Abstract
Diacylglycerol kinase α (DGKα) regulates diacylglycerol levels, catalyzing its conversion into phosphatidic acid. The α isoform is central to immune response regulation; it downmodulates Ras-dependent pathways and is necessary for establishment of the unresponsive state termed anergy. DGKα functions are regulated in part at the transcriptional level although the mechanisms involved remain poorly understood. Here, we analyzed the 5′ end structure of the mouse DGKα gene and detected three binding sites for forkhead box O (FoxO) transcription factors, whose function was confirmed using luciferase reporter constructs. FoxO1 and FoxO3 bound to the 5′ regulatory region of DGKα in quiescent T cells, as well as after interleukin-2 (IL-2) withdrawal in activated T cells. FoxO binding to this region was lost after complete T cell activation or IL-2 addition, events that correlated with FoxO phosphorylation and a sustained decrease in DGKα gene expression. These data strongly support a role for FoxO proteins in promoting high DGKα levels and indicate a mechanism by which DGKα function is downregulated during productive T cell responses. Our study establishes a basis for a causal relationship between DGKα downregulation, IL-2, and anergy avoidance.
INTRODUCTION
The diacylglycerol kinases (DGK) phosphorylate diacylglycerol (DAG) into phosphatidic acid (PA), modulating the levels of these two lipid second messengers, which have several key functions in cells. DAG propagates signals by membrane recruitment of cytosolic proteins containing C1 domains, such as protein kinase C and D (PKC and PKD, respectively), the Ras-guanine nucleotide exchange factor (GEF) RasGRP1, and the Rac-GTPase-activating protein (GAP) chimaerins (3). DAG deregulation is linked to tumorigenesis, metastasis, diabetes, heart disease, and altered immune responses (9, 13, 45, 53). PA binds and activates proteins involved in cell growth, survival, vesicular trafficking, and cytoskeletal remodeling, and its altered metabolism is also linked to disease onset (7, 14, 40). Interest in the DGK as key modulators of DAG and PA function has increased in recent years as better understanding of DGK regulatory mechanisms offers opportunities for the development of novel strategies to modulate lipid metabolism for therapeutic purposes (for reviews, see references 32 and 44).
DGK function attracted special attention following the characterization of its role in T lymphocyte activation. Productive activation of T lymphocytes requires the integration of the pathways regulated by Ras/mitogen-activated protein kinase (MAPK)/AP1 and Ca2+/nuclear factor of activated T cells (NFAT). Failure to trigger an adequate balance of these signals, due, for example, to lack of costimulation, drives T cells into a nonresponsive state termed anergy, in which cells survive for long periods in the absence of proliferation (1). DGKα is a type I DGK particularly abundant in thymus and mature T lymphocytes (55), and early studies showed its function as a negative modulator of the Ras/MAPK pathway. DGKα limits DAG-mediated membrane localization and activation of the Ras GEF RasGRP1 following T cell receptor (TCR) triggering and is subjected to precise transcriptional regulation throughout T cell activation. Naive T cells express high DGKα levels, which diminish rapidly following T cell encounter with antigen-presenting cells (47). DGKα downregulation permits adequate DAG-mediated activation of the RasGRP1/Ras/MAPK/AP1 pathway, essential for productive T cell responses. DGKα was identified as a gene upregulated under anergic conditions, confirming a causal relationship between high DGKα expression and failure to complete a productive T cell response (28). Studies in mouse models further confirmed the contribution of DGKα to nonproliferative T cell states; its overexpression in mouse T lymphocytes impaired T cell activation and expansion, whereas T cells from DGKα-deficient mice were anergy resistant (38, 56).
Despite identification of DGKα as an anergy-induced gene and its acute downregulation during T cell activation, the basic mechanisms that regulate DGKα expression in T lymphocytes remain unknown. The earliest attempt to dissect DGKα-regulatory elements came from Fujikawa et al. for the human DGKα (hDGKα) gene (10); they reported an initiator element (Inr), no TATA box, and putative binding sites (BS) for transcription factors Ets-1, Sp1, and Ap-2. Although it is induced by anergic conditions, DGKα expression is not regulated by NFAT, regardless of the critical role of this transcription factor in the control of other anergy-induced genes (49).
Here, we report the initial characterization of the 5′-end structure of the mouse DGKα (mDGKα) gene. Analysis of this region revealed several conserved binding sites for various transcription factors and suggests the presence of at least two putative alternative promoters. DGKα mRNA levels are high in quiescent lymphocytes but decrease after TCR activation. We observed that the magnitude and duration of this decrease correlated with the intensity of activation, that they were enhanced by costimulation, and that they were further maintained by interleukin-2 (IL-2) addition. Our data strongly support the concept that elevated DGKα expression in quiescent, nonactivated cells is regulated by three FoxO-binding sites, identified at the distal 5′ end region of the gene and conserved in mammals. Our studies identify a mechanism in T cells by which DGKα function is downstream of the AKT/FoxO axis. This mechanism provides a plausible explanation for the causal relation between “weak” TCR stimulation and anergy induction and the capacity of IL-2 to rescue anergic cells.
MATERIALS AND METHODS
Mice, tissue preparation, cell lines, and cell culture.
Mouse tissues were isolated from 6- to 12-week-old BALB/c or C57BL/6J mice according to protocols approved by the CNB/CSIC Ethics Committee on Animal Experimentation. C57BL/6 DGKα−/− mice have been reported previously (38). C57BL/6-Tg (TcraTcrb) 1100Mjb/J (OT-1) mice were purchased from The Jackson Laboratory and crossed with DGKα−/− mice to generate O-T1 DGKα−/− mice in the Centro Nacional de Biotecnología/Consejo Superior de Investigaciones Científicas (CNB/CSIC) facilities. NIH 3T3, HEK293, F9, M1, and EL4 cell lines were obtained from the ATCC. Pro-B BaF/3 lymphoid cells, engineered to express human IL-2 receptor β (IL-2Rβ), were cultured in WEHI.3B cell-conditioned medium as an IL-3 source (33).
BaF/3 cells in exponential growth were synchronized by deprivation of conditioned medium for 20 h, after which cell cycle entry was induced by IL-2 addition (50 U/ml). Cells were harvested at different times and processed for RNA extraction.
RNA isolation, RT-PCR, full-length cDNA amplification, and real-time PCR.
Total RNA extraction was performed with TRIzol reagent (TriReagent; Sigma). Primers used are listed in Table S1 in the supplemental material. For reverse transcription-PCR (RT-PCR) amplification, 1 μg of total RNA was processed to eliminate DNA, followed by first-strand cDNA synthesis using oligo(dT)12–18 (Invitrogen). To amplify mDGKα full-length cDNA (end-to-end RT-PCR amplification), cDNA synthesis was performed with oligo(dT) and total DNase I-treated RNA from adult BALB/c mouse thymus. cDNA was treated with RNase H (Invitrogen) to eliminate the parental strand, and primer a or c, together with the 3′ end primer g, was used for long PCR amplification with the Expand long template system (Roche). The product (∼2.8 kb) was excised from agarose gel, purified using QIAquick columns (Qiagen, Germany), and cloned in the pGEM-T Easy vector (Promega). This plasmid was introduced into competent cells; several clones were picked, grown, and purified on Qiagen columns. All clones were sequenced. PCR amplification was performed in a MyCycler thermal cycler (Bio-Rad Laboratories).
For real-time PCR, we used cDNA as the template and Fast SYBR green master mix (Applied Biosystems) on an ABI Prism 7900HT iCycler (Applied Biosystems). All data are expressed as the ratio of target gene expression to that of the housekeeping gene 36B4. For DGKα amplification, we used an optimized version of routinely used primers (see Table S1 in the supplemental material). Agarose gel analysis and melt curves were analyzed to ensure amplification of specific target sequences. For other genes, primers used have been reported previously (36B4, alias Rplp0 [41], DGKζ [38], p27 [4], l-selectin [21], and perforin [29]).
PE assay.
Primer extension (PE) was performed according to Phelps et al. (42). We used 90 μg of total RNA from tissues (thymus and liver) and cell lines (F9 teratocarcinoma and M1 acute myeloid leukemia) or from torulla yeast (Candida utilis Y660) (Ambion) as a negative control and primer d located in exon 1 of the mDGKα transcript (NM_016811). The primer was 5′ end labeled with T4 polynucleotide kinase (New England BioLabs) and [32P]dCTP and purified on a G-25 column (Bio-Rad).
Isolation and stimulation of mouse T lymphocytes.
Six- to 12-week-old mice were sacrificed according to national and European Union (EU) guidelines, and T cells were isolated from spleen or thymus according to standard protocols (34). For TCR stimulation experiments, T cells were purified from spleen using negative selection (Dynabeads; Invitrogen). Cells (107) were processed or seeded in 3-cm-diameter plates (precoated with anti-CD3 antibody as indicated in Fig. 2) in complete medium (RPMI medium with 10% fetal bovine serum [FBS], 2 mM glutamine, β-mercaptoethanol) with anti-CD3 or -CD28 antibody or both (1.0 or 0.2 μg/ml, respectively); cells were also concanavalin A (ConA) stimulated (2 μg/ml; Sigma) for different times. Cells were harvested and processed for RNA isolation, Western blotting, or chromatin immunoprecipitation (ChIP) analysis. Spleen CD8+ T cells were purified from DGKα−/− mice and wild-type (wt) littermates, stimulated as indicated in Fig. 5 and then stained and processed for flow cytometry analysis. For analysis of OT-1 transgenic T cells, total splenocytes (2 × 107) were cultured in medium containing a cognate peptide (SIINFEKL; at 2 μM for 2 days). Cells were washed extensively and starved for 2 days or maintained with IL-2 (50 U/ml) alone or with 1 μM AKT inhibitor (AKTi) VIII (AKTi-1/2; Calbiochem).
Fig 2.
DGKα mRNA levels decrease in response to activation stimulus. Spleen T lymphocytes were cultured in medium containing anti-CD3 antibody (CD3 soluble) or on CD3-coated plates in medium containing anti-CD28 antibody (CD3 plate + CD28). Cells were collected at the indicated times, RNA was extracted, and real-time PCR was performed. (A) Relative expression of the transcripts derived from DGKα (all versions with internal primers [Fig. 1A] and the two promoters), DGKζ and p27. Data indicate fold expression relative to that of each gene under basal conditions. (B) Spleen T lymphocytes were cultured on plate-bound CD3 in medium alone or with anti-CD28 antibody and processed as described for panel A. Data indicate fold expression relative to plate-bound CD3 at 24 h. (C) IL-2 was added to BaF/3 cells after 24 h of deprivation of cytokines and serums. Cells were collected at indicated times, RNA was isolated, and transcripts were amplified by RT-PCR. Bcl-X expression, which increases during BaF/3 cell proliferation, was monitored as a control. One representative experiment is shown for each case (n = 3). A 100-bp DNA ladder was used for size markers (lane M).
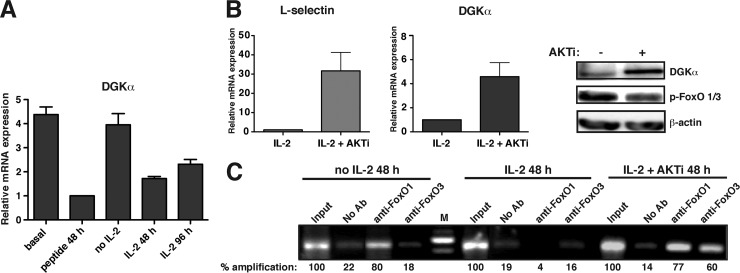
Fig 5.
DGKα expression is under the control of the AKT-FoxO axis. CD8+ T lymphocytes from OT-1 transgenic mice were incubated with the cognate peptide (48 h), washed extensively, and treated as indicated. (A) Cells were rested for 48 h or IL-2 challenged at 48 and 96 h. DGKα expression was then evaluated by real-time PCR using internal primers. Data indicate fold expression relative to that of peptide stimulation conditions. (B) Cells were IL-2 challenged (48 h) in the presence of the inhibitor AKTi. DGKα and l-selectin expression were determined by real-time PCR, using the internal primers in the case of DGKα. Data indicate fold expression relative to that of IL-2 stimulation conditions (left and center). DGKα levels were also evaluated by Western blotting (right). (C) Cells were rested (48 h) in medium (RPMI medium) or IL-2 stimulated (48 h) alone or in the presence of AKTi. ChIP of DGKα FoxO BS2 was performed using the indicated anti-FoxO antibody. Amplification products were analyzed by gel electrophoresis and quantified using ImageJ. Values are represented as a percentage relative to the ChIP input fraction, considered 100% amplification. In all cases, results are representative of at least three independent experiments.
For secondary antigen challenge, CD8+ T cells were mixed 1:1 with EL4 cells, unloaded or preloaded with peptide-specific ligands (SIINFEKL for high affinity and SIIQFEHL for low affinity), incubated (20 min at 37°C), and processed for analysis.
Plasmids and construction of luciferase reporter vectors.
The mouse bacterial artificial chromosome (BAC) clone RP24-341N10 (BACPAC Resources; Children's Hospital, Oakland Research Institute, Oakland, CA) containing the entire mouse DGKα locus was used as the template. Two distinct promoter fragments were generated by PCR amplification using primers containing BglII or BamHI sequences. Amplified fragments were doubly digested with these enzymes, cloned into the luciferase-encoding, promoterless pGL3 basic plasmid (Promega), and verified by sequencing (Unidad de Genómica, Parque Científico/UAM, Madrid, Spain). The pGL3DGKα short promoter (pGL3DGKαshort) covered positions −370 to +41, while the pGL3 DGKα long promoter (pGL3DGKαwt) covered positions −1827 to +41 and contained the three FoxO BS (see Fig. 3A for relative positions). Point mutations were introduced into pGL3DGKα using mutant primers specific for each of the three FoxO BS (see Fig. 3B; also see Table S1 in the supplemental material), and a QuikChange site-directed mutagenesis kit (Stratagene) was used to generate plasmids with mutations in distinct FoxO BS. The pRL-null vector (Promega), which encodes Renilla luciferase, was used as an internal transfection control in luciferase activity experiments. Plasmids containing human FoxO1 (pCDNA FoxO1) and FoxO3a (pECE-FoxO3a) genes were purchased from Addgene.
Fig 3.
FoxO transcription factors enhance DGKα promoter activity. (A) Putative regulatory elements in the mouse DGKα gene. Transcription initiation sites are indicated (arrows). Promoter and regulatory regions (Pr and Rr, respectively) are depicted (boxes). The +1 position was assigned in the Inr element. Putative binding sites for transcription factors are indicated by rectangles; FoxO sites are gray. (B) Sequence of the three FoxO-binding sites (BS1, BS2, and BS3) of the mouse DGKα gene. The mouse core consensus sequence (see reference 2) is in bold, and mutated nucleotides are indicated (stars). (C) Luciferase activity of NIH 3T3 cells transfected with the indicated reporter constructs, alone (gray bars) or with pECE-FoxO3 (black bars). Relative light units (RLU) indicate the luciferase/Renilla ratio (mean ± standard deviation of three independent experiments). Lane M, size markers. (D) Luciferase activity of NIH 3T3 cells transfected with the indicated reporter constructs. Data indicate fold induction of luciferase activity for each construct after cotransfection with pECE-FoxO3. Data are representative of three independent experiments (mean ± standard deviation). mut, mutant.
Transient cell transfections and luciferase reporter assays.
NIH 3T3 cells, cultured in 6-cm-diameter plates, were transfected with the luciferase reporter constructs (1.5 μg), Renilla luciferase vector (0.5 μg), and, when indicated in Fig. 3, with the FoxO-encoding plasmid (1.5 μg), using Lipofectamine 2000 (Invitrogen); after 48 h, cells were starved and processed using a dual-luciferase reporter assay system (Promega). Firefly and Renilla luciferase activities were measured in a plate luminometer (Infinite plate reader; Tecan). A fraction of each sample was analyzed by Western blotting to confirm FoxO protein overexpression.
Nuclear cell extracts.
Cells were harvested, washed with phosphate-buffered saline (PBS), and resuspended in 1 volume of cold NAR-A buffer (10 mM HEPES, pH 7.9, 10 mM KCl, 0.1 mM EDTA) containing 1 mM dithiothreitol (DTT) and protease and phosphatase inhibitors. The cell suspension was maintained on ice (10 min), one volume of 1% NP-40 was added, and the mixture was incubated (for 3 min at room temperature). Samples were mixed on a vortex for at least 10 s until a white pellet appeared and then centrifuged (5,000 × g for 2 min at 4°C). The supernatant corresponded to cytoplasmic extracts. A volume of NAR-C buffer (20 mM HEPES, pH 7.9, 0.4 M NaCl, 1 mM EDTA) containing protease and phosphatase inhibitors was then added to the pellet, which corresponded to cell nuclei, and the mixture was incubated (30 to 60 min at 4°C, with strong agitation). The suspension was then centrifuged (15,000 × g for 5 min at 4°C), and the supernatants, which corresponded to nuclear extracts, were processed to determine protein concentration.
EMSA.
We used three pairs of complementary primers covering the putative FoxO BS of the mDGKα gene promoter (see Table S1 in the supplemental material). The forward primer in each case was biotin labeled at the 3′ end (Sigma). To obtain the heteroduplex DNA probe, the corresponding forward and reverse primers were mixed at a 1:1 molar ratio, heated to 70°C, and allowed to cool slowly at room temperature. Nuclear extracts (10 μg) were incubated with 100 pmol of DNA probe in electrophoretic mobility shift assay (EMSA) binding buffer (30 min at room temperature), using an EMSA accessory kit (Novagen). Protein-DNA complexes were separated in 6% acrylamide nondenaturing gels and transferred to a positively charged nylon membrane (Hybond N+; Amersham Biosciences), and DNA probe complexes were detected by incubating membranes with streptavidin-peroxidase (overnight at 4°C) and developed with enhanced chemiluminescence (ECL).
Cell lysis and Western blot analysis.
Cells were harvested, rinsed once with cold PBS, and lysed with p70 buffer (10 mM HEPES, pH 7.5, 15 mM KCl, 1 mM EDTA, 1 mM EGTA, 10% glycerol, 0.2% NP-40). For blots in Fig. 6 and 7, radioimmunoprecipitation assay (RIPA) buffer was used instead of p70. Protease and phosphatase inhibitors were freshly added to buffers. Cell lysates (30 to 50 μg) were separated by SDS-PAGE, transferred to polyvinylidene difluoride (PVDF) membranes (Bio-Rad), and analyzed by Western blotting. Anti-phospho-FoxO1 and -FoxO3 (pFoxO1/3), anti-FoxO1, and anti-FoxO3a antibodies were from Cell Signaling, anti-β-actin was from Sigma, and anti-p27 was from MBL; DGKα was detected with a rabbit polyclonal antibody generated in our laboratory (47).
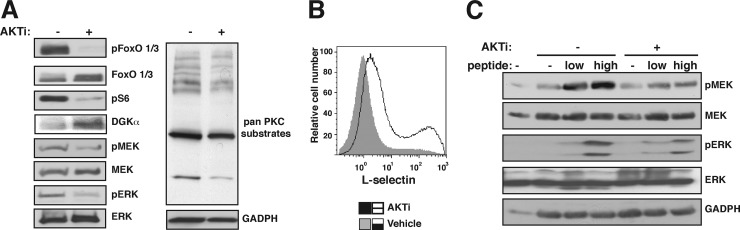
Fig 6.
AKT/FoxO axis control DAG-meditated responses. CD8+ T lymphocytes from OT-1 transgenic mice were incubated with the cognate peptide (48 h), washed extensively, and IL-2 challenged (48 h) in the presence of AKTi. (A) Phosphorylation (p-) levels of FoxO, S6, MEK, ERK, and PKC substrates were evaluated by Western blotting. (B) Surface expression of l-selectin determined by flow cytometry in vehicle- and AKTi-treated cells. (C) T cells were mixed with EL4 cells alone or loaded with low- or high-affinity peptide (20 min). The cell mixture was lysed for Western blot analysis. MEK and ERK phosphorylation levels were evaluated. Lysate of EL4 cells was included as an input control (first lane). Results are representative of at least three independent experiments. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Fig 7.
DGKα regulates IL-2-mediated responses. (A) CD8+ T lymphocytes from wt and DGKα-deficient mice were stimulated with plate-bound anti-CD3 antibody and soluble anti-CD28 antibody and, where indicated, with IL-2 for 72 h. Proliferation was determined by carboxyfluorescein succinimidyl ester dilution analysis. (B) Wild-type and DGKα-deficient mouse CD8+ T lymphocytes from OT-1 transgenic mice were incubated with the cognate peptide (48 h) in the presence of AKTi. Phosphorylation levels of FoxO, MEK, and ERK were evaluated by Western blotting and quantified using ImageJ. Relative phosphorylation is indicated beneath each blot. FoxO phosphorylation was normalized to total FoxO levels, and MEK and ERK phosphorylation levels were normalized to the GAPDH level. In each case, the percentage was determined considering phosphorylation of wt cells without inhibitor as 100%. (C) Wild-type and DGKα-deficient mouse CD8+ T lymphocytes from OT-1 transgenic mice were incubated with the cognate peptide (peptide 48 h), washed extensively, and rested for 48 h (no IL-2) or IL-2 challenged for 96 h (IL-2 96 h). Expression levels of l-selectin and the cytotoxic marker perforin were evaluated by real-time quantitative PCR. In all cases, the data are representative of at least three independent experiments. (D) Model of FoxO-mediated transcriptional regulation of the DGKα gene. In quiescent lymphocytes, FoxO transcription factors bind to the DGKα promoter, leading to elevated DGKα expression. Weak TCR activation leads to a transitory downregulation of the protein. Strong TCR activation together with costimulation enhances PI3K/AKT pathway activation, leading to sustained FoxO phosphorylation and preventing FoxO binding to the DGKα promoter. Under these conditions DGK levels drop. Via modulation of the AKT/FoxO pathway, IL-2 also promotes a sustained drop in DGKα expression, allowing activation of Ras-derived pathways.
Chromatin immunoprecipitation.
Endogenous binding of FoxO transcription factors to the mDGKα gene was determined as described in Sandoval et al. (46), with minor modifications. Nuclei isolated from formaldehyde-cross-linked cells were lysed, and cross-linked chromatin was sonicated to yield 400- to 800-bp fragments. Diluted soluble chromatin fragments were precleared with blocked protein G coupled to Dynabeads (Invitrogen). FoxO-DNA complexes were immunoprecipitated from 50 μg of DNA using FoxO1 or FoxO3 antibody (2 μg; Cell Signaling). Immune complexes were isolated with blocked protein G coupled to Dynabeads, washed extensively, and chromatin eluted by heating. A control aliquot of cross-linked chromatin was treated as above, in the absence of antibody. The supernatant of this immunoprecipitation was used as a positive control (input) to amplify FoxO BS; the eluted fraction was used as a negative control (no-antibody control). DNA from all samples was purified with a PCR purification kit (Qiagen) and used for PCR analysis with specific primers covering the three putative FoxO BS (see Table S1 in the supplemental material). PCR fragments were separated by 1% agarose gel electrophoresis, stained with ethidium bromide, and quantified using ImageJ software.
Bioinformatic analyses.
Sequence searches and analysis were performed using the University of California at Santa Cruz (UCSC) Genome Browser (http://genome.ucsc.edu/index.html) (11), regulatory Vista (rVista), version 2.0 (27), and Blat (20) programs. Sequence alignments were performed using CLUSTALW (http://www.genome.jp/tools/clustalw/).
Nucleotide sequence accession numbers.
The sequence data from this study have been deposited in the NCBI GenBank database under accession numbers JN832677, JN832678, JN832679, JN832680, JN832681, JN832682, and JN832683.
RESULTS
Alternative transcription at the 5′ end of the mDGKα gene.
The mouse DGKα gene (Dgka) covers 28.29 kb and maps to the reverse strand of chromosome 10. The reported mouse DGKα mRNA reference, NM_016811, suggests that transcription starts in a region approximately 200 bp before the end of exon 1. The first ATG is located in exon 2; this transcription start site (TSS) thus renders a 370-bp 5′ untranslated region (UTR) (Fig. 1A). This organization differs from that reported for the human DGKα gene, in which the TSS was mapped relative to an Inr element and is more distant from the first ATG (10). Based on these observations, we tested whether the Inr element is also conserved in the mouse genome (UCSC Genome Browser, mouse, July 2007; NCB137/mm9). We identified the Inr sequence position and observed a high degree of identity (>85%) between mouse and human sequences (see Fig. S1 in the supplemental material). In mouse, the Inr is located more than 700 bp upstream of the 5′ end of DGKα mRNA (NM_016811) (Fig. 1A), suggesting the existence of an alternative TSS.
Fig 1.
Analysis of the mouse DGKα 5′ end and transcripts generated. (A) Position of promoter regions, transcription initiation sites (arrows), exons, Inr, primers, splicing events, and the three DGKα 5′ end transcripts detected. (B) RT-PCR amplification of transcript fragments from thymus poly(A)+ RNA. (C) PE analysis of RNA from tissues (thymus and liver) and cell lines (F9 teratocarcinoma and M1 acute myeloid leukemia). Arrowheads indicate the sizes of amplified transcripts. A 100-bp DNA ladder (B) or a DraI digestion of pUC18 plasmid (C) was used for size markers (lanes M).
To determine whether the Inr is functional, we performed RT-PCR using RNA poly(A)+ from mouse thymus, with one primer downstream of the Inr (primer a) and another located in exon 2 (primer b) (Fig. 1; see also Table S1 in the supplemental material). Two products were obtained, one of 384 and one of 336 bp (Fig. 1B, left). Sequence analysis of the two amplicons showed that they diverge in a small (50 bp) internal region and that both differ from the NM_016811 sequence at the 5′ end (see Fig. S2 in the supplemental material). RT-PCR amplification using primer c, positioned a few bases upstream of the beginning of the NM_016811 transcript sequence, and primer b yielded a 371-bp fragment that corresponds in sequence to the NM_016811 transcript (Fig. 1B, right; see also Fig. 2). This indicates that the transcript generated at this region has a 5′ UTR longer than that previously described.
Complete gene transcripts were amplified using primer a or c in combination with primer g, located at the 3′ end of DGKα mRNA (see Table S1 and Fig. S3 in the supplemental material), further confirming the existence of mDGKα mRNAs with alternative 5′ end regions. Our data showed alternative first exons at the mDGKα gene locus. Alternative processing at the 5′ end of the gene coincided with several expressed sequence tags (EST) and exon-intron boundaries in the region (see Tables S2 and S3 in the supplemental material). We propose a nomenclature for the additional 5′ end exons (Fig. 1A) and show the organization of the transcripts generated (see Fig. S3).
To corroborate variation in transcription initiation, we used primer extension (PE) analysis with a labeled primer (Fig. 1A, primer d) and RNA from several sources, including yeast RNA as a negative control and RNA from the murine F9 and M1 cell lines. We obtained several extension products, indicating possible distinct TSSs, with a major band of ∼200 bp that corresponds to that derived from the Inr region and includes exon 1b (Fig. 1C).
The distance between the two TSSs (approximately 700 bp) and the variation in the use of the first exons suggest alternative promoter regions in the mDGKα gene. Such separation between TSSs due to alternation in promoters is common and widely reported for mammalian genes (22, 25). The existence of alternative promoter elements is further supported by the characteristic organization of the regions surrounding each TSS (described below). Combined in silico analysis and experimental RT-PCR of different mouse tissues and cell lines thus indicate two promoter regions in the mDGKα gene, which we designated promoter region 1 (Pr1) and promoter region 2 (Pr2) (Fig. 1A), and suggest regulatory complexity in the control of DGKα expression.
Quiescent T cells have high mDGKα mRNA levels.
In T lymphocytes, translated DGKα mRNA levels decrease in response to TCR engagement, which correlates with the decrease in DGKα protein during the T cell activation process (47). We used real-time PCR to test variation in mDGKα gene-derived transcript levels in murine T cells following productive and nonproductive stimulation (Fig. 2A). Transcripts derived from both promoters were amplified in purified naïve spleen T cells, which are in G0 cell cycle phase and thus quiescent. As a control, we amplified an internal common region for all transcripts (Fig. 1A, oligonucleotides e and f). To induce “weak” TCR cell stimulation, cells were stimulated with medium containing anti-TCR antibody (anti-CD3), which induced a sharp, although transitory, decrease in DGKα mRNA transcript levels at 24 h; at 48 h, transcript levels were near 80% of initial values (Fig. 2A). To induce strong, productive T cell activation, cells were cultured on anti-CD3 antibody-coated plates, and an antibody to the costimulatory molecule CD28 was added to the medium (anti-CD3/CD28). This stimulation also decreased transcript levels, but in a difference from weak stimulation, this reduction was persistent. There were no differences in the activities of the two DGKα promoters, which showed a pattern similar to that of the common internal region. Expression levels of the other major T cell-expressed DGK, the ζ isoform, and the cell cycle inhibitor p27 were also evaluated under these stimulation conditions (Fig. 2A). DGKζ expression decreased with TCR stimulation, but no gross differences were observed among the different stimulation conditions. In contrast, p27 mRNA levels followed a tendency similar to that of DGKα mRNA; the decrease was transitory during weak TCR activation but remained low during productive T cell activation.
We tested variation in mDGKα gene transcription in response to costimulation. In this case, cells were cultured on anti-CD3-coated plates in medium alone or with anti-CD28 antibody (Fig. 2B). At 48 h, the effect of TCR stimulation on the DGKα transcription rate was modestly enhanced by costimulation and affected both promoters.
TCR triggering leads to synthesis and secretion of the cytokine IL-2, which elicits T cell proliferation in an autocrine and paracrine fashion (31). Some studies have shown DGKα downregulation as a result of IL-2 binding to its high-affinity receptor in activated T cells (52). We tested IL-2-mediated changes in DGKα mRNA levels using BaF/3 pro-B cells ectopically expressing the IL-2 receptor, a model used to assess IL-2-triggered signals independently of those elicited by the TCR (33). Quiescent cells expressed the three DGKα transcripts, whose levels dropped sharply after cell stimulation and then increased at 24 h (Fig. 2C); again, amplification patterns were similar for the two transcripts and the common internal region. Bcl-X gene expression, which increases transiently during the cell cycle, was analyzed as a control. These results indicate that in lymphocytes both of the DGKα promoters show elevated activity levels in quiescent cells that decrease in response to TCR triggering or IL-2 stimulation.
The mDGKα distal 5′ end has conserved functional sites for FoxO transcription factor binding.
Our experiments indicated similar controls for all mDGKα transcripts during cell proliferation. To identify the transcription factors responsible for this regulation, we designated the +1 start site at the Pr1 to coincide with the Inr element in the human DGKα gene (10) and used regulatory Vista (rVista), version 2.0, in conjunction with the TRANSFAC database (27). This analysis detected potential binding sites for STAT5a/b, Lef1 (lymphoid enhancer-binding factor 1), p53, Egr (early growth response), AP2, Ets-1, Sp1, and CREB (cyclic AMP [cAMP] response element binding protein). CG enrichment (CpG islands) in the Pr2 suggested epigenetic control of DGKα expression (Fig. 3A). Although most binding sites were located in the vicinity of the promoter regions, others were found in two regions we designated regulatory regions 1 and 2 (Fig. 3A). All four regions are highly conserved among species, including mouse and humans (see Fig. S4 to S6 in the supplemental material).
Three highly conserved FoxO-binding sequences were found in regulatory region 1 (Fig. 3A and B; see also Fig. S6 and Table S4 in the supplemental material). We concentrated on these sites since distal regions are potent modulators of gene expression, which could affect both promoters. In addition, FoxO factors regulate genes implicated in quiescence, and their activity is greatly reduced in response to mitogenic stimulation (17). The mammalian FoxO family includes FoxO1, FoxO3, FoxO4, and FoxO6 proteins, which are similar in structure and DNA-binding preferences (30).
To evaluate the transcription-promoting capacity of these FoxO-binding sites, we generated luciferase reporter constructs. pGL3-DGKαwt contained the 5′ upstream regulatory region of the DGKα gene, including the three FoxO-binding sites; pGL3-DGKαshort has a shorter promoter fragment that lacks the FoxO sites (Fig. 3C). The constructs were transfected into NIH 3T3 cells, alone or with an expression vector encoding FoxO3a, and luciferase activity was evaluated. Both constructs showed luciferase activity although only pGL3-DGKαwt showed increased promoter activity when FoxO3a was overexpressed. Point mutations in nucleotides critical for FoxO binding were generated in the pGL3-DGKαwt construct to obtain the mutant pGL3-DGKαmt (Fig. 3B). In this case, promoter activity was not upregulated when cells were cotransfected with the FoxO3a-encoding vector (Fig. 3D). These data indicate that the FoxO-binding sites in mDGKα regulatory region 1 are functional and enhance transcription in the presence of FoxO factors.
In vivo binding of FoxO transcription factors to DGKα regulatory region 1 correlates with high DGKα gene expression.
These experiments suggested that FoxO factors promote transcriptional activity by binding to the distal region of the mDGKα gene. To rule out indirect effects, we tested direct FoxO factor binding to the mDGKα gene regulatory region. Using an electrophoretic mobility shift assay (EMSA), we tested whether the putative mDGKα FoxO-binding sites bound nuclear proteins. DNA heteroduplexes for each of the three FoxO-binding sites were incubated with nuclear extracts of HEK293 cells, which express high FoxO3 levels. In all cases, we detected a slow-migrating band suggestive of protein binding to the heteroduplex; the nonspecific DNA control probe migrated normally (Fig. 4A). Similar experiments with a DNA heteroduplex for FoxO-binding site 1 (BS1) and spleen or thymus T cell nuclear extracts yielded a slow-migrating band (Fig. 4B).
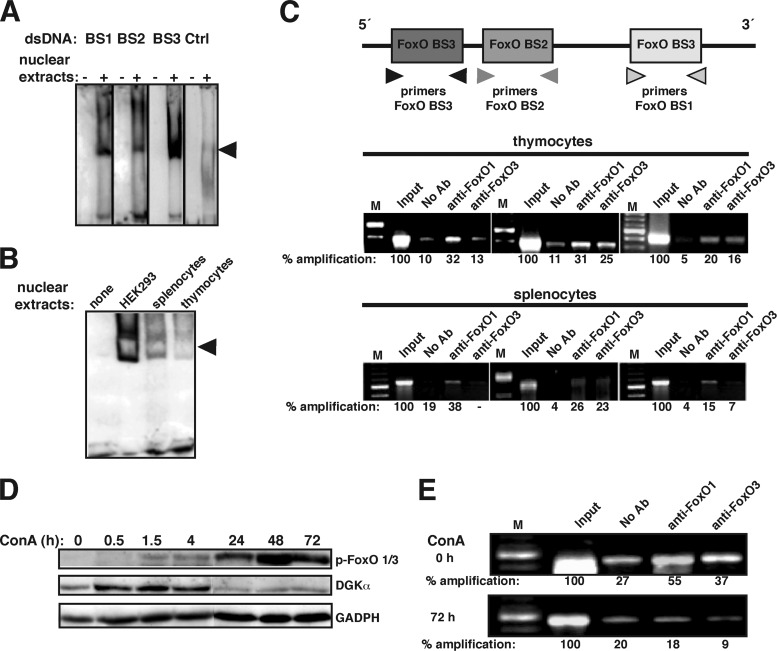
Fig 4.
FoxO transcription factors bind to the DGKα promoter. (A) Biotinylated DNA probes bearing the distinct FoxO BS of the mouse DGKα promoter or a nonspecific control probe (Ctrl) were used in EMSAs with nuclear extracts of HEK293 cells. Slow-migrating bands (arrowhead) were observed for the three BS but not the control. dsDNA, double-stranded DNA. (B) The experiment is as described for panel A, using nuclear extracts of freshly isolated splenocytes and thymocytes. The EMSA for BS1 is shown; results were similar for the other BS. (C) ChIP of the three DGKα locus FoxO BS in thymocytes or splenocytes. Anti-FoxO1 or -FoxO3 antibodies are indicated. Amplification products were analyzed by gel electrophoresis and quantified using ImageJ. Values are represented as a percentage relative to the ChIP input fraction, considered 100% amplification. Ab, antibody. (D) Freshly isolated splenocytes were stimulated with ConA (1 μg/ml) for the times indicated, and FoxO1/3 protein phosphorylation (p-FoxO1/3) and DGKα levels were evaluated. (E) ChIP of DGKα FoxO BS2 using nuclear extracts of splenocytes, untreated (top) or stimulated with ConA for 72 h (bottom). Anti-FoxO1 or -FoxO3 antibodies are indicated. Amplification products were analyzed by gel electrophoresis and quantified as described for panel C. In all cases, results are representative of at least three independent experiments. A 100-bp DNA ladder was used for size markers (lanes M).
We used a chromatin immunoprecipitation (ChIP) assay in mouse thymocytes and splenocytes to test in vivo FoxO binding. For ChIP experiments, we used specific antibodies to FoxO1 and FoxO3, the T cell-expressed family members. We immunoprecipitated chromatin-bound FoxO3 and FoxO1 protein from cell extracts and PCR amplified the promoter regions encompassing each of the three FoxO-binding sites (Fig. 4C, top). In thymocytes, FoxO1 and FoxO3 bound to the three BS although BS3 showed limited FoxO3 binding (Fig. 4C, center). In splenocytes, FoxO1 bound to the three BS, whereas FoxO3 was detected only in BS2 (Fig. 4C, bottom). Similar experiments using anti-FoxO3 antibody in HEK293 cells showed FoxO binding to all three BS; FoxO binding to the DGKα gene in murine and human cells concurs with FoxO BS conservation in DGKα (see Fig. S6 in the supplemental material).
FoxO DNA binding is controlled by phosphorylation. Although several kinases can modify the FoxO phosphorylation status, AKT-mediated phosphorylation is the best characterized. AKT-mediated phosphorylation of FoxO factors leads to nuclear exit, which blocks their transactivating function (51). The phosphatidylinositol 3-kinase (PI3K)/AKT pathway is activated during TCR and IL-2 signaling, suggesting a shared DGKα-regulatory mechanism. We assessed whether DGKα expression correlates with FoxO phosphorylation levels in splenocytes stimulated for different times with concanavalin A (ConA), a mitogenic lectin for lymphocytes (Fig. 4D). FoxO proteins showed little phosphorylation in response to initial stimulation (1.5 h), which increased to high levels at later times after ConA stimulation (24 h). FoxO phosphorylation preceded a reduction in DGKα levels, as indicated by transcriptional regulation of this enzyme by FoxO factors. ChIP analysis of FoxO BS2 on the DGKα gene showed constitutive association of FoxO1 and FoxO3 proteins in untreated cells, which was lost following ConA treatment (Fig. 4E). These experiments indicate a direct correlation between loss of FoxO binding and DGKα downregulation in activated T cells.
During T cell activation, productive TCR stimulation is followed by IL-2 secretion. We used a well-characterized model of transgenic major histocompatibility complex (MHC) class I-restricted TCR, the OT-1 mouse (18), to evaluate transcriptional regulation of DGKα in a more physiological context. Spleen CD8+ T cells were pulsed with a specific ovalbumin (OVA)-derived peptide ligand (SIINFEKL); after 48 h, cells were washed extensively and cultured alone or with exogenous IL-2 for a further 48 h. Enzyme expression was determined by real-time PCR; at 48 h of peptide treatment, we found a pronounced decrease in DGKα mRNA levels, which corroborated our observations using anti-TCR antibodies. DGKα mRNA levels remained low only when cells were cultured with IL-2; in contrast, DGKα expression increased to nearly basal levels when cells were cultured alone (Fig. 5A). These data are in agreement with our findings in BaF/3 cells (Fig. 2D) and indicate that the reduction in DGKα levels is transitory in the absence of IL-2 signaling.
The IL-2/AKT/FoxO pathway controls DGKα gene expression.
IL-2 is a cytokine with pleiotropic effects; in addition to its mitogenic properties in T cells, it has pivotal functions in the lymphocyte differentiation program. In CD4+ T cells, IL-2 modulates effector cell differentiation via regulation of cytokine receptor expression (26). In CD8+ T cells, IL-2 promotes differentiation into effector or memory cytolytic lymphocytes (CTL) (19). IL-2-mediated activation of the PI3K/AKT axis is crucial for promoting the CTL differentiation program, as well as for regulating the homing and migratory properties of these cells. By repressing FoxO function, IL-2 downregulates expression of certain receptor and adhesion molecules such as IL-7Rα, CXCR7, and l-selectin (29).
To examine the contribution of the IL-2/PI3K/AKT axis to DGKα regulation, we analyzed its expression in CD8+ T cells after treatment with an AKT inhibitor (AKT inhibitor VIII, AKTi-1/2). Inhibitor-treated cells showed a significant increase in DGKα mRNA and protein levels (Fig. 5B); treatment also increased mRNA levels of the FoxO target l-selectin. We also measured FoxO factor binding to the BS2 of the gene in cells that were IL-2 deprived or in exponential growth in medium containing IL-2 or IL-2 plus AKTi (Fig. 5C). In IL-2-deprived cells, only FoxO1 bound to the promoter region. FoxO factors did not bind to the DGKα gene in cells cultured with IL-2 alone, but FoxO1 and FoxO3 bound to the promoter region when the inhibitor was added. The inhibitor also enhanced the activity of both DGKα promoters (see Fig. S9 in the supplemental material). This confirms that IL-2 suppresses DGKα expression via a PI3K/AKT-dependent mechanism.
Regulation of DGKα expression by IL-2 suggests that this cytokine modulates DAG-mediated Ras/extracellular signal-regulated kinase (ERK) activation via AKT/FoxO/DGKα. We assessed the contribution of the PI3K/AKT pathway to DAG-related pathways in CD8+ T cells treated with AKTi. FoxO1/3 and S6 proteins were phosphorylated in cells growing exponentially in IL-2, as were the Ras effectors MEK and ERK and PKC substrates; these last three phosphorylation profiles indicate active DAG-promoted signaling. AKT inhibition led not only to dephosphorylation of FoxO factors and S6 but also to decreased phosphorylation of MEK, ERK, and PKC substrates. This reduction in DAG-sensitive phosphorylation was paralleled by an increase in DGKα expression (Fig. 6A). As reported previously (29), AKT inhibition also increased cell surface expression of l-selectin (Fig. 6B).
We tested whether the DGKα upregulation promoted by AKT inhibition affected secondary antigen encounter. For use as antigen-presenting cells, EL4 lymphoma cells were preloaded with OVA-derived peptide (SIINFEKL or SIIQFEHL) and then mixed with AKTi-treated CD8+ cells; TCR-mediated activation of the Ras/ERK pathway was then evaluated. Control cells showed clear increases in MEK and ERK phosphorylation levels, which were small in AKTi-treated cells, as predicted for cells with high DGKα levels (Fig. 6C).
DGKα regulates IL-2-mediated responses.
Our results indicate that IL-2 activation of the PI3K/AKT pathway coordinates DAG-mediated regulation of the Ras/ERK pathway, in part by repressing DGKα expression. This suggests that DGKα, a negative regulator of TCR-mediated Ras activation, could also act as a repressor of IL-2-mediated functions. To explore the DGKα contribution to IL-2 functions, we examined IL-2-mediated responses in primary CD8+ T cells from DGKα-deficient mice. Lack of DGKα increased cell proliferation in response to TCR and costimulatory signals (Fig. 7A, top); this increase was not due to enhanced IL-2 production as it was similar in wild-type and knockout mice (38). The effect appears to be the result of enhanced IL-2 sensitivity in DGKα-deficient cells since the mitogenic effect of the exogenous cytokine was more potent, as determined by cell numbers after each division (Fig. 7A, bottom).
To assess IL-2-dependent responses in CD8+ DGKα-deficient cells, we backcrossed DGKα−/− mice with OT-1 transgenic mice. OT-1 DGKα−/− mice were born at predicted Mendelian rates and appeared normal. Percentages of T cell populations in thymus and the periphery showed no gross alterations relative to wild-type littermates although there was a small reduction in lymph node CD8+ T cell numbers. Naïve, memory, and effector cell ratios were similar to those of wild-type OT-1 mice (see Fig. S8 in the supplemental material). To test the contribution of DGKα in IL-2-mediated DAG-Ras signaling, we evaluated MEK and ERK phosphorylation in CD8+ T cells from OT-1 DGKα−/− mice. As reported for TCR-mediated stimulation (38), CD8+ T cells from DGKα−/− mice showed hyperactivation of the Ras pathway in response to IL-2 (Fig. 7B). In DGKα-deficient cells, AKTi treatment led to a decrease in Ras signaling, which was much less pronounced than in wild-type cells. This indicates that although DGKα might not be the only negative regulator of DAG whose activity is reduced by the IL-2/AKT pathway, DGKα activity regulates basal DAG signaling during IL-2 activation.
To compare the capacity of IL-2 to induce differentiation in CD8+ T cells derived from OT-1 DGKα−/− mice, we examined the expression levels of l-selectin and perforin, two effector CD8+ T cell markers. l-Selectin expression decreased in response to activation and decreased further after IL-2 addition. In contrast, IL-2 withdrawal promoted a slight increase in l-selectin expression. In accordance with the strong IL-2 sensitivity, we found that in DGKα−/− CD8+ T cells l-selectin expression was lower than in wild-type cells. The sensitivity to IL-2 was also translated into strong, sustained perforin expression in DGKα−/− cells (Fig. 7C). These results are in agreement with negative modulation of IL-2 responses by DGKα.
In summary, our data show the importance of PI3K/AKT/FoxO pathway control in the transcriptional regulation of DGKα. This regulation allows T cells to activate Ras through a mechanism that integrates the intensity of TCR- and IL-2-mediated signals.
DISCUSSION
Here, we identify a role for the PI3K/AKT/FoxO pathway in the transcriptional regulation of DGKα in mice. Interest in this lipid kinase increased after its detection in a screening for anergy-regulated genes (28). This finding and the anergy-resistant phenotype of DGKα−/− mice suggested that DGKα has important role in Ras/ERK/AP1 pathway control during T cell activation. FoxO-mediated regulation of DGKα explains its rapid attenuation in response to full activation signals, as well as the reversion of anergy by addition of IL-2. PI3K/AKT/FoxO pathway control of DGKα, combined with DGKα regulation of the Ras/ERK axis, provides T cells with an extremely sensitive, unique mechanism that coordinates the intensity of TCR- and cytokine-mediated signals with biological effects.
Analysis of the 5′ upstream genomic region of the mDGKα gene allowed us to characterize alternative mRNA transcripts and identify three FoxO-binding sites that might drive mDGKα expression in vivo. The data also indicate the presence of binding sites for additional transcription factors such as p53, Egr, and STAT, in accordance with microarray data (31). This complex organization concurs with the strict regulation of this gene and suggests that many different signaling pathways converge to fine-tune DGKα levels. The existence of alternative promoters to drive DGKα expression is intriguing and offers several possibilities. Generation of mRNAs with alternative 5′ ends that precede the coding region suggest, through similarity to other genes (24), modulation in nascent mRNA processing. This would give rise to mature mRNAs with distinct stability and/or ribosomal recruitment and/or generation of different proteins. The full-length transcript derived from the Pr1 region lacked exon 10 (see Fig. S3 in the supplemental material), whereas we distinguished two distinct Pr2-derived transcripts, one corresponding to the NM_016811 sequence and the other with longer exons 13 and 21 than those in NM_016811 (see Fig. S3). These two differently processed transcripts potentially give rise to truncated proteins. Alternative transcripts are predicted by bioinformatic analyses (NCBI AceView, alternative splicing database project [http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/]), and smaller DGKα forms were found in a study of neutrophils from localized aggressive periodontitis patients (accession number AY930112.1) and in a recent study in liver (35); additional studies are nonetheless needed to address the function of these DGKα protein variants.
Real-time PCR analysis showed higher relative expression of Pr1 than that of Pr2 (≈17-fold). In primary T cells and the BaF/3 B cell line, both promoters consistently showed high activity levels under resting conditions, which decreased following TCR triggering or IL-2 addition (Fig. 2 and 5; see also Fig. S9 in the supplemental material). Although we observed similar behaviors between the two DGKα promoters under the conditions studied here, we cannot rule out possible differences in activity levels in specific circumstances such as embryonic development or cell differentiation.
Our results strongly support the idea that DGKα is expressed via FoxO transcription factors. This FoxO-mediated regulation helps us to understand how the enzyme levels are coupled to TCR stimulation intensity and to the presence of costimulation. FoxO phosphorylation is downstream of the PI3K/AKT axis, whose activity increases with signal intensity. TCR stimulation elicits PI3K activity through its p110δ catalytic isoform, leading to weak, transitory FoxO phosphorylation (37). Costimulation results in the recruitment and activation of additional PI3K isoforms, enhancing FoxO phosphorylation (12). Unlike other AKT targets, FoxO is phosphorylated only after AKT is phosphorylated by mammalian target of rapamycin (mTOR) at Ser473 via mTOR complex 2 (16). mTOR activation is a requisite for cell cycle progression, which might explain the coordination between DGKα decrease and cell cycle progression. This concurs with the data showing that the reduced DGKα expression observed during cell cycle entry of naïve T cells mirrors that of the cell cycle inhibitor p27 (Fig. 2B) and with our previous observation that DGKα protein levels remain high if the TCR is stimulated in the presence of rapamycin (31). Results were similar in the fibroblast-derived NIH 3T3 cell line, in which DGKα gene expression was reduced in response to serum-induced cell cycle entry (our unpublished results).
T lymphocytes lacking DGKα show increased IL-2-triggered proliferation, confirming DGKα contribution as a negative regulator of cell cycle progression whose levels should decrease to allow cell expansion. This role of DGKα correlates with its well-characterized function as a negative regulator of the Ras/ERK axis in T cells and its overexpression during anergy. Although our data demonstrate the concerted regulation of cell cycle entry and DGKα downregulation, they do not prove that cell cycle progression regulates DGKα transcriptional regulation. Addressing this question awaits more detailed studies. Our data indicate that the PI3K/AKT pathway can also couple DGKα transcriptional regulation to processes other than proliferation, such as IL-2-promoted CTL differentiation; in this case and unlike naïve T cells, DGKα and p27 expression patterns were not similar (see Fig. S9 in the supplemental material).
ChIP experiments demonstrated FoxO1 and FoxO3 binding to DGKα regulatory region 1 in resting T lymphocytes. FoxO factors are crucial for correct T cell homeostasis, with FoxO1 function predominating over that of FoxO3 in peripheral lymphoid organs. Whereas in thymocytes both FoxO isoforms bound to all three FoxO BS of DGKα, in splenocytes FoxO1, but not FoxO3, bound the three FoxO BS (Fig. 4). The identification of FoxO-binding sites in the DGKα regulatory region facilitates interpretation of data from microarray analyses, which consistently show high DGKα expression in response to quiescence-inducing signals, differentiation, or apoptosis (31). The description of DGKα as a FoxO target suggests that it participates in the control of nonproliferative or differentiated cell states and correlates well with DGKα function in T lymphocytes (31).
Our study shows that DGKα behavior is similar to that of other well-characterized FoxO target genes in peripheral T lymphocytes. Either directly and/or through regulation of the Krüppel-like transcription factor (KLF-2), FoxO1 controls expression of trafficking molecules such as l-selectin, CCR7, and the sphingosine-1-phosphate receptor (15, 21, 39), as well as that of IL-7Rα, which governs homeostatic survival of naive T cells (50). Expression of all these molecules is reduced after PI3K/AKT pathway triggering as a result of TCR/IL-2 activation (54). DGKα expression decreased following IL-2 treatment, which was reversed by pharmacological inhibition of AKT or IL-2 withdrawal. Increased DGKα expression due to AKT inhibition coincided with higher FoxO1 and FoxO3 recruitment to the promoter (Fig. 5). Our observations are in accordance with data from a recent microarray study that shows DGKα upregulation by AKT inhibition (29).
PI3K/AKT- and DAG-induced pathways must work in concert during T cell activation and cytokine-promoted differentiation to guarantee correct T cell outcomes. TCR-mediated ERK phosphorylation requires PI3K function since ERK phosphorylation is reduced in PI3K p110δ−/− mice. In addition, ERK-mediated l-selectin shedding is impaired in these mice (36), as well as in wild-type mice after treatment with a PI3Kδ inhibitor (29). The mechanism is not known but is unlikely to be a result of changes in DGKα transcription or protein stability since the TCR-induced decrease in DGKα protein levels is a late event (Fig. 4D). DGKα downregulation is maintained by IL-2-mediated PI3K/AKT activation, an observation that defines an additional function for DGKα as a negative regulator of Ras/ERK activation during cytokine-promoted effects. Accordingly, lymphocytes from DGKα-deficient mice show increased IL-2-dependent proliferation, increased Ras/ERK activation, and an enhanced IL-2-mediated cytotoxic program. IL-2/AKT/FoxO transcription control of DGK activity appears specific to the DGKα isoform since the DGKζ expression pattern is distinct. DGKζ mRNA levels decreased during TCR activation, although in a manner different from that of DGKα (Fig. 2), and AKT inhibition did not lead to increased DGKζ mRNA levels (see Fig. S9C in the supplemental material).
By negatively regulating RasGRP1, the DGK control the intensity of TCR-derived signals. RasGRP1 supplies T cells with a mechanism with which to regulate basal Ras levels. The level of Ras signals permits rapid transformation of an analogue input (changes in antigen affinities) into an analogic output (response versus nonresponse) (6). This DGK control of TCR signal intensity is suggested to have a critical role in the regulation of effector versus memory T cell differentiation. IL-2 regulation of DGKα levels strongly suggests that, as for naïve T cells, activated T cells have a mechanism that allows regulation of homeostatic Ras activation, which would help to discriminate the intensity of signals received by cells, in this case, from the cytokine milieu. This is consistent with the differential roles of IL-2 and IL-15 in the onset of cell memory, attributed to the greater capacity of IL-2 to trigger PI3K signaling (5). Transcriptional regulation of DGKα thus exemplifies how the PI3K/AKT/FoxO axis is coupled to Ras-mediated signals, a mechanism that links T cell differentiation with the intensity threshold. Not surprisingly, in a mouse model of lymphocytic choriomeningitis virus infection, DGKα deficiency promotes CD8+ T cell expansion but decreases memory CD8+ responses (48).
T cell nonresponsiveness to antigen, or anergy, is regulated by a transcriptional program controlled by the factors NFAT and Egr2 (23, 28, 43); DGKα has conserved binding regions for these two factors (Fig. 3; see also the supplemental material). It remains unclear whether these factors are directly responsible for DGKα upregulation in models of anergy induction. Unlike the situation with other anergy genes such as Grail and Caspase3, NFAT homodimers do not promote DGKα overexpression (49). It is unlikely that FoxO factors alone support the DGKα increases observed in some anergy induction models (28, 56), but their binding to a DGKα promoter might be decisive in maintaining these active transcription conditions. IL-2-mediated DGKα downregulation strongly suggests that FoxO factors mediate rescue of anergic cells via this cytokine (8).
In summary, DGKα regulation by the PI3K/AKT/FoxO pathway (Fig. 7) identifies important, unreported functions for this enzyme and helps to explain its precise regulation at the onset of T cell activation. FoxO-mediated DGKα expression in quiescent cells might enhance DAG metabolism, preventing naïve T cell activation in response to incomplete stimulation. Loss of FoxO activity during productive T cell activation reduces DGKα levels, leading to activation of DAG-mediated functions. IL-2-induced signals would prolong DGKα downregulation, providing signals that facilitate expansion and differentiation of CTL populations.
Supplementary Material
ACKNOWLEDGMENTS
We thank I.M.'s group members for helpful discussion, H. Reyburn for input on the manuscript, and C. Mark for editorial assistance.
M.M.-M. is supported by a Juan de la Cierva fellowship, D.S. holds an FPI fellowship, and P.T.-A. and E.A. hold FPU fellowships, all from the Spanish Ministry of Education; A.A.-F. is supported by the Spanish Anti-Cancer Association (AECC). This work was supported in part by grants from the Spanish Ministry of Health (Instituto de Salud Carlos III; RD067002071035), from the Spanish Ministry of Education (BFU2010-21138) and the Madrid regional government (S-SAL-0311) to IM.
We declare that we do not have any conflicts of interest.
Footnotes
Published ahead of print 13 August 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Bandyopadhyay S, Soto-Nieves N, Macian F. 2007. Transcriptional regulation of T cell tolerance. Semin. Immunol. 19:180–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Biggs WH, III, Cavenee WK, Arden KC. 2001. Identification and characterization of members of the FKHR (FOX O) subclass of winged-helix transcription factors in the mouse. Mamm. Genome. 12:416–425 [DOI] [PubMed] [Google Scholar]
- 3. Carrasco S, Merida I. 2007. Diacylglycerol, when simplicity becomes complex. Trends Biochem. Sci. 32:27–36 [DOI] [PubMed] [Google Scholar]
- 4. Cellot S, et al. 2007. Sustained in vitro trigger of self-renewal divisions in Hoxb4hiPbx1(10) hematopoietic stem cells. Exp. Hematol. 35:802–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cornish GH, Sinclair LV, Cantrell DA. 2006. Differential regulation of T-cell growth by IL-2 and IL-15. Blood 108:600–608 [DOI] [PubMed] [Google Scholar]
- 6. Daniels MA, et al. 2006. Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature 444:724–729 [DOI] [PubMed] [Google Scholar]
- 7. Di Paolo G, Kim TW. 2011. Linking lipids to Alzheimer's disease: cholesterol and beyond. Nat. Rev. Neurosci. 12:284–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dure M, Macian F. 2009. IL-2 signaling prevents T cell anergy by inhibiting the expression of anergy-inducing genes. Mol. Immunol. 46:999–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fu Y, Rubin CS. 2011. Protein kinase D: coupling extracellular stimuli to the regulation of cell physiology. EMBO Rep. 12:785–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fujikawa K, Imai S, Sakane F, Kanoh H. 1993. Isolation and characterization of the human diacylglycerol kinase gene. Biochem. J. 294:443–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fujita PA, et al. 2011. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 39:D876–D882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Garcon F, et al. 2008. CD28 provides T-cell costimulation and enhances PI3K activity at the immune synapse independently of its capacity to interact with the p85/p110 heterodimer. Blood 111:1464–1471 [DOI] [PubMed] [Google Scholar]
- 13. Griner EM, Kazanietz MG. 2007. Protein kinase C and other diacylglycerol effectors in cancer. Nat. Rev. Cancer. 7:281–294 [DOI] [PubMed] [Google Scholar]
- 14. Grkovich A, Dennis EA. 2009. Phosphatidic acid phosphohydrolase in the regulation of inflammatory signaling. Adv. Enzyme Regul. 49:114–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gubbels Bupp MR, et al. 2009. T cells require Foxo1 to populate the peripheral lymphoid organs. Eur. J. Immunol. 39:2991–2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guertin DA, et al. 2006. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCα, but not S6K1. Dev. Cell 11:859–871 [DOI] [PubMed] [Google Scholar]
- 17. Hedrick SM. 2009. The cunning little vixen: Foxo and the cycle of life and death. Nat. Immunol. 10:1057–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hogquist KA, et al. 1994. T cell receptor antagonist peptides induce positive selection. Cell 76:17–27 [DOI] [PubMed] [Google Scholar]
- 19. Kalia V, et al. 2010. Prolonged interleukin-2Rα expression on virus-specific CD8+ T cells favors terminal-effector differentiation in vivo. Immunity 32:91–103 [DOI] [PubMed] [Google Scholar]
- 20. Kent WJ. 2002. BLAT–the BLAST-like alignment tool. Genome Res. 12:656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kerdiles YM, et al. 2009. Foxo1 links homing and survival of naive T cells by regulating l-selectin, CCR7 and interleukin 7 receptor. Nat. Immunol. 10:176–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kimura K, et al. 2006. Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 16:55–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kowalski J, Drake C, Schwartz RH, Powell J. 2004. Non-parametric, hypothesis-based analysis of microarrays for comparison of several phenotypes. Bioinformatics 20:364–373 [DOI] [PubMed] [Google Scholar]
- 24. Kwan T, et al. 2008. Genome-wide analysis of transcript isoform variation in humans. Nat. Genet. 40:225–231 [DOI] [PubMed] [Google Scholar]
- 25. Landry JR, Mager DL, Wilhelm BT. 2003. Complex controls: the role of alternative promoters in mammalian genomes. Trends Genet. 19:640–648 [DOI] [PubMed] [Google Scholar]
- 26. Liao W, Lin JX, Leonard WJ. 2011. IL-2 family cytokines: new insights into the complex roles of IL-2 as a broad regulator of T helper cell differentiation. Curr. Opin. Immunol. 23:598–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Loots GG, Ovcharenko I. 2004. rVISTA 2.0: evolutionary analysis of transcription factor binding sites. Nucleic Acids Res. 32:W217–W221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Macian F, et al. 2002. Transcriptional mechanisms underlying lymphocyte tolerance. Cell 109:719–731 [DOI] [PubMed] [Google Scholar]
- 29. Macintyre AN, et al. 2011. Protein kinase B controls transcriptional programs that direct cytotoxic T cell fate but is dispensable for T cell metabolism. Immunity 34:224–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Maiese K, Chong ZZ, Shang YC. 2008. OutFOXOing disease and disability: the therapeutic potential of targeting FoxO proteins. Trends Mol. Med. 14:219–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Merida I, et al. 2009. Diacylglycerol kinase alpha, from negative modulation of T cell activation to control of cancer progression. Adv. Enzyme Regul. 49:174–188 [DOI] [PubMed] [Google Scholar]
- 32. Merida I, Avila-Flores A, Merino E. 2008. Diacylglycerol kinases: at the hub of cell signalling. Biochem. J. 409:1–18 [DOI] [PubMed] [Google Scholar]
- 33. Merida I, Williamson P, Kuziel WA, Greene WC, Gaulton GN. 1993. The serine-rich cytoplasmic domain of the interleukin-2 receptor beta chain is essential for interleukin-2-dependent tyrosine protein kinase and phosphatidylinositol-3-kinase activation. J. Biol. Chem. 268:6765–6770 [PubMed] [Google Scholar]
- 34. Merino E, et al. 2008. Lck-dependent tyrosine phosphorylation of diacylglycerol kinase alpha regulates its membrane association in T cells. J. Immunol. 180:5805–5815 [DOI] [PubMed] [Google Scholar]
- 35. Nakano T, et al. 2012. Altered expression of diacylglycerol kinase isozymes in regenerating liver. J. Histochem. Cytochem. 60:130–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Okkenhaug K, et al. 2002. Impaired B and T cell antigen receptor signaling in p110δ PI 3-kinase mutant mice. Science 297:1031–1034 [DOI] [PubMed] [Google Scholar]
- 37. Okkenhaug K, et al. 2006. The p110δ isoform of phosphoinositide 3-kinase controls clonal expansion and differentiation of Th cells. J. Immunol. 177:5122–5128 [DOI] [PubMed] [Google Scholar]
- 38. Olenchock BA, et al. 2006. Disruption of diacylglycerol metabolism impairs the induction of T cell anergy. Nat. Immunol. 7:1174–1181 [DOI] [PubMed] [Google Scholar]
- 39. Ouyang W, Li MO. 2011. Foxo: in command of T lymphocyte homeostasis and tolerance. Trends Immunol. 32:26–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Peng X, Frohman MA. 2011. Mammalian phospholipase D physiological and pathological roles. Acta Physiol. (Oxf.) 204:219–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Peterson TR, et al. 2011. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell 146:408–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Phelps DE, et al. 1998. Coupled transcriptional and translational control of cyclin-dependent kinase inhibitor p18INK4c expression during myogenesis. Mol. Cell. Biol. 18:2334–2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Powell JD. 2006. The induction and maintenance of T cell anergy. Clin. Immunol. 120:239–246 [DOI] [PubMed] [Google Scholar]
- 44. Sakane F, Imai S, Kai M, Yasuda S, Kanoh H. 2008. Diacylglycerol kinases as emerging potential drug targets for a variety of diseases. Curr. Drug Targets 9:626–640 [DOI] [PubMed] [Google Scholar]
- 45. Samuel VT, Petersen KF, Shulman GI. 2010. Lipid-induced insulin resistance: unravelling the mechanism. Lancet 375:2267–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sandoval J, et al. 2004. RNAPol-ChIP: a novel application of chromatin immunoprecipitation to the analysis of real-time gene transcription. Nucleic Acids Res. 32:e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sanjuan MA, et al. 2003. T cell activation in vivo targets diacylglycerol kinase alpha to the membrane: a novel mechanism for Ras attenuation. J. Immunol. 170:2877–2883 [DOI] [PubMed] [Google Scholar]
- 48. Shin J, O'Brien TF, Grayson JM, Zhong XP. 2012. Differential regulation of primary and memory CD8 T cell immune responses by diacylglycerol kinases. J. Immunol. 188:2111–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Soto-Nieves N, et al. 2009. Transcriptional complexes formed by NFAT dimers regulate the induction of T cell tolerance. J. Exp. Med. 206:867–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Takada K, Jameson SC. 2009. Naive T cell homeostasis: from awareness of space to a sense of place. Nat. Rev. Immunol. 9:823–832 [DOI] [PubMed] [Google Scholar]
- 51. Tzivion G, Dobson M, Ramakrishnan G. 2011. FoxO transcription factors; regulation by AKT and 14-3-3 proteins. Biochim. Biophys. Acta 1813:1938–1945 [DOI] [PubMed] [Google Scholar]
- 52. Verdeil G, Chaix J, Schmitt-Verhulst AM, Auphan-Anezin N. 2006. Temporal cross-talk between TCR and STAT signals for CD8 T cell effector differentiation. Eur. J. Immunol. 36:3090–3100 [DOI] [PubMed] [Google Scholar]
- 53. Wattenberg BW, Raben DM. 2007. Diacylglycerol kinases put the brakes on immune function. Sci. STKE 2007:pe43 doi:10.1126/stke.3982007pe43 [DOI] [PubMed] [Google Scholar]
- 54. Waugh C, Sinclair L, Finlay D, Bayascas JR, Cantrell D. 2009. Phosphoinositide (3,4,5)-triphosphate binding to phosphoinositide-dependent kinase 1 regulates a protein kinase B/Akt signaling threshold that dictates T-cell migration, not proliferation. Mol. Cell. Biol. 29:5952–5962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yamada K, Sakane F, Kanoh H. 1989. Immunoquantitation of 80 kDa diacylglycerol kinase in pig and human lymphocytes and several other cells. FEBS Lett. 244:402–406 [DOI] [PubMed] [Google Scholar]
- 56. Zha Y, et al. 2006. T cell anergy is reversed by active Ras and is regulated by diacylglycerol kinase-alpha. Nat. Immunol. 7:1166–1173 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.