvard Medical School, and Ashok Balasubramanyam from Baylor Medical College for valuable discussion; and Ashley Kong, Andrea Takemoto, and Elizabeth Nguyen-Wu for additional assistance.
L.A.S. designed and performed research, analyzed data, and wrote the manuscript. A.C.H. performed research, analyzed data, and contributed with analytical tools. S.K., A.S., C.L.G., and A.V.R. performed research and discussed data. F.P.B. contributed with analytical tools and discussed data. M.J.B. mentored in all research steps. All authors reviewed, edited, and discussed the manuscript.
We declare that we have no conflicts of interest.
Footnotes
Published ahead of print 13 August 2012
REFERENCES
- 1. Bjorbaek C, El-Haschimi K, Frantz JD, Flier JS. 1999. The role of SOCS-3 in leptin signaling and leptin resistance. J. Biol. Chem. 274:30059–30065 [DOI] [PubMed] [Google Scholar]
- 2. Boitani C, Puglisi R. 2008. Selenium, a key element in spermatogenesis and male fertility. Adv. Exp. Med. Biol. 636:65–73 [DOI] [PubMed] [Google Scholar]
- 3. Burk RF, Hill KE. 2009. Selenoprotein P-expression, functions, and roles in mammals. Biochim. Biophys. Acta 1790:1441–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Burk RF, Hill KE. 2005. Selenoprotein P: an extracellular protein with unique physical characteristics and a role in selenium homeostasis. Annu. Rev. Nutr. 25:215–235 [DOI] [PubMed] [Google Scholar]
- 5. Burk RF, et al. 2007. Deletion of apolipoprotein E receptor-2 in mice lowers brain selenium and causes severe neurological dysfunction and death when a low-selenium diet is fed. J. Neurosci. 27:6207–6211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chanoine JP, Wong AC, Lavoie JC. 2004. Selenium deficiency impairs corticosterone and leptin responses to adrenocorticotropin in the rat. Biofactors 20:109–118 [DOI] [PubMed] [Google Scholar]
- 7. Chen J, Berry MJ. 2003. Selenium and selenoproteins in the brain and brain diseases. J. Neurochem. 86:1–12 [DOI] [PubMed] [Google Scholar]
- 8. Clarke NF, et al. 2006. SEPN1: associated with congenital fiber-type disproportion and insulin resistance. Ann. Neurol. 59:546–552 [DOI] [PubMed] [Google Scholar]
- 9. Cohen JC, Horton JD, Hobbs HH. 2011. Human fatty liver disease: old questions and new insights. Science 332:1519–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Collins R, et al. 2012. Biochemical discrimination between selenium and sulfur 1: a single residue provides selenium specificity to human selenocysteine lyase. PLoS One 7:e30581 doi:10.1371/journal.pone.0030581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Combs GF, Jr, et al. 2011. Determinants of selenium status in healthy adults. Nutr. J. 10:75 doi:10.1186/1475-2891-10-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Copeland PR, Fletcher JE, Carlson BA, Hatfield DL, Driscoll DM. 2000. A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J. 19:306–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cypess AM, Kahn CR. 2010. The role and importance of brown adipose tissue in energy homeostasis. Curr. Opin. Pediatr. 22:478–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Davies BS, Beigneux AP, Fong LG, Young SG. 2012. New wrinkles in lipoprotein lipase biology. Curr. Opin. Lipidol 23:35–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Deagen JT, Butler JA, Beilstein MA, Whanger PD. 1987. Effects of dietary selenite, selenocystine and selenomethionine on selenocysteine lyase and glutathione peroxidase activities and on selenium levels in rat tissues. J. Nutr. 117:91–98 [DOI] [PubMed] [Google Scholar]
- 16. Du JL, et al. 2008. Association of SelS mRNA expression in omental adipose tissue with Homa-IR and serum amyloid A in patients with type 2 diabetes mellitus. Chin. Med. J. (Engl.) 121:1165–1168 [PubMed] [Google Scholar]
- 17. Eckel RH, et al. 2011. Obesity and type 2 diabetes: what can be unified and what needs to be individualized? J. Clin. Endocrinol. Metab. 96:1654–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Esaki N, Nakamura T, Tanaka H, Soda K. 1982. Selenocysteine lyase, a novel enzyme that specifically acts on selenocysteine. Mammalian distribution and purification and properties of pig liver enzyme. J. Biol. Chem. 257:4386–4391 [PubMed] [Google Scholar]
- 19. Flohe L. 2007. Selenium in mammalian spermiogenesis. Biol. Chem. 388:987–995 [DOI] [PubMed] [Google Scholar]
- 20. Gao Y, et al. 2007. Secretion of the glucose-regulated selenoprotein SEPS1 from hepatoma cells. Biochem. Biophys. Res. Commun. 356:636–641 [DOI] [PubMed] [Google Scholar]
- 21. Ghosh S, et al. 2010. Mice with null mutation of Ceacam I develop nonalcoholic steatohepatitis. Hepat. Med. 2010:69–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hill KE, et al. 2007. The selenium-rich C-terminal domain of mouse selenoprotein P is necessary for the supply of selenium to brain and testis but not for the maintenance of whole body selenium. J. Biol. Chem. 282:10972–10980 [DOI] [PubMed] [Google Scholar]
- 23. Hill KE, et al. 2003. Deletion of selenoprotein P alters distribution of selenium in the mouse. J. Biol. Chem. 278:13640–13646 [DOI] [PubMed] [Google Scholar]
- 24. Hoffmann FW, et al. 2010. Dietary selenium modulates activation and differentiation of CD4+ T cells in mice through a mechanism involving cellular free thiols. J. Nutr. 140:1155–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hoffmann PR, et al. 2007. The selenoproteome exhibits widely varying, tissue-specific dependence on selenoprotein P for selenium supply. Nucleic Acids Res. 35:3963–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hwang JT, et al. 2006. Selenium regulates cyclooxygenase-2 and extracellular signal-regulated kinase signaling pathways by activating AMP-activated protein kinase in colon cancer cells. Cancer Res. 66:10057–10063 [DOI] [PubMed] [Google Scholar]
- 27. Kim KH. 1997. Regulation of mammalian acetyl-coenzyme A carboxylase. Annu. Rev. Nutr. 17:77–99 [DOI] [PubMed] [Google Scholar]
- 28. Kim SS, et al. 2011. Exercise training and selenium or a combined treatment ameliorates aberrant expression of glucose and lactate metabolic proteins in skeletal muscle in a rodent model of diabetes. Nutr. Res. Pract. 5:205–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Klein EA, et al. 2011. Vitamin E and the risk of prostate cancer: the selenium and vitamin E cancer prevention trial (SELECT). JAMA 306:1549–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kurokawa S, et al. 2011. Mammalian selenocysteine lyase is involved in selenoprotein biosynthesis. J. Nutr. Sci. Vitaminol. (Tokyo) 57:298–305 [DOI] [PubMed] [Google Scholar]
- 31. Ledesma MC, et al. 2011. Selenium and vitamin E for prostate cancer: post-SELECT (Selenium and Vitamin E Cancer Prevention Trial) status. Mol. Med. 17:134–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lei XG, Cheng WH. 2005. New roles for an old selenoenzyme: evidence from glutathione peroxidase-1 null and overexpressing mice. J. Nutr. 135:2295–2298 [DOI] [PubMed] [Google Scholar]
- 33. Lei XG, Vatamaniuk MZ. 2011. Two tales of antioxidant enzymes on beta cells and diabetes. Antioxid. Redox. Signal. 14:489–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Leinninger GM. 2009. Location, location, location: the CNS sites of leptin action dictate its regulation of homeostatic and hedonic pathways. Int. J. Obes. (Lond.) 33(Suppl. 2): S14–S17 [DOI] [PubMed] [Google Scholar]
- 35. Lippman SM, et al. 2009. Effect of selenium and vitamin E on risk of prostate cancer and other cancers: the Selenium and Vitamin E Cancer Prevention Trial (SELECT). JAMA 301:39–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Low SC, Berry MJ. 1996. Knowing when not to stop: selenocysteine incorporation in eukaryotes. Trends Biochem. Sci. 21:203–208 [PubMed] [Google Scholar]
- 37. Marsili A, et al. 2011. Mice with a targeted deletion of the type 2 deiodinase are insulin resistant and susceptible to diet induced obesity. PLoS One 6:e20832 doi:10.1371/journal.pone.0020832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McClung JP, et al. 2004. Development of insulin resistance and obesity in mice overexpressing cellular glutathione peroxidase. Proc. Natl. Acad. Sci. U. S. A. 101:8852–8857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Medina MC, et al. 2011. The thyroid hormone-inactivating type III deiodinase is expressed in mouse and human β-cells and its targeted inactivation impairs insulin secretion. Endocrinology 152:3717–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Merino B, Diez-Fernandez C, Ruiz-Gayo M, Somoza B. 2006. Choroid plexus epithelial cells co-express the long and short form of the leptin receptor. Neurosci. Lett. 393:269–272 [DOI] [PubMed] [Google Scholar]
- 41. Mihara H, Kurihara T, Watanabe T, Yoshimura T, Esaki N. 2000. cDNA cloning, purification, and characterization of mouse liver selenocysteine lyase. Candidate for selenium delivery protein in selenoprotein synthesis. J. Biol. Chem. 275:6195–6200 [DOI] [PubMed] [Google Scholar]
- 42. Misu H, et al. 2010. A liver-derived secretory protein, selenoprotein P, causes insulin resistance. Cell Metab. 12:483–495 [DOI] [PubMed] [Google Scholar]
- 43. Muecke R, Schomburg L, Buentzel J, Kisters K, Micke O. 2010. Selenium or no selenium—that is the question in tumor patients: a new controversy. Integr. Cancer Ther. 9:136–141 [DOI] [PubMed] [Google Scholar]
- 44. Mueller AS, et al. 2009. Regulation of the insulin antagonistic protein tyrosine phosphatase 1B by dietary Se studied in growing rats. J. Nutr. Biochem. 20:235–247 [DOI] [PubMed] [Google Scholar]
- 45. Mueller AS, et al. 2008. Redox regulation of protein tyrosine phosphatase 1B by manipulation of dietary selenium affects the triglyceride concentration in rat liver. J. Nutr. 138:2328–2336 [DOI] [PubMed] [Google Scholar]
- 46. Omi R, et al. 2010. Reaction mechanism and molecular basis for selenium/sulfur discrimination of selenocysteine lyase. J. Biol. Chem. 285:12133–12139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Raman AV, et al. 2012. Absence of selenoprotein P but not selenocysteine lyase results in neurological dysfunction. Genes Brain Behav. 11:601–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rayman MP. 2008. Food-chain selenium and human health: emphasis on intake. Br. J. Nutr. 100:254–268 [DOI] [PubMed] [Google Scholar]
- 49. Rayman MP. 2000. The importance of selenium to human health. Lancet 356:233–241 [DOI] [PubMed] [Google Scholar]
- 50. Ricart-Jane D, Cejudo-Martin P, Peinado-Onsurbe J, Lopez-Tejero MD, Llobera M. 2005. Changes in lipoprotein lipase modulate tissue energy supply during stress. J. Appl. Physiol. 99:1343–1351 [DOI] [PubMed] [Google Scholar]
- 51. Riese C, et al. 2006. Selenium-dependent pre- and posttranscriptional mechanisms are responsible for sexual dimorphic expression of selenoproteins in murine tissues. Endocrinology 147:5883–5892 [DOI] [PubMed] [Google Scholar]
- 52. Schomburg L, et al. 2003. Gene disruption discloses role of selenoprotein P in selenium delivery to target tissues. Biochem. J. 370:397–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schweizer U, Brauer AU, Kohrle J, Nitsch R, Savaskan NE. 2004. Selenium and brain function: a poorly recognized liaison. Brain Res. Brain Res. Rev. 45:164–178 [DOI] [PubMed] [Google Scholar]
- 54. Small-Howard A, et al. 2006. Supramolecular complexes mediate selenocysteine incorporation in vivo. Mol. Cell. Biol. 26:2337–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Speckmann B, et al. 2008. Selenoprotein P expression is controlled through interaction of the coactivator PGC-1α with FoxO1a and hepatocyte nuclear factor 4α transcription factors. Hepatology 48:1998–2006 [DOI] [PubMed] [Google Scholar]
- 56. Stranges S, et al. 2007. Effects of long-term selenium supplementation on the incidence of type 2 diabetes: a randomized trial. Ann. Intern. Med. 147:217–223 [DOI] [PubMed] [Google Scholar]
- 57. Tamura T, et al. 2004. Selenophosphate synthetase genes from lung adenocarcinoma cells: Sps1 for recycling l-selenocysteine and Sps2 for selenite assimilation. Proc. Natl. Acad. Sci. U. S. A. 101:16162–16167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tobe R, Mihara H, Kurihara T, Esaki N. 2009. Identification of proteins interacting with selenocysteine lyase. Biosci. Biotechnol. Biochem. 73:1230–1232 [DOI] [PubMed] [Google Scholar]
- 59. Wakil SJ, Abu-Elheiga LA. 2009. Fatty acid metabolism: target for metabolic syndrome. J. Lipid Res. 50(Suppl.): S138–S143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang KT, Wang J, Li LF, Su XD. 2009. Crystal structures of catalytic intermediates of human selenophosphate synthetase 1. J. Mol. Biol. 390:747–759 [DOI] [PubMed] [Google Scholar]
- 61. Xu XM, et al. 2007. Biosynthesis of selenocysteine on its tRNA in eukaryotes. PLoS Biol. 5:e4 doi:10.1371/journal.pbio.0050004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Xu XM, et al. 2010. Targeted insertion of cysteine by decoding UGA codons with mammalian selenocysteine machinery. Proc. Natl. Acad. Sci. U. S. A. 107:21430–21434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yang JG, Hill KE, Burk RF. 1989. Dietary selenium intake controls rat plasma selenoprotein P concentration. J. Nutr. 119:1010–1012 [DOI] [PubMed] [Google Scholar]
- 64. Yang SJ, et al. 2011. Serum selenoprotein P levels in patients with type 2 diabetes and prediabetes: implications for insulin resistance, inflammation, and atherosclerosis. J. Clin. Endocrinol. Metab. 96:E1325–E1329 [DOI] [PubMed] [Google Scholar]
- 65. Yeh JY, Ou BR, Gu QP, Whanger PD. 1998. Influence of gender on selenoprotein W, glutathione peroxidase and selenium in tissues of rats. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 119:151–155 [DOI] [PubMed] [Google Scholar]
- 66. Zhao LF, et al. 2010. Hormonal regulation of acetyl-CoA carboxylase isoenzyme gene transcription. Endocr. J. 57:317–324 [DOI] [PubMed] [Google Scholar]