Abstract
Accumulation of excess lipid in nonadipose tissues is associated with oxidative stress and organ dysfunction and plays an important role in diabetic complications. To elucidate molecular events critical for lipotoxicity, we used retroviral promoter trap mutagenesis to generate mutant Chinese hamster ovary cell lines resistant to lipotoxic and oxidative stress. A previous report of a mutant from this screen demonstrated that under lipotoxic conditions, small nucleolar RNAs (snoRNAs) in the rpL13a gene accumulate in the cytosol and serve as critical mediators of lipotoxic cell death. We now report a novel, independent mutant in which a single provirus disrupted one allele of the gene encoding the spliceosomal protein SmD3, creating a model of haploinsufficiency. We show that snoRNA expression and the abundance of snoRNA-containing intron lariats are decreased in SmD3 mutant cells, even though haploinsufficiency of SmD3 supports pre-mRNA splicing. The mechanism through which SmD3 regulates the expression of intronic snoRNAs likely involves effects of SmD3 on the levels of small nuclear RNAs (snRNAs) U4 and U5. Our data implicate SmD3 as a critical determinant in the processing of intronic noncoding RNAs in general and as an upstream mediator of metabolic stress response pathways through the regulation of snoRNA expression.
INTRODUCTION
Elevations in serum triglycerides and free fatty acids (FA) play an important role in the pathogenesis of diabetic complications. Under physiological conditions, mammalian adipose cells internalize and store large quantities of lipid. However, under pathophysiological conditions, accumulation of fatty acids in nonadipose tissues causes cell dysfunction and cell death that lead to impaired organ function (43). This phenomenon, known as lipotoxicity, contributes to the pathogenesis of heart failure, renal dysfunction, steatohepatitis, and progressive pancreatic insufficiency (1, 17, 37, 38).
In vitro models in which the medium of cultured cells is supplemented with excess fatty acid have been used to probe metabolic and signaling pathways involved in the cellular response to lipid overload. In a time- and dose-dependent manner, long-chain saturated fatty acids induce apoptosis in a variety of cell types (6, 7, 21, 24, 45), and this response is enhanced by high glucose (8). Although lipid overload in nonadipose cells is initially buffered by cytoprotective triglyceride stores (20, 23), when the limited capacity for neutral lipid storage in nonadipose cells is exceeded, excess saturated fatty acids initiate several cellular stress response pathways. Fatty acid-induced endoplasmic reticulum stress can result in reactive oxygen species (ROS) generation (40). Independently, oxidative stress is induced in a variety of cell types through activation of NADPH oxidase, mitochondrial dysfunction due to remodeling of organelle membranes, and excessive cycles of oxidative phosphorylation (16, 31, 41). Administration of antioxidants to cultured cells and animal models of lipotoxicity mitigate against lipotoxic cell death (4, 5, 19, 21), suggesting a central role for oxidative stress in lipotoxicity.
Our laboratory has used promoter trap mutagenesis and a loss-of-function genetic screen in Chinese hamster ovary (CHO) cells to gain new insights into the lipotoxic pathway. Previously, we identified three intronic small nucleolar RNAs (snoRNAs) within the ribosomal protein L13a (rpL13a) locus that are necessary for the propagation of ROS during lipotoxicity (27). Most vertebrate snoRNAs are encoded within introns and cotranscribed with their host genes (42). Canonically, box C/D and box H/ACA snoRNAs guide modification of rRNA by 2′-O-methylation and pseudouridylation, respectively (26). While the box C/D rpL13a snoRNAs are predicted to direct 2′-O-methylation of rRNAs (28), putative rRNA targets of these snoRNAs are unaltered during lipotoxicity in wild-type (WT) or rpL13a-haploinsufficient cells. The observations that rpL13a snoRNAs rapidly accumulate in the cytosol during metabolic stress and are required for lipotoxic cell death suggest that cytoplasmic RNAs may be their primary targets and that efficient processing of these intronic elements is important for the lipotoxic response. Studies from other groups have demonstrated that intronic box C/D snoRNP protein assembly occurs at the C1 complex stage of splicing (13), with subsequent lariat formation at the C2 complex stage of splicing, debranching, and exonucleolytic trimming (18, 29, 33). However, the precise molecular mechanisms through which snoRNAs are induced and regulated during lipotoxicity remain to be elucidated.
In the present study, we characterize an independent mutant from this genetic screen. This novel mutant cell line is haploinsufficient for SmD3, a core component of the spliceosome. We demonstrate that SmD3 participates in the lipotoxic response through regulation of intron lariat abundance and biogenesis of intron-encoded rpL13a snoRNAs. We also provide evidence linking the expression of SmD3 to the levels of critical small nuclear RNA (snRNA) components of the spliceosome and generalized production of intronic noncoding RNAs (ncRNAs). Our results extend the known function of SmD3 in splicing to a specific role within individual snRNPs essential for the biogenesis of intronic ncRNAs.
MATERIALS AND METHODS
Materials.
Palmitate was from Nu-Chek Prep. [14C]palmitate and [α-32P]UTP were from PerkinElmer Life Sciences. Camptothecin, actinomycin D, and hygromycin were from Calbiochem. Staurosporine, H2O2, menadione, phloretin, and clotrimazole were from Sigma-Aldrich. Fatty acid-free bovine serum albumin (BSA) was from SeraCare. Propidium iodide and 5 (and-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate acetyl ester (CM-H2DCFDA) were from Invitrogen. All synthetic oligonucleotides were from IDT. Restriction enzymes were from New England BioLabs.
Cell culture.
CHO-K1 cells (American Type Culture Collection) and CHO-derived cell lines were maintained in high-glucose (4.5 mg/ml Dulbecco's modified Eagle's medium and Ham's F-12 nutrient mixture [1:1]) medium with 5% noninactivated fetal bovine serum, 2 mM l-glutamine, 50 units/ml penicillin G sodium, 50 units/ml streptomycin sulfate, and 1 mM sodium pyruvate. NIH 3T3 cells (American Type Culture Collection) were maintained in high-glucose (4.5 mg/ml Dulbecco's modified Eagle's medium) medium with 10% bovine calf serum, 2 mM l-glutamine, 50 units/ml penicillin G sodium, and 50 units/ml streptomycin sulfate. For lipotoxicity experiments, cell culture medium was supplemented with 500 μM (WT CHO, mutant 6H2, control, and small interfering RNA [siRNA]-treated 3T3 cells) or 250 μM (short hairpin RNA [shRNA]-transfected cells) palmitate complexed to BSA at a 2:1 molar ratio, as described previously (21). For ROS induction, media were supplemented with 2.3 mM H2O2 or 50 μM menadione.
Generation of CHO cell mutants.
Vesicular stomatitis virus G protein-pseudotyped murine retrovirus encoding the ROSAβgeo retroviral promoter trap was generated as described previously (9, 30). CHO cells were transduced with retrovirus at a low multiplicity of infection (an average of 1 integration per 10 genomes), and mutants were isolated as described previously (3). The number of retroviral insertions within the mutant-cell genome was assessed by Southern blotting. Genomic DNA was digested with restriction enzymes, separated by agarose gel electrophoresis, transferred to nylon membranes, and probed with a 32P-labeled probe corresponding to the ROSAβgeo proviral sequence.
Cell death assays.
Cell death was assessed by membrane permeability to propidium iodide (PI) staining and flow cytometry (21). Following treatments, cells were harvested by trypsinization and stained with 1 μM PI. The percentage of PI-positive cells was determined by flow cytometry, quantifying 104 cells/sample.
Identification of the trapped gene.
The endogenous gene disrupted by retroviral insertion was identified by 5′ rapid amplification of cDNA ends (RACE) using an oligonucleotide tag and ROSAβgeo sequences (Smart RACE cDNA amplification kit; Clontech). The 5′ RACE product was TA cloned, sequenced, and tested for sequence similarity by NCBI BLAST. PCR to verify retroviral integration within the snrpD3 gene used snrpD3 forward and either snrpD3 reverse or ROSAβgeo reverse primers.
Quantitative real-time PCR (qRT-PCR).
RNA was isolated using TRIzol or TRIzol LS reagent (Invitrogen) and reverse transcribed to cDNA using the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen) following the manufacturer's instructions. Reverse transcription was performed with oligo(dT) to detect mRNA or random hexamers to detect pre-mRNA and intron lariats. For quantification of snoRNAs and pre-microRNAs (miRNAs), RNA was isolated using TRIzol LS (Invitrogen). cDNA synthesis was primed with hairpin stem-loop oligonucleotides as previously described (27), with overhang complementarity to the 3′ end of the processed snoRNA or pre-miRNA. cDNA was amplified for 40 PCR cycles using SYBR green PCR master mixture (Applied Biosystems) and 100 nM template-specific primers in an ABI Prism 7500 Fast real-time PCR system. Relative quantification of gene expression was performed using the comparative threshold method as described by the manufacturer.
Generation of SmD3 antibody.
For immunoblot detection of SmD3, polyclonal rabbit antipeptide antibody was generated by ProSci Incorporated. Animals were immunized with peptide NH2-CTGEVYRGKLIEAED-OH (murine sequence) conjugated to keyhole limpet hemocyanin (KLH). The animals received six rounds of immunization, followed by affinity purification of serum.
Immunoblot analyses.
Whole-cell protein lysates were prepared using RIPA buffer (50 mM Tris-Cl, 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, and 5 mM EDTA) containing 1 mM phenylmethylsulfonyl fluoride and 1× Protease Complete inhibitor mixture (Roche). Subcellular fractions were isolated by sequential detergent solubilization as described previously (15). Proteins (40 μg) were resolved by 15% (for SmD3) or 12% (for hsp90, fibrillarin, and β-actin) SDS-PAGE and transferred to nitrocellulose membranes (Whatman). The membranes were probed with antibodies to SmD3 (1:500), β-actin (A 2066; 1:5,000; Sigma), hsp90 (SPA-846; 1:2,000; Stressgen), fibrillarin (MMS-581S; 1:500; Covance), and SmB (S0698; 1:1,000; Sigma). Proteins were visualized using appropriate horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories; 1:10,000) and chemiluminescence reagents (PerkinElmer Life Sciences). Band intensities were quantified by densitometry (Bio-Rad Image Laboratory Software).
Generation of SmD3 shRNA clones.
Hamster snrpD3 cDNA sequence was used to design siRNA oligonucleotides using Ambion's siRNA Target Finder Program. shRNA oligonucleotides were designed from siRNA sequences that conferred effective knockdown in transient-transfection assays, and each was cloned into a pSilencer 4.1-CMV hygro vector (Ambion) containing a hygromycin resistance cassette. shRNA vectors were transfected into CHO cells with Lipofectamine 2000 reagent (Invitrogen). The cells were plated at limiting dilutions and treated with 80 μg/ml hygromycin. Clonal lines were isolated, and SmD3 knockdown was assessed by immunoblotting.
Fatty acid uptake.
Cells (2 × 106) were resuspended in 1 ml phosphate-buffered saline (PBS) containing 500 μM [14C]palmitate complexed to 250 μM BSA and incubated for 1 min at 37°C. The cells were washed with 10 ml PBS containing 0.1% BSA and 500 μM phloretin and filtered, and cell-associated 14C was quantified by scintillation counting. A parallel aliquot of cells was used for quantification of protein by bicinchoninic acid assay (Pierce).
Detection of reactive oxygen species.
Cells (2 × 105) were plated in 12-well plates 24 h prior to various treatments. The cells were rinsed with PBS and incubated with PBS containing 0.5 mM MgCl2, 0.92 mM CaCl2, and 3 μM CM-H2DCFDA (Invitrogen) in the dark at 37°C for 1 h. The cells were then rinsed with PBS, harvested by trypsinization, and quenched with culture medium. Mean fluorescence was determined by flow cytometry on 104 cells/sample.
snoRNA probe synthesis and RNase digestion.
Hamster-specific snoRNA probes were generated with a Megashortscript kit (Ambion). Double-stranded RNA (dsRNA) templates were generated for probes for each rpL13a snoRNA by PCR amplification of cloned hamster rpL13a genomic sequence templates using primers containing the T7 RNA polymerase promoter and used for in vitro RNA transcription of 32P-labeled snoRNA probes. miR-16 probes were synthesized using templates from the mirVana miRNA detection kit (Ambion). RNA was isolated from cells using a mirVana miRNA isolation kit (Ambion) and hybridized to 32P-labeled RNA probes (mirVana miRNA detection kit) overnight at 42 to 52°C, followed by RNase digestion and ethanol precipitation. RNA was separated by 10% or 15% polyacrylamide gel electrophoresis and visualized by autoradiography.
Luciferase plasmids and transient transfection.
The split luciferase vector containing a β-globin intron was as described previously (48). All constructs generated by PCR or QuikChange (Stratagene) mutagenesis were confirmed by sequencing. The β-globin intron in the split luciferase reporter was replaced with rpL13a intron 2 containing snoRNA U32a or the rpL13a intron 2 lacking the 83-nucleotide U32a snoRNA sequence. Cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol and assayed 20 h posttransfection.
Microarray sample preparation and data analysis.
NIH 3T3 cells (2 × 105) were plated in 6-well plates 24 h pretransfection. The cells were transfected with 40 pmol locked nucleic acid (LNA)/DNA oligonucleotides specifically targeting green fluorescent protein (GFP) or SmD3 (Exiqon) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol, and total RNA was harvested 23 h posttransfection using TRIzol reagent (Invitrogen). The resulting RNA was quantified by A260 and A280 readings using a Nanodrop spectrophotometer (Nanodrop Technologies) and qualitatively assessed using a BioAnalyzer 2100 (Agilent Technolgies). cDNA was prepared using the NuGen Ovation System, and microarray data were generated with Affymetrix GeneChip Mouse Exon 1.0 ST arrays containing 266,200 probe sets in the Siteman Cancer Center Molecular and Genomic Analysis Core (Washington University School of Medicine). Partek Genomics Suite 6.5 was used to calculate probe set intensities from Affymetrix intensity (CEL) files using the robust multiarray average (RMA) algorithm with default settings at both the gene level and the probe set level. Probe sets with RMA intensities below 3 across all samples were excluded to eliminate probe sets with low expression levels. Alternative splicing multiway analysis of variance (ANOVA) was applied using Partek Genomics Suite defaults to identify alternative splicing events with a false-discovery rate (FDR) of <0.1. Core exon level analysis was also applied at the exon level to determine differential expression of exons not grouped by transcript.
Luciferase detection.
Cells (3 × 104) were plated in triplicate in 96-well plates. Following transfection of luciferase reporters as described above, the cells were lysed with Dual-Glo luciferase reagent (Promega) according to the manufacturer's protocol. Luciferase was detected using a Tecan Infinite M200 microplate reader and Magellan software.
Transient knockdown.
For SmD3 knockdown, chimeric LNA/DNA oligonucleotides (Exiqon) were designed to specifically target GFP or the murine snrpD3 sequences. A BLAT search of the mouse genome was performed using the UCSC Genome Browser for each LNA/DNA oligonucleotide. The GFP LNA/DNA oligonucleotide showed no sequence homology to the mouse genome, and the SmD3 LNA/DNA oligonucleotide displayed alignment only to the snrpD3 gene. NIH 3T3 cells (2 × 105) were plated in 6-well plates 24 h pretransfection. Cells were transfected with 40 pmol LNA/DNA oligonucleotides using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol, and RNA was harvested 23 h posttransfection. For SmB knockdown, NIH 3T3 cells (1 × 105) were plated in 6-well plates 24 h pretransfection. The cells were transfected with 50 pmol control (Ambion) or snrpb (s74100; Ambion) Silencer Select siRNA using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's protocol, and RNA was harvested 24 h posttransfection.
Immunoprecipitation.
For snRNA immunoprecipitation, NIH 3T3 cells were plated and transfected as performed for SmD3 transient knockdown. Twenty-three hours posttransfection, cells were harvested in lysis buffer (50 mM Tris-Cl, pH 8, 150 mM NaCl, 0.5% Nonidet P-40) containing 1 mM phenylmethylsulfonyl fluoride and 1× Protease Complete inhibitor mixture (Roche) and incubated on ice for 30 min. The cells were sonicated with five 5-s pulses using a Branson Sonifier 250. The cells were incubated on ice for 20 s between pulses. The cell lysates were centrifuged at 15,000 × g for 30 min at 4°C to remove insoluble material and immunoprecipitated using anti-Sm Y12 antibody (ab3138; 1:100; Abcam) or IgG control. RNA was isolated from immunoprecipitated samples using TRIzol (Invitrogen) and reverse transcribed to cDNA using the SuperScript III first-strand synthesis system for RT-PCR (Invitrogen) following the manufacturer's instructions. Quantification of anti-Sm-immunoprecipitated snRNA expression relative to control IgG-precipitated snRNA was performed by qRT-PCR.
RESULTS
SmD3 haploinsufficiency confers resistance to palmitate-induced cell death.
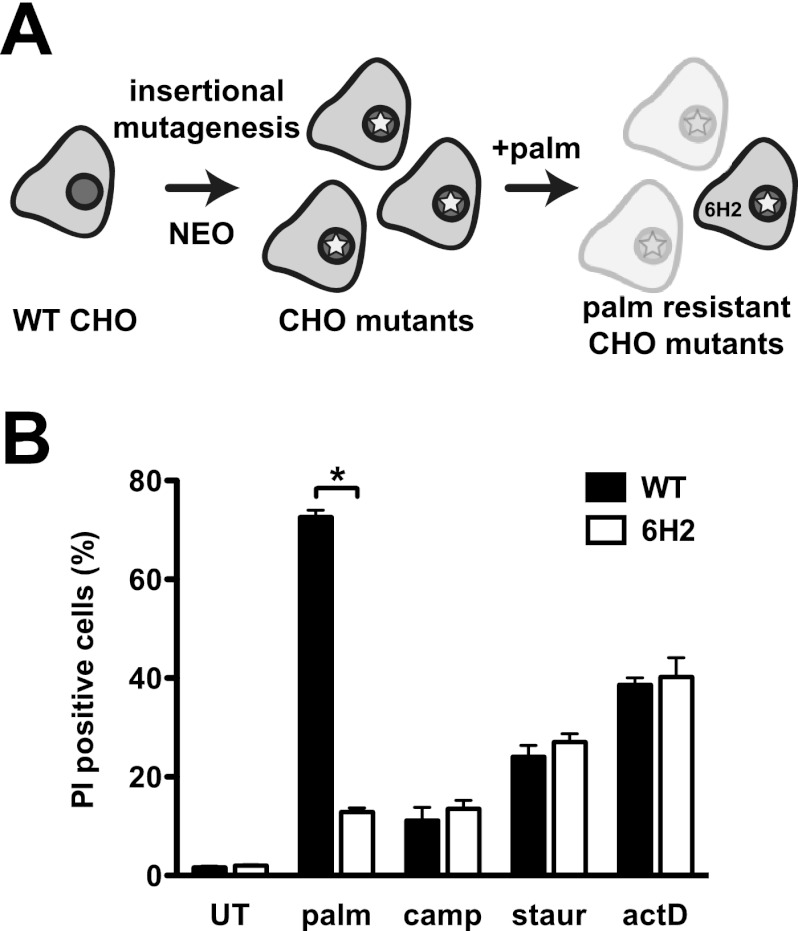
To identify genes critical for the lipotoxic response, we carried out a genetic screen in CHO cells using the ROSAβgeo retroviral promoter trap, as previously described, to generate cells with one proviral integration (3) (Fig. 1A). Since the provirus contains only a splice acceptor, a promoterless β-galactosidase–neomycin phosphotransferase cassette, and polyadenylation sequences, a fusion transcript is generated and leads to antibiotic resistance only following insertion downstream of an active promoter. CHO mutants were selected in medium containing G418 and then incubated in medium supplemented with 500 μM palmitate, which kills WT cells. The palmitate-resistant 6H2 mutant cell line was isolated from this screen.
Fig 1.
6H2 cells are resistant to palmitate-induced cell death. (A) WT CHO cells were transduced with the ROSAβgeo retrovirus, leading to integration of the provirus containing a splice acceptor, promoterless β-galactosidase–neomycin resistance cassette, and polyadenylation sequences. Promoter trapping and gene disruption at the site of integration were selected for by growth in neomycin (NEO), and palmitate-resistant mutants were selected by growth in medium with 500 μM palmitate (palm) for 48 h. (B) WT and palmitate-resistant 6H2 mutant cells were incubated with 500 μM palmitate for 48 h or 10 μM camptothecin (camp), 80 nM staurosporine (staur), or 2 μM actinomycin D (actD) for 24 h. Cell death was quantified by PI staining and flow cytometry. The data are expressed as mean fluorescence and standard error (SE) for 3 independent experiments with 104 cells/sample. *, P < 0.005 for 6H2 versus WT. UT, untreated.
To quantify the degree of palmitate resistance, WT and 6H2 cells were treated with palmitate for 48 h, and cell death was quantified by PI staining and flow cytometric analysis. Compared to WT cells, 6H2 cells were significantly protected from palmitate-induced death (Fig. 1B). In contrast, treatment of WT and 6H2 cells with the general apoptosis inducers camptothecin, staurosporine, and actinomycin D revealed no differences in sensitivity. Therefore, palmitate-resistant 6H2 cells are not generally resistant to cell death.
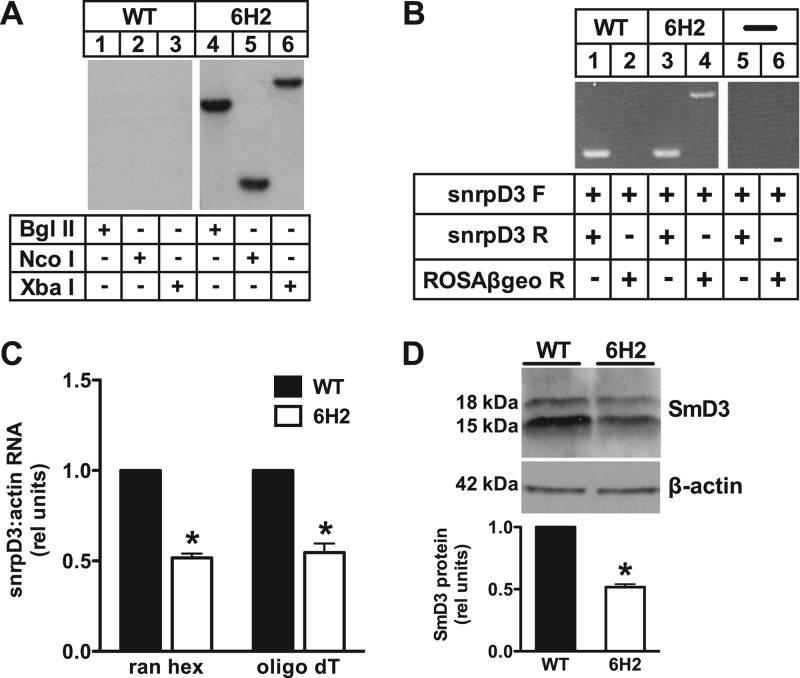
Promoter trap mutagenesis facilitated identification of the disrupted gene because of the unique fusion transcript produced upon a single productive integration. To confirm the presence of a single retroviral integration, Southern blot analysis of 6H2 genomic DNA was performed, probing for the ROSAβgeo sequence (Fig. 2A). The presence of a single hybridizing band in DNA digested with multiple different restriction enzymes is consistent with a single retroviral integration. To identify the disrupted gene in the mutant 6H2 cells, mRNA was isolated and used for 5′ RACE. Unique sequence from the RACE product was analyzed using NCBI BLAST, which revealed that the site of integration was the snrpD3 gene encoding the small nuclear ribonucleoprotein SmD3. SmD3 is a component of the spliceosome that, together with other Sm family proteins, forms a heteroheptameric ring around snRNAs U1, U2, U4, and U5 to generate snRNPs essential for the removal of introns from pre-mRNA (46). PCR was performed to confirm integration of the ROSAβgeo sequence into snrpD3 (Fig. 2B). Reactions with forward and reverse primers designed to snrpD3 resulted in PCR products for both WT and 6H2 cDNA, indicating that each cell type maintains at least one intact allele. As expected, no product was detectable in WT cells when snrpD3 forward and ROSAβgeo reverse primers were used, but this set of primers produced the expected PCR product from the fusion transcript in 6H2 cells. These PCR results confirm our 5′ RACE identification of the disrupted gene and suggest that 6H2 cells are haploinsufficient for snrpD3. Consistent with this model, qRT-PCR revealed an ∼50% reduction in relative snrpD3 mRNA (Fig. 2C) and Western blotting revealed a corresponding ∼50% reduction of SmD3 protein (a doublet at 15 kDa and 18 kDa) in 6H2 relative to WT cells (Fig. 2D). Thus, expression of snrpD3 in 6H2 cells is consistent with a model in which integration of the ROSAβgeo provirus disrupted one of two alleles for snrpD3.
Fig 2.
6H2 cells are haploinsufficient for SmD3. (A) Autoradiogram showing Southern blot analysis of WT (lanes 1 to 3) and 6H2 (lanes 4 to 6) genomic DNA digested with the restriction enzyme BglII, NcoI, or XbaI. The blot was probed with a 32P-labeled fragment corresponding to the ROSAβgeo sequence. (B) PCR was performed on cDNA from WT (lanes 1 and 2) and 6H2 (lanes 3 and 4) cells with reaction mixtures containing no cDNA as controls (lanes 5 and 6). Forward (F) and reverse (R) primers for snrpD3 were designed to detect endogenous snrpD3 (lanes 1, 3, and 5). The forward snrpD3 primer and reverse primer for the proviral sequence were used to detect fusion transcript (lanes 2, 4, and 6). (C) RNA was isolated from WT and 6H2 cells and reverse transcribed using either random hexamers (ran hex) to prime total RNA or oligo(dT) to prime mRNA. snrpD3 expression was determined by qRT-PCR and normalized to β-actin expression. (D) Protein expression in WT and 6H2 cells was determined by Western blotting and quantified by densitometry (bands at 15 kDa and 18 kDa likely reflect known posttranslational modification of SmD3). A representative blot is shown for SmD3 and β-actin control. On the bar graphs, the data are expressed as means and SE for three independent experiments. *, P < 0.05 for 6H2 versus WT.
Targeted knockdown of SmD3 recapitulates the 6H2 phenotype.
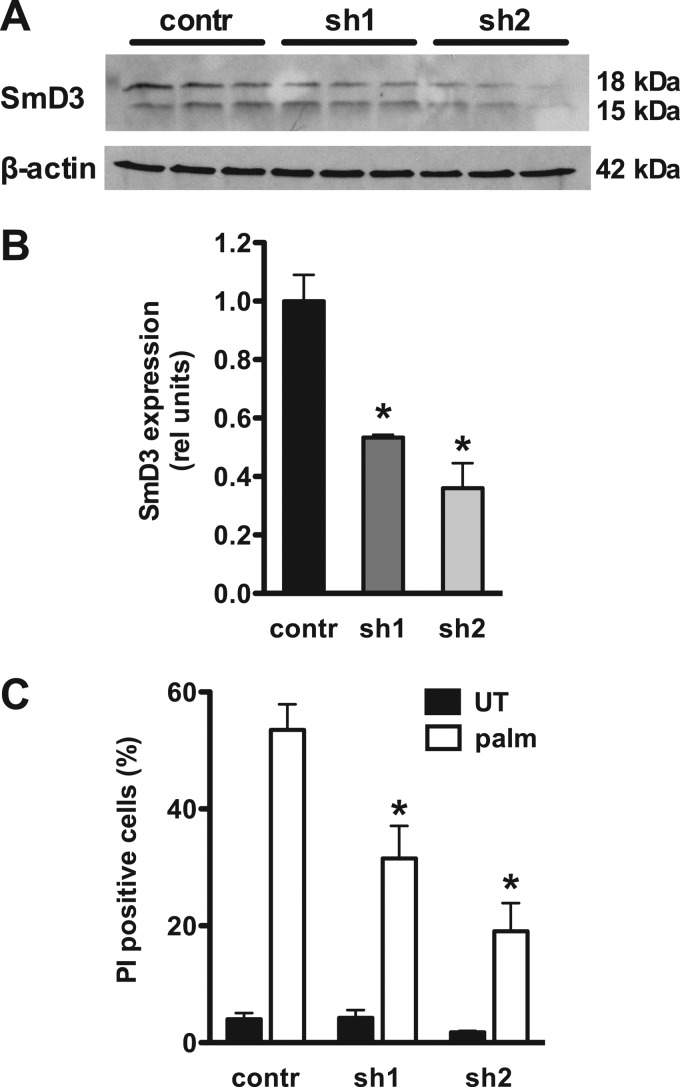
To confirm that the palmitate-resistant phenotype in 6H2 cells is due to diminished SmD3 protein expression, we used shRNA to knock down SmD3 in WT CHO cells and tested for associated changes in palmitate sensitivity. SmD3 protein levels were measured by Western blotting following isolation of individual stable clonal knockdown lines. Two independently isolated clonal lines showed 47% (sh1) and 64% (sh2) knockdown relative to scrambled-shRNA-transfected cells (Fig. 3A and B). Knockdown clones were protected from palmitate-induced death as measured by PI staining, and the degree of protection was proportional to the degree of knockdown (Fig. 3C). These data provide independent genetic evidence that loss of function of SmD3 protects against lipotoxicity.
Fig 3.
Targeted knockdown of SmD3 confers palmitate resistance. Stable clonal cell lines were generated following transfection with a scrambled (contr) or snrpD3-targeting (sh1 and sh2) shRNA. (A) SmD3 and β-actin protein expression was determined by Western blotting. The blot shows three independent protein samples from each respective cell line. (B) SmD3 expression relative to β-actin was quantified by densitometry of blots as shown in panel A. The graph shows means and SE for three independent samples. *, P < 0.05 for knockdown versus scrambled. rel, relative. (C) Scrambled and knockdown cells were treated with palmitate for 48 h, and cell death was assessed by PI staining and flow cytometry. All data are expressed as mean fluorescence and SE for three independent experiments with 104 cells/sample. *, P < 0.005 for knockdown versus scrambled.
SmD3 disruption protects cells from generalized oxidative stress induction.
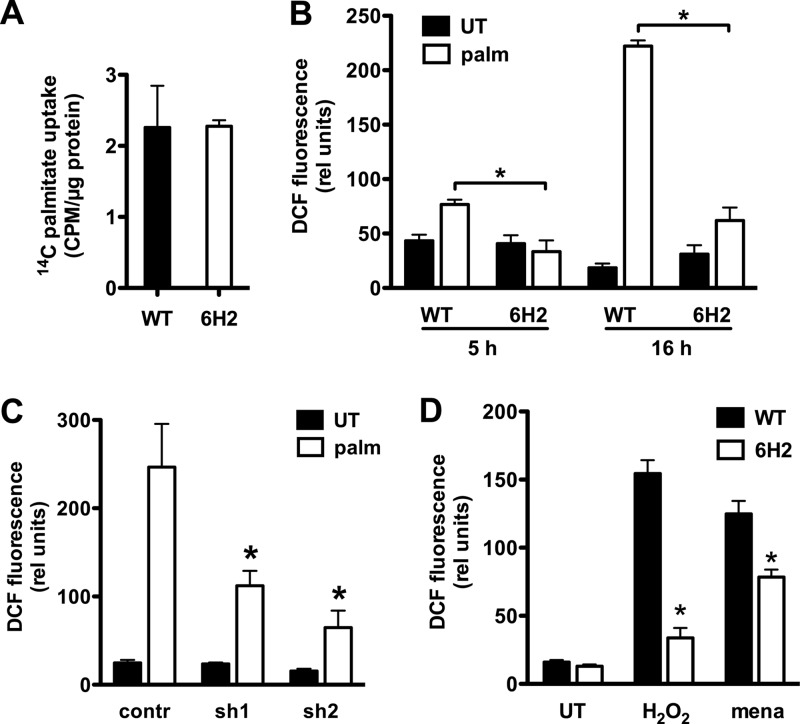
Lipotoxicity is known to involve FA import and the generation of oxidative stress (4, 20). To test whether 6H2 cells acquired resistance through diminished capacity to take up palmitate, initial rates of FA uptake were quantified in WT and 6H2 cells. There was no significant difference between WT and 6H2 cells (Fig. 4A), indicating that resistance to lipotoxicity in 6H2 cells did not result from failure to take up exogenous FA. To probe downstream aspects of the lipotoxic response, we quantified palmitate-induced ROS in WT and 6H2 cells by CM-H2DCFDA staining and flow cytometric analysis. At 5 and 16 h following palmitate supplementation, ROS induction was significantly blunted in 6H2 cells (Fig. 4B). SmD3 knockdown clones were also protected from palmitate-induced ROS (Fig. 4C). More direct induction of oxidative stress following exposure to H2O2 or menadione also resulted in blunted ROS levels in 6H2 cells compared to WT cells (Fig. 4D), indicating 6H2 cells are protected, not only from palmitate-induced ROS, but also from generalized oxidative stress induction or amplification.
Fig 4.
SmD3 disruption protects cells from palmitate-induced and generalized oxidative stress induction. (A) WT and 6H2 cells were incubated with [14C]palmitate under lipotoxic conditions (500 μM palmitate). The mean initial rates of palmitate uptake per μg protein (+SE) are expressed for 3 independent experiments. (B, C, and D) WT and 6H2 cells were incubated with palmitate (5 h and 16 h) (B), scrambled (contr) and knockdown (sh1 and sh2) cells were treated with palmitate (16 h) (C), or WT and 6H2 cells were untreated (UT) or treated with menadione (mena) or H2O2 (2 h) (D). ROS induction was assessed by CM-H2DCFDA labeling and flow cytometry. The graphs show mean fluorescence and SE for 3 independent experiments with 104 cells/sample. *, P < 0.05 for 6H2 versus WT or for knockdown versus scrambled.
SmD3 regulates intronic noncoding RNA expression.
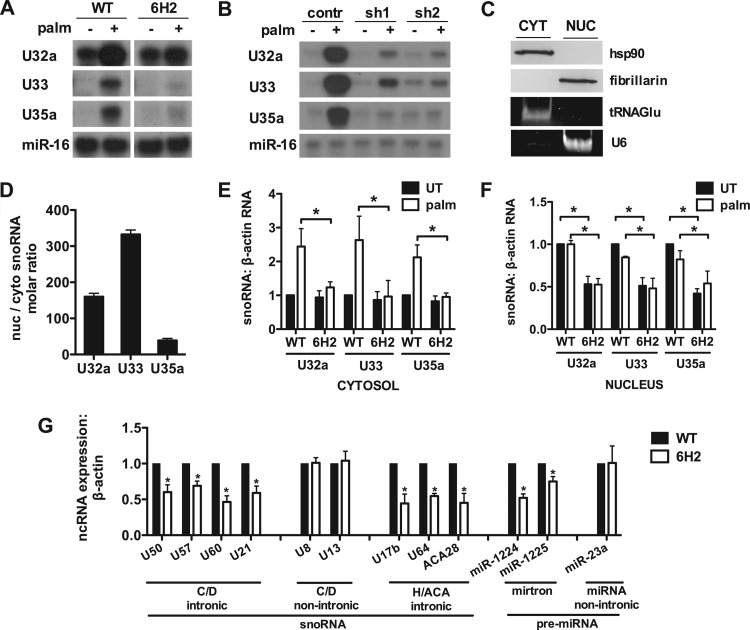
The ROS resistance phenotype observed in 6H2 cells is similar to that of another mutant isolated from the same genetic screen, in which snoRNAs U32a, U33, and U35a, embedded within introns of the rpL13a gene, were shown to function as mediators of lipotoxicity (27). Given the related phenotypes of these two mutants and the well-known interactions of SmD3 with RNA, we assayed for the expression of the rpL13a snoRNAs in 6H2 cells by RNase protection assay. Following palmitate treatment, WT cells show the expected increase in snoRNA expression by RNase protection (Fig. 5A). Under the same conditions, snoRNA induction is blunted in 6H2 cells. Similarly, SmD3 knockdown clones were impaired in rpL13a snoRNA induction relative to the control (Fig. 5B). While snoRNAs are thought to be produced in the nucleus and canonical box C/D snoRNAs function in that location, our prior study demonstrated that rpL13a snoRNAs accumulate in the cytosol during metabolic stress. Fractionation of WT and 6H2 cells by sequential detergent solubilization revealed that under basal conditions snoRNAs are detectable in the nucleus and the cytosol but are substantially more abundant in the nucleus (Fig. 5C and D). qRT-PCR analysis of these fractions revealed reduced rpL13a snoRNAs in the cytosol in 6H2 cells under palmitate-treated conditions and reduced levels of these snoRNAs in the nucleus under both basal and palmitate-treated conditions (Fig. 5E and F). The observation that haploinsufficiency of SmD3 caused impairment of basal expression and lipotoxic cytosolic accumulation of the intronic rpL13a snoRNAs, a deficit known to cause resistance to lipotoxicity, is consistent with the ROS-resistant phenotype observed in 6H2 cells. Furthermore, the observation that nuclear levels of the rpL13a snoRNAs are decreased in 6H2 cells under basal, as well as lipotoxic, conditions implies a defect in the nuclear production of the snoRNAs.
Fig 5.
Intronic noncoding RNA expression is disrupted in 6H2 cells. (A and B) WT and 6H2 cells (A) and scrambled control and knockdown cells (B) were untreated or supplemented with palmitate for 48 h. Small RNA was harvested and used in an RNase protection assay with 32P-labeled rpL13a snoRNA probes or miR-16 probe as a control. The protected probe was analyzed by autoradiography. (C, D, E, and F) WT and 6H2 cells were untreated or treated with palmitate for 9 h. The cells were separated into cytosolic (CYT) and nuclear (NUC) fractions by sequential detergent solubilization. (C) Fractions were analyzed by Western blotting or PCR visualization for cytosolic markers, hsp90 and tRNAGlu, and for nuclear markers, fibrillarin and U6 RNA. (D) Molar quantities of rpL13a snoRNAs from cytosolic and nuclear fractions were quantified relative to a standard curve. The plot represents the relative molar ratio. (E and F) Total RNA was prepared from the cytosolic (E) and nuclear (F) fractions and analyzed for rpL13a snoRNA abundance relative to β-actin by qRT-PCR. (G) Total RNA was prepared from nuclear fractions of WT and 6H2 cells and analyzed for intronic and nonintronic box C/D snoRNAs, intronic box H/ACA snoRNAs, and splicing-dependent/Drosha-independent intronic and nonintronic miRNAs. All data are expressed as means and SE for three independent experiments. *, P < 0.05 for 6H2 versus WT.
To test whether 6H2 cells have a general defect in expression of intronic snoRNAs, we measured basal nuclear expression of intronic box C/D snoRNAs U50, U57, U60, and U21 and intronic box H/ACA snoRNAs U17b, U64, and ACA28 (Fig. 5G). Expression of each of these snoRNAs was reduced in the nuclei of 6H2 versus WT cells. Furthermore, we measured the expression of intronic splicing-dependent/Drosha-independent pre-miRNAs to probe an unrelated class of intronic ncRNAs (2, 35). The mirtrons miR-1224 and miR-1225 showed reduced expression in 6H2 cells. In contrast, independently transcribed, splicing-independent, nonintronic snoRNAs U8 and U13 and pre-miR-23a showed no difference in expression between WT and 6H2 cells. Taken together, these data suggest that WT levels of SmD3 are generally required for effective expression of splicing-dependent intronic ncRNAs.
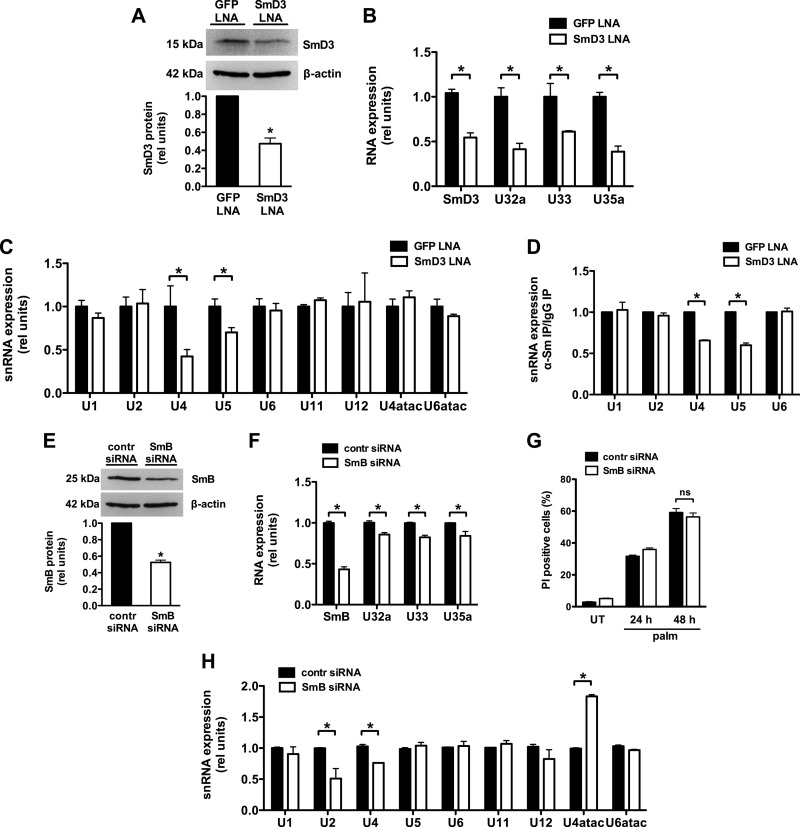
SmD3 knockdown perturbs snRNP biogenesis.
Disruption of multiple components of the snRNP assembly pathway has previously been shown to alter snRNP expression (39, 49). To test whether reduced levels of SmD3 affect snRNA expression, we used a chimeric LNA/DNA oligonucleotide to specifically knock down SmD3 in murine fibroblasts. Following knockdown of SmD3 to ∼50% of control SmD3 levels, we observed reductions in rpL13a snoRNAs similar to those in our mutant (Fig. 6A and B). In SmD3 knockdown cells, qRT-PCR revealed that U4 and U5 snRNA expression was decreased, but levels of other snRNAs were indistinguishable from those of the control (Fig. 6C). Since unbound snRNAs are unstable compared to snRNAs in snRNP complexes, quantification of snRNA levels provides insight into snRNP integrity (36, 50). Consistent with this, U4 and U5 snRNAs were reduced following immunoprecipitation with the anti-Sm protein Y12 antibody, indicating that U4 and U5 snRNPs are also diminished (Fig. 6D). In contrast, 50% knockdown of SmB, another Sm protein family member, produced more modest decreases in snoRNA production (Fig. 6E and F). SmB knockdown cells remained sensitive to palmitate-induced death (Fig. 6G) and displayed a different pattern of alteration of snRNA expression, characterized by decreases in U2 and U4 and increase in U4atac (Fig. 6H). Our data suggest that a reduction in SmD3 expression disrupts a specific set of snRNPs. While there is some overlap with the effects of SmB knockdown on snRNA levels, the pattern with SmD3 is distinct.
Fig 6.
SmD3 knockdown disrupts U4 and U5 snRNPs. (A, B, C, and D) NIH 3T3 cells were transfected with LNA/DNA oligonucleotides specifically targeting GFP or SmD3, achieving SmD3 knockdown as shown in panel A. (A) Protein expression in GFP- and SmD3-transfected cells was determined by Western blotting and quantified by densitometry. A representative blot is shown for SmD3 and β-actin control. (B) Nuclear RNA was isolated and analyzed for SmD3 mRNA and rpL13a snoRNA expression by qRT-PCR relative to 36B4. (C) Total RNA was isolated and analyzed for snRNA expression relative to 36B4 by qRT-PCR. (D) Cell lysates were immunoprecipitated using anti-Sm Y12 antibody or IgG control. RNA was isolated following immunoprecipitation and analyzed by qRT-PCR for anti-Sm-immunoprecipitated snRNA relative to control (IgG)-precipitated snRNA. (E, F, G, and H) NIH 3T3 cells were transfected with control (contr) siRNA or siRNA targeting SmB. (E) Protein expression in control and SmB siRNA cells was determined by Western blotting and quantified by densitometry. A representative blot is shown for SmB and β-actin control. (F) Nuclear RNA was isolated and analyzed for SmB and rpL13a snoRNA expression by qRT-PCR relative to 36B4. (G) Control and SmB siRNA cells were incubated with 500 μM palmitate for 24 h or 48 h. Cell death was quantified by PI staining and flow cytometry. (H) Total RNA was isolated and analyzed for snRNA expression by qRT-PCR relative to 36B4. The data are expressed as means and SE for three independent experiments. *, P < 0.05 for SmD3 versus GFP or SmB versus control; ns, not significant.
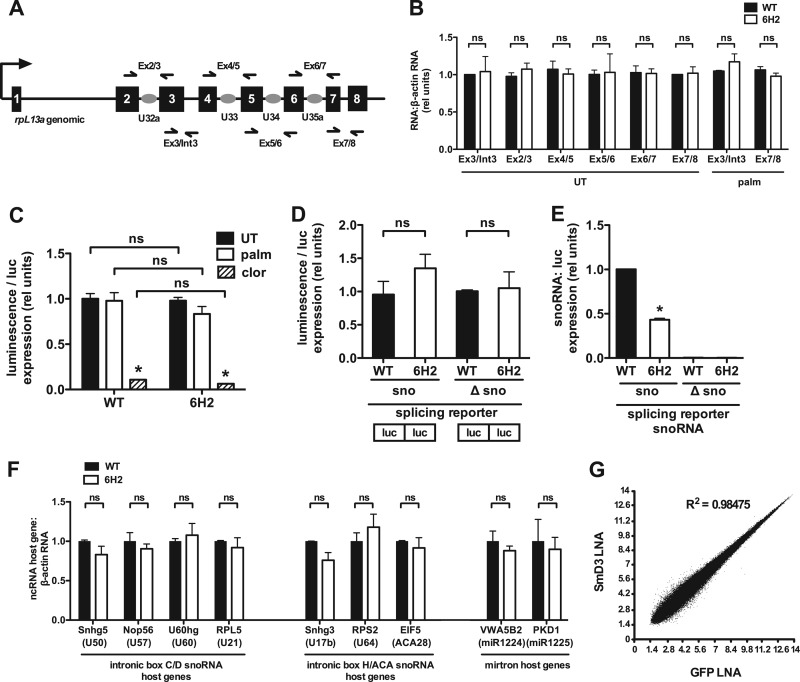
6H2 cells display normal splicing efficiency.
Given that SmD3 levels affect snRNP expression, we hypothesized that differences in intronic ncRNA expression caused by SmD3 haploinsufficiency in the 6H2 cells could be explained by a defect in splicing. To assess splicing related to the rpL13a snoRNAs, we measured the expression of endogenous rpL13a splicing precursors and spliced products. There was no difference in rpL13a pre-mRNA levels between WT and 6H2 cells under basal or palmitate-treated conditions, as assessed by amplifying random-hexamer-primed cDNA with primer pairs designed across the junction between exon 3 and intron 3 (Ex3/Int3) (Fig. 7A and B). Quantification of the endogenous rpL13a mRNA, using oligo(dT)-primed cDNA and primers designed across the splice junction between exons 7 and 8, showed similar expression in WT and 6H2 cells (Ex7/8) (Fig. 7A and B). Furthermore, priming across four splice junctions formed by the removal of snoRNA-encoding introns (Ex 2/3, 4/5, 5/6, and 6/7) displayed similar expression in WT and 6H2 cells (Fig. 7A and B).
Fig 7.
Host gene expression is normal in 6H2 cells. (A) The rpL13a locus is shown, with the noncoding region (black lines), exons (black boxes), and snoRNAs (gray ovals). The full arrow indicates transcription start. Half arrows indicate locations of primers for qRT-PCR analysis of pre-mRNA and mRNA. (B) WT and 6H2 cells were untreated (UT) or treated with palmitate for 9 h. For analysis of rpL13a pre-mRNA expression, total RNA was reverse transcribed using random hexamers and amplified using primers that span the junction between exon 3 and intron 3. For analysis of rpL13a mRNA expression, total RNA was transcribed using oligo(dT) and amplified using primers that span exon-exon junctions. Primer pairs are as indicated in panel A. (C) WT and 6H2 cells were transfected with a split luciferase reporter containing a β-globin intron; 24 h posttransfection, the cells were left untreated or treated with palmitate or clotrimazole (clor) for 4 h. Luminescence was measured and normalized to luciferase pre-mRNA expression by qRT-PCR. (D and E) WT and 6H2 cells were transfected with a split luciferase construct containing the intact U32a intron (sno) or the U32a intron lacking the 83-nucleotide U32a snoRNA (Δsno). Total RNA was analyzed for luciferase pre-mRNA and U32a snoRNA expression. Luminescence (D) and U32a snoRNA (E) were normalized to luciferase pre-mRNA expression. Note that differences between the murine intronic sequences in the reporter construct and endogenous hamster sequences enabled discrimination between exogenous murine and endogenous hamster snoRNAs using species-specific PCR primers. (F) Total RNA was prepared from WT and 6H2 cells and analyzed for host genes of endogenous intron-encoded snoRNAs and mirtrons by qRT-PCR relative to β-actin mRNA. (G) Exon array analysis was used to predict differences in alternative splicing. Relative probe set intensity values from exon arrays were plotted for SmD3 LNA- versus GFP LNA-transfected cells using data from three independent samples/arrays for each condition. All data are expressed as means and SE for three independent experiments. *, P < 0.05; ns, not significant.
To assess splicing efficiency more broadly, we transfected WT and 6H2 cells with a previously validated reporter construct for expression of firefly luciferase from two exons separated by a β-globin intron (48). Functional splicing results in the formation of a luciferase mRNA encoding a protein with measurable luminescence and a processed β-globin intron lariat. In the absence of splicing, a truncated luciferase protein without enzymatic activity is formed due to multiple in-frame stop codons. Following transfection of this construct, no difference in luciferase production was detected between WT and 6H2 cells under untreated or palmitate-treated conditions or in the presence of clotrimazole, a known splicing inhibitor (48) (Fig. 7C). To test whether the presence of an intronic snoRNA affected splicing of flanking exons in 6H2 cells, we replaced the intron in the split luciferase vector with the murine rpL13a intron 2 containing U32a. Following transfection with this construct, there was no difference in luciferase levels between WT and 6H2 cells, although expression of the exogenous murine U32a was reduced in 6H2 cells (Fig. 7D and E). Similarly, we detected no difference in luminescence using a Δsno split luciferase splicing reporter containing mutated rpL13a intron 2 sequences lacking the entire 83-nucleotide U32a, and pre-mRNA from the reporters with the intact U32a intron and the Δsno intron were comparable (data not shown). Furthermore, we quantified mRNA expression of the host genes containing the intronic noncoding elements quantified in Fig. 5G. We detected no differences in mRNA levels from any of these host genes between WT and 6H2 cells (Fig. 7F). Together, these data indicate that differences in intronic noncoding RNA levels are not attributable to defective splicing of exons in 6H2 cells.
SmD3 knockdown does not alter alternative splicing.
Previously, it was reported that 85% or more knockdown of the survival of motor neuron (SMN) protein disrupts the snRNP assembly pathway and leads to widespread differences in snRNA levels and alternative splicing in a number of tissues (49). To test whether reduced levels of SmD3 affected snoRNA processing through broad alterations in alternative splicing, exon utilization was analyzed using the Affymetrix GeneChip mouse exon 1.0 ST microarray. Due to the lack of publically available exon microarrays containing hamster sequences, we performed this analysis in SmD3 knockdown murine fibroblasts that phenocopy the mutant hamster 6H2 cells (Fig. 6A and B). RNA was harvested from three independent samples of GFP and SmD3 knockdown cells. Among the 266,200 probe sets supported by putative full-length mRNA, ∼190,000 probe sets with significant signals above background and representing exons of ∼16,000 out of ∼30,000 genes in the mouse genome were included in the analysis. With a false-discovery rate set at less than 0.1, no genes were identified as having potential splicing pattern changes (change, ≥1.5-fold). Plotting probe set intensity values from control (GFP LNA) samples against SmD3 knockdown samples revealed a highly linear relationship (R2 = 0.98475), consistent with little variance between the two groups (Fig. 7G). These data show that a 50% reduction in SmD3 expression does not have global effects on alternative splicing.
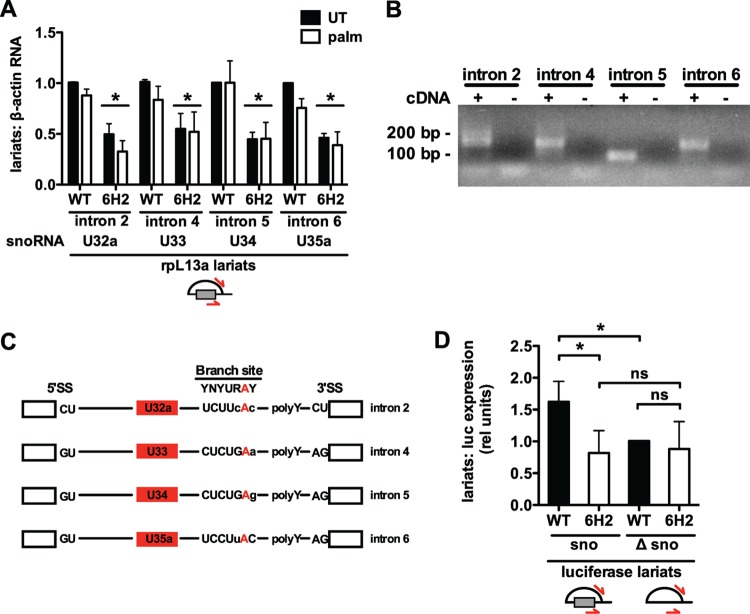
SmD3 controls intron lariat abundance.
Box C/D snoRNAs, box H/ACA snoRNAs, and mirtrons are each defined by unique consensus sequences, protein assembly factors, and positions within the intron (2, 14, 26, 34, 35). Nonetheless, these intronic noncoding RNAs all require pre-mRNA splicing, intron lariat formation, debranching, and exonucleolytic trimming prior to formation of a functional ribonucleoprotein. We used qRT-PCR to probe snoRNA precursors to determine the level of processing at which 6H2 cells are defective. Intron lariats from each rpL13a snoRNA-containing intron were quantified by qRT-PCR primers reading across the branch point with a sense primer designed to a 3′ region of the intron and an antisense primer to a 5′ region (44). This approach revealed a 38 to 63% decrease in lariats from each of the snoRNA-containing rpL13a introns under untreated and palmitate-treated conditions in 6H2 cells (Fig. 8A). The validity of this approach was confirmed by the observation of a single PCR product for each primer pair (Fig. 8B) and sequence analysis of the PCR products (Fig. 8C). Each primer pair read across the branch point and allowed mapping of branch sites in rpL13a introns, each defined by an adenosine and 6 of 7 nucleotides correlating with the consensus major spliceosome branch site. Interestingly, we were unable to detect PCR products for endogenous rpL13a intron lariats lacking snoRNAs, a finding consistent with recent work by others showing that intronic sequences lacking snoRNAs are degraded more quickly than introns containing snoRNAs (47). On the other hand, we were able to quantify both snoRNA-containing and non-snoRNA-containing introns when they were overexpressed in cells transfected with the split luciferase vector construct containing the U32a intron and the snoRNA-deleted U32a sequence, respectively. In WT cells, the presence of the intronic snoRNA was associated with increased lariat abundance, whereas this apparent increase was not observed in 6H2 cells (Fig. 8D). These findings suggest that SmD3 contributes to expression of intronic snoRNAs by enhancing intron lariat formation or stability.
Fig 8.
snoRNA-containing intron lariats are decreased in 6H2 cells. (A) WT and 6H2 CHO cells were untreated or treated with palm for 9 h. Nuclear RNA was isolated and reverse transcribed using random hexamers. rpL13a intron lariat abundance was determined by qRT-PCR using primers reading across branch points. Quantification of the lariat PCR product was normalized to β-actin mRNA. (B and C) PCR products generated by intron lariat primers in CHO WT cells were visualized on a 2% agarose gel (B) or cloned and sequenced (C). (B) Lanes with cDNA (+) contained reverse transcription product as the template. Lanes without cDNA (−) were negative-control PCRs with no reverse transcription product provided as the template. (C) Sequencing revealed the branch site adenosine (red) for each intron. Branch site nucleotides showing conservation (uppercase) are indicated. (D) WT and 6H2 cells were transfected with a split luciferase construct containing the intact murine U32a intron (sno) or the U32a intron lacking the 83-nucleotide U32a snoRNA (Δsno). Intron lariat abundance was determined by qRT-PCR using primers specific for the murine lariat sequences and normalized to luciferase pre-mRNA expression. All data are expressed as means and SE for three independent experiments. *, P < 0.05; ns, not significant.
DISCUSSION
Through the use of a genetic screen in CHO cells, our laboratory has identified loci involved in lipid-induced cell death, a process in which oxidative stress is a central feature. In previously published work, we have shown that lipotoxic and oxidative stress critically involve cytosolic expression of intronic snoRNAs from the rpL13a genomic locus (27). The present study provides new insight into the underlying mechanisms of metabolic stress responses through the characterization of a mutant cell line with disruption of one allele of the gene encoding SmD3. Our data show that reduced cellular levels of SmD3 decrease the propagation of oxidative stress and protect cells from palmitate-induced death. Although mutant 6H2 cells are distinct from previously described palmitate-resistant mutants from our screen, mutation at the SmD3 locus confers a related molecular phenotype in that 6H2 cells fail to induce rpL13a snoRNAs under lipotoxic stress. We show that SmD3, beyond its known role in splicing, regulates expression of the intronic snoRNAs. Compared to WT levels of SmD3, reduced levels of SmD3 lead to decreased intron lariat abundance and decreases in U4 and U5 snRNPs necessary for intron lariat formation. These perturbations decrease the basal levels of the rpL13a intronic snoRNAs and subsequently blunt their induction during lipotoxicity.
SmD3, together with six other Sm proteins, forms a heptameric ring around the uridine-rich Sm-binding site found in snRNAs. These snRNPs form the major building blocks of the spliceosome. Sequential assembly of the hetero-oligomers of SmD1-SmD2, SmE-SmF-SmG, and SmB/B′-SmD3 is a highly regulated process (10, 32). Although these proteins are critical for proper snRNP assembly, our data indicate that haploinsufficiency of SmD3 is sufficient to maintain splicing of a specific endogenous pre-mRNA and an exogenously provided splicing reporter under normal growth conditions. Consistent with an ability to support overall wild-type capacity for mRNA splicing, the growth of mutant 6H2 cells is indistinguishable from that of parental wild-type cells. Our studies suggest that excision of introns during the catalytic stage of splicing goes to completion, and haploinsufficient levels of SmD3 in these cells is not limiting for production of mRNAs. Furthermore, ∼50% knockdown of SmD3 does not cause global changes in exon utilization. Together, these findings indicate that the phenotype of SmD3 haploinsufficiency is not related to global defects in formation of mRNAs.
On the other hand, haploinsufficiency for SmD3 unmasks a previously unsuspected role for the protein in the production of intronic snoRNAs. The precise mechanisms through which SmD3 regulates intronic snoRNA expression remain to be determined. One possibility is that SmD3 has a direct role in snoRNA biogenesis unrelated to its function in snRNPs. Unlike SmD3 haploinsufficient cells, SmB knockdown cells remain sensitive to palmitate-induced death, suggesting that SmD3 has a function distinct from those of other Sm protein family members. Additionally, SmD3 has been observed to function independently of the Sm core in the cytoplasm (12). SmD3 has also been previously shown to bind small Cajal body RNAs (scaRNAs), a class of snoRNAs specifically localizing to Cajal bodies, and human telomerase (hTR) via a CAB box site (11). However, H/ACA snoRNAs U17b and U64, which are affected by haploinsufficiency of SmD3, both lack CAB box sequences. Future studies will be required to decipher whether SmD3 can function independently of the Sm core during snoRNA biogenesis.
Our data are most consistent with a model in which SmD3 does not directly interact with the maturing snoRNA during processing but rather controls processing indirectly through a role in snRNP assembly and/or stabilization (Fig. 9). Knockdown of SmD3 results in decreased expression of U4 and U5 snRNPs that are required for precatalytic spliceosome formation and initiation of an active spliceosome capable of intron lariat formation (25). Reduced expression of snoRNA lariat precursors, resulting from decreased lariat production or stability, leads to decreased abundance of the mature intronic snoRNAs and failure to support their induction during lipotoxicity in 6H2 cells. These differences in intron lariat abundance and snRNP expression suggest that the phenotype observed in 6H2 cells is related to spliceosomal machinery, even though haploinsufficiency of SmD3 is sufficient to maintain production of mRNAs. We also observed that wild-type levels of SmD3 are critical for the ability to express a number of intronic noncoding RNA elements, including other box C/D snoRNAs, H/ACA snoRNAs, and mirtrons. These ncRNAs do not share significant sequence similarities, and each assembles with a unique set of proteins during processing, making it unlikely that SmD3 directly recognizes and associates with the nascent RNP. It has been shown that the C-terminal tail of SmD3 interacts with the central Tudor domain of splicing factor SPF30 (22), indicating that SmD3 may be important for the assembly of splicing factors specific to individual snRNPs. Individual Sm proteins may be critical for the assembly of specific factors within different snRNPs, consistent with the different snRNA expression profiles observed following SmD3 knockdown versus SmB knockdown. In future studies, it will be of interest to determine whether Sm protein-splicing factor interactions are critical for SmD3's role in snRNP assembly and/or stabilization and may provide insights into the distinct but overlapping roles of SmD3 and SmB in assembly and stabilization of snRNPs.
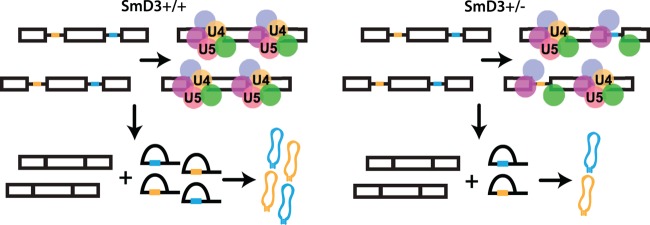
Fig 9.
Role of SmD3 in snoRNA production. A model for the SmD3 role in snoRNA production is shown. The open boxes represent exons, the black lines represent intronic sequences, and snoRNA sequences are shown in yellow and blue. The colored circles represent snRNPs. In the presence of wild-type levels of SmD3, the complement of snRNAs is sufficient to support splicing of pre-mRNAs into mature mRNAs and intron lariats that are sufficiently long-lived to produce snoRNAs. While haploinsufficiency of SmD3 is able to support wild-type levels of mRNA production from splicing, associated decreases in the abundance of the of U4 and U5 snRNPs results in decreased intron lariat abundance and decreased levels of intronic snoRNAs.
Genetic screens provide powerful approaches to dissect cell biological phenomena. Inherent in this approach is the advantage that genes are identified on the basis of their functional contributions to the pathway of study. Since multiple mutations may affect different components in the pathway, genetic approaches also have the ability to elucidate interacting elements. Our screen for mutations that render cells resistant to lipotoxicity identified SmD3 as an upstream regulatory element necessary for the expression of rpL13a snoRNAs. To our knowledge, this is the first report of the loss-of-function phenotype for SmD3. The finding of a second mutation that reduces rpL13a snoRNA expression and leads to resistance to lipotoxicity further highlights the important role of these ncRNAs in metabolic stress responses. More importantly, our study provides novel insights into the broader molecular cell biology of intronic noncoding RNA elements. Future studies of the precise molecular interactions of SmD3 within individual snRNPs are likely to elucidate mechanisms through which the protein regulates noncoding RNA biogenesis.
ACKNOWLEDGMENTS
We are grateful to Gideon Dreyfuss for providing the luciferase splicing reporter. We acknowledge the Alvin J. Siteman Cancer Center at Washington University School of Medicine and Barnes-Jewish Hospital (NCI Cancer Center Support grant P30 CA91842) for use of the Center for Biomedical Informatics and Multiplex Gene Analysis Genechip Core Facility.
This work was supported by grants from the NIH (R01 DK064989 to J.E.S.; R01 HL103001 to D.S.O.; T32 HL07275 to B.S.S.) and the Burroughs Wellcome Foundation (1005935 to J.E.S.).
Footnotes
Published ahead of print 6 August 2012
REFERENCES
- 1. Angulo P. 2002. Nonalcoholic fatty liver disease. N. Engl. J. Med. 346:1221–1231 [DOI] [PubMed] [Google Scholar]
- 2. Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. 2007. Mammalian mirtron genes. Mol. Cell 28:328–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Borradaile NM, et al. 2006. A critical role for eukaryotic elongation factor 1A-1 in lipotoxic cell death. Mol. Biol. Cell 17:770–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Borradaile NM, et al. 2006. Disruption of endoplasmic reticulum structure and integrity in lipotoxic cell death. J. Lipid Res. 47:2726–2737 [DOI] [PubMed] [Google Scholar]
- 5. Brookheart RT, Michel CI, Listenberger LL, Ory DS, Schaffer JE. 2009. The non-coding RNA gadd7 is a regulator of lipid-induced oxidative and endoplasmic reticulum stress. J. Biol. Chem. 284:7446–7454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cacicedo JM, Benjachareowong S, Chou E, Ruderman NB, Ido Y. 2005. Palmitate-induced apoptosis in cultured bovine retinal pericytes. Diabetes 54:1838–1845 [DOI] [PubMed] [Google Scholar]
- 7. de Vries JE, et al. 1997. Saturated but not mono-unsaturated fatty acids induce apoptotic cell death in neonatal rat ventricular myocytes. J. Lipid Res. 38:1384–1394 [PubMed] [Google Scholar]
- 8. El-Assaad W, et al. 2003. Saturated fatty acids synergize with elevated glucose to cause pancreatic β-cell death. Endocrinology 144:4154–4163 [DOI] [PubMed] [Google Scholar]
- 9. Friedrich G, Soriano P. 1991. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 5:1513–1523 [DOI] [PubMed] [Google Scholar]
- 10. Friesen WJ, Massenet S, Paushkin S, Wyce A, Dreyfuss G. 2001. SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol. Cell 7:1111–1117 [DOI] [PubMed] [Google Scholar]
- 11. Fu D, Collins K. 2006. Human telomerase and Cajal body ribonucleoproteins share a unique specificity of Sm protein association. Genes Dev. 20:531–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gonsalvez GB, Rajendra TK, Wen Y, Praveen K, Matera AG. 2010. Sm proteins specify germ cell fate by facilitating oskar mRNA localization. Development 137:2341–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hirose T, Shu MD, Steitz JA. 2003. Splicing-dependent and -independent modes of assembly for intron-encoded box C/D snoRNPs in mammalian cells. Mol. Cell 12:113–123 [DOI] [PubMed] [Google Scholar]
- 14. Hirose T, Steitz JA. 2001. Position within the host intron is critical for efficient processing of box C/D snoRNAs in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 98:12914–12919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Holden P, Horton WA. 2009. Crude subcellular fractionation of cultured mammalian cell lines. BMC Res. Notes 2:243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Inoguchi T, et al. 2000. High glucose level and free fatty acid stimulate reactive oxygen species production through protein kinase C-dependent activation of NAD(P)H oxidase in cultured vascular cells. Diabetes 49:1939–1945 [DOI] [PubMed] [Google Scholar]
- 17. Jiang T, et al. 2005. Diet-induced obesity in C57BL/6J mice causes increased renal lipid accumulation and glomerulosclerosis via a sterol regulatory element-binding protein-1c-dependent pathway. J. Biol. Chem. 280:32317–32325 [DOI] [PubMed] [Google Scholar]
- 18. Kiss T, Filipowicz W. 1995. Exonucleolytic processing of small nucleolar RNAs from pre-mRNA introns. Genes Dev. 9:1411–1424 [DOI] [PubMed] [Google Scholar]
- 19. Lee Y, et al. 2006. Alpha-lipoic acid prevents lipotoxic cardiomyopathy in acyl CoA-synthase transgenic mice. Biochem. Biophys. Res. Commun. 344:446–452 [DOI] [PubMed] [Google Scholar]
- 20. Listenberger LL, et al. 2003. Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc. Natl. Acad. Sci. U. S. A. 100:3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Listenberger LL, Ory DS, Schaffer JE. 2001. Palmitate-induced apoptosis can occur through a ceramide-independent pathway. J. Biol. Chem. 276:14890–14895 [DOI] [PubMed] [Google Scholar]
- 22. Little JT, Jurica MS. 2008. Splicing factor SPF30 bridges an interaction between the prespliceosome protein U2AF35 and tri-small nuclear ribonucleoprotein protein hPrp3. J. Biol. Chem. 283:8145–8152 [DOI] [PubMed] [Google Scholar]
- 23. Liu L, et al. 2009. DGAT1 expression increases heart triglyceride content but ameliorates lipotoxicity. J. Biol. Chem. 284:36312–36323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Maedler K, Oberholzer J, Bucher P, Spinas GA, Donath MY. 2003. Monounsaturated fatty acids prevent the deleterious effects of palmitate and high glucose on human pancreatic beta-cell turnover and function. Diabetes 52:726–733 [DOI] [PubMed] [Google Scholar]
- 25. Makarov EM, et al. 2002. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298:2205–2208 [DOI] [PubMed] [Google Scholar]
- 26. Matera AG, Terns RM, Terns MP. 2007. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 8:209–220 [DOI] [PubMed] [Google Scholar]
- 27. Michel CI, et al. 2011. Small nucleolar RNAs U32a, U33, and U35a are critical mediators of metabolic stress. Cell Metab. 14:33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nicoloso M, Qu LH, Michot B, Bachellerie JP. 1996. Intron-encoded, antisense small nucleolar RNAs: the characterization of nine novel species points to their direct role as guides for the 2′-O-ribose methylation of rRNAs. J. Mol. Biol. 260:178–195 [DOI] [PubMed] [Google Scholar]
- 29. Ooi SL, Samarsky DA, Fournier MJ, Boeke JD. 1998. Intronic snoRNA biosynthesis in Saccharomyces cerevisiae depends on the lariat-debranching enzyme: intron length effects and activity of a precursor snoRNA. RNA 4:1096–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ory DS, Neugeboren BA, Mulligan RC. 1996. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc. Natl. Acad. Sci. U. S. A. 93:11400–11406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ostrander DB, Sparagna GC, Amoscato AA, McMillin JB, Dowhan W. 2001. Decreased cardiolipin synthesis corresponds with cytochrome c release in palmitate-induced cardiomyocyte apoptosis. J. Biol. Chem. 276:38061–38067 [DOI] [PubMed] [Google Scholar]
- 32. Pellizzoni L, Yong J, Dreyfuss G. 2002. Essential role for the SMN complex in the specificity of snRNP assembly. Science 298:1775–1779 [DOI] [PubMed] [Google Scholar]
- 33. Petfalski E, Dandekar T, Henry Y, Tollervey D. 1998. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol. Cell. Biol. 18:1181–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Richard P, Kiss AM, Darzacq X, Kiss T. 2006. Cotranscriptional recognition of human intronic box H/ACA snoRNAs occurs in a splicing-independent manner. Mol. Cell. Biol. 26:2540–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ruby JG, Jan CH, Bartel DP. 2007. Intronic microRNA precursors that bypass Drosha processing. Nature 448:83–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sauterer RA, Feeney RJ, Zieve GW. 1988. Cytoplasmic assembly of snRNP particles from stored proteins and newly transcribed snRNA's in L929 mouse fibroblasts. Exp. Cell Res. 176:344–359 [DOI] [PubMed] [Google Scholar]
- 37. Sharma S, et al. 2004. Intramyocardial lipid accumulation in the failing human heart resembles the lipotoxic rat heart. FASEB J. 18:1692–1700 [DOI] [PubMed] [Google Scholar]
- 38. Shimabukuro M, Zhou YT, Levi M, Unger RH. 1998. Fatty acid-induced beta cell apoptosis: a link between obesity and diabetes. Proc. Natl. Acad. Sci. U. S. A. 95:2498–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shpargel KB, Matera AG. 2005. Gemin proteins are required for efficient assembly of Sm-class ribonucleoproteins. Proc. Natl. Acad. Sci. U. S. A. 102:17372–17377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Song B, Scheuner D, Ron D, Pennathur S, Kaufman RJ. 2008. Chop deletion reduces oxidative stress, improves β cell function, and promotes cell survival in multiple mouse models of diabetes. J. Clin. Invest. 118:3378–3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sunny N, Parks E, Browning J, Burgess S. 2011. Excessive hepatic mitochondrial TCA cycle and gluconeogenesis in humans with nonalcoholic fatty liver disease. Cell Metab. 14:804–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tycowski KT, Shu MD, Steitz JA. 1993. A small nucleolar RNA is processed from an intron of the human gene encoding ribosomal protein S3. Genes Dev. 7:1176–1190 [DOI] [PubMed] [Google Scholar]
- 43. Unger RH. 1995. Lipotoxicity in the pathogenesis of obesity-dependent NIDDM. Genetic and clinical implications. Diabetes 44:863–870 [DOI] [PubMed] [Google Scholar]
- 44. Vogel J, Hess WR, Börner T. 1997. Precise branch point mapping and quantification of splicing intermediates. Nucleic Acids Res. 25:2030–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wei Y, Wang D, Topczewski F, Pagliassotti MJ. 2006. Saturated fatty acids induce endoplasmic reticulum stress and apoptosis independently of ceramide in liver cells. Am. J. Physiol. Endocrinol. Metab. 291:E275–E281 [DOI] [PubMed] [Google Scholar]
- 46. Will CL, Lührmann R. 2001. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 13:290–301 [DOI] [PubMed] [Google Scholar]
- 47. Windhager L, et al. 26 April 2012, posting date. Ultra short and progressive 4sU-tagging reveals key characteristics of RNA processing at nucleotide resolution. Genome Res. doi:10.1101/gr.131847.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Younis I, et al. 2010. Rapid-response splicing reporter screens identify differential regulators of constitutive and alternative splicing. Mol. Cell. Biol. 30:1718–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang Z, et al. 2008. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell 133:585–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zieve GW, Sauterer RA, Feeney RJ. 1988. Newly synthesized small nuclear RNAs appear transiently in the cytoplasm. J. Mol. Biol. 199:259–267 [DOI] [PubMed] [Google Scholar]