LETTER
Streptococcus pyogenes is capable of causing a wide spectrum of diseases, ranging from throat and skin infection to life-threatening invasive diseases (2). The sequelae of streptococcal infections, such as acute rheumatic fever, represent a big health hazard (1). Recently, the emergence of invasive streptococcal diseases in India has been reported (5). Streptococcal emm types of the Indian subcontinent differ from those of other countries (7, 8). emm1, which is one of the most prevalent invasive types of Western countries, is rare in India. Instead, another type, designated emm1-2, is prevalent in India (8), and a subtype, emm1-2.2, is associated with invasive disease (5). The development of antibiotic resistance of S. pyogenes in India is a serious problem. High resistance rates were observed for trimethoprim sulfamethoxazole, which is commonly prescribed in rural settings of India (3, 6). In this study, we performed whole-genome sequencing of a clinical isolate of type emm1-2 and detected for the first time the trimethoprim resistance determinant dihydrofolate reductase gene, dfrG. The invasive emm1-2.2 S. pyogenes strain A1085 was collected during a survey in India. We identified an integrated sequence, not present in the genomes of S. pyogenes reference strains available in the NCBI database. BLAST analysis of a 3.3-kb integration element showed 99% to 100% identity to genomic DNA of Staphylococcus aureus TW20 (GenBank accession no. FN433596.1) and SAV0404 (GenBank accession no. AB205645.1). Within the integration sequence, three open reading frames (ORFs) were predicted (Fig. 1). ORF_01 was identified as dfrG, encoding the trimethoprim-resistant dihydrofolate reductase. ORF_02 and ORF_03 encode hypothetical proteins identical to annotated proteins in Staphylococcus aureus TW20. Flanking regions of the S. pyogenes emm1-2.2 integration element were compared with regions of strain SF370 (emm1), which led to identification of SPy_1769 as the integration site, which was confirmed by PCR with primers specific for conserved regions. The amplification product of strain A1085 was sequenced, which confirmed the results of the genome sequencing. The dfrG gene was amplified and sequenced (Fig. 2; Table 1). The integration element of 3.3 kb and the specific 500-bp PCR product of dfrG were detected in all tested emm1-2.2 strains from different sites. Strain SF370 (4) showed no integration element, and dfrG could not be amplified (Fig. 2; Table 1).
Fig 1.
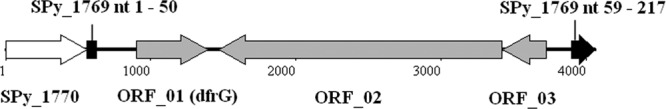
Genetic map of the dfrG gene locus in S. pyogenes emm1-2 isolates. Genome sequencing of the invasive emm1-2.2 S. pyogenes strain A1085 identified an integrated sequence of 3.3 kb, not present in the genomes of S. pyogenes reference strains. Within the integration sequence, three open reading frames were predicted, which are indicated by gray arrows, with the direction of transcription shown by the arrowhead. ORF_01 was identified as dfrG, encoding the trimethoprim-resistant dihydrofolate reductase. ORF_02 and ORF_03 encode hypothetical proteins. Flanking regions of the integration element were compared with S. pyogenes SF370 (emm1), and SPy_1769 was identified as the integration site. The first 50 nucleotides (nt) of SPy_1769 are shown as a black box, followed by the integration element. Nucleotides 59 to 217 of SPy_1769 are upstream of the integration element, depicted as a black arrow. The white arrow corresponds to the gene Spy_1770.
Fig 2.
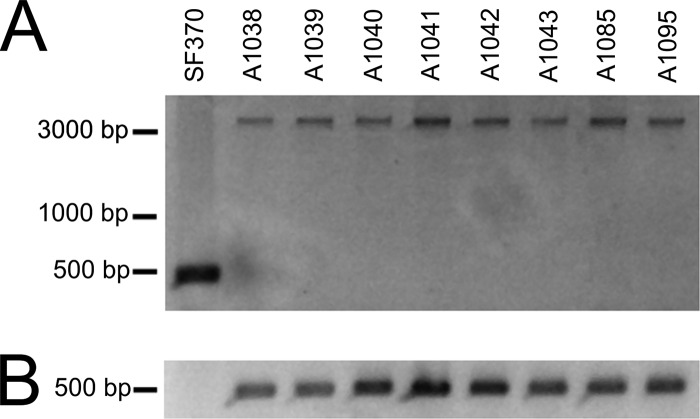
PCR detection of the insertion sequence (A) and dfrG (B). (A) The presence of the integration element was analyzed by PCR with primers specific for conserved regions flanking the integration site. S. pyogenes emm1-2 strains showed an ∼3.8-kb amplification product, indicating the integration element. As expected, S. pyogenes SF370 (negative control) showed no integration element, which is indicated by a 500-bp PCR product. (B) Specific PCR products of dfrG (500 bp) were detected in all tested emm1-2.2 strains, whereas SF370 was negative for these products.
Table 1.
Trimethoprim resistance in S. pyogenes emm1-2.2
| Isolate | Relevant properties | MIC of trimethoprim (μg/ml) | Reference |
|---|---|---|---|
| SF370 | emm1 Tmps, dfrG mutant | ≤2 | 4 |
| A1038 | Skin isolate, emm1-2.2 Tmpr dfrG+ | >512 | 8 |
| A1039 | Skin isolate, emm1-2.2 Tmpr dfrG+ | >512 | 8 |
| A1040 | Skin isolate, emm1-2.2 Tmpr dfrG+ | >512 | 8 |
| A1041 | Throat isolate, emm1-2.2 Tmpr dfrG+ | >512 | 8 |
| A1042 | Throat isolate, emm1-2.2 Tmpr dfrG+ | >512 | 8 |
| A1043 | Throat isolate, emm1-2.2 Tmpr dfrG+ | >512 | 8 |
| A1085 | Invasive isolate, emm1-2.2 Tmpr dfrG+ | >512 | 5 |
| A1095 | Invasive isolate, emm1-2.2 Tmpr dfrG+ | >512 | 5 |
The resistance to trimethoprim of the emm1-2.2 strains was demonstrated by the disk diffusion test. The MIC of trimethoprim was determined by the agar dilution method. The MIC was recorded as the lowest concentration of trimethoprim that inhibited visible growth after 18 h of incubation at 37°C (Table 1). MIC values for emm1-2.2 strains were high (>512 μg/ml), in contrast to those for the susceptible control strain SF370 (MIC ≤ 2 μg/ml).
Although trimethoprim resistance has been observed with beta-hemolytic streptococci, the underlying molecular mechanisms were not elucidated. S. pyogenes emm1-2.2 is an important emm type in northern India associated with skin, throat, and invasive infections. The detection of the trimethoprim resistance determinant dfrG in all clinical isolates belonging to emm1-2.2 is a cause of serious concern that underlines the need for longitudinal surveillance of isolates from different parts of India.
ACKNOWLEDGMENTS
We thank the European Commission FP6 Assist (http://www.helmholtz-hzi.de/en/research/research_projects/view/projekt/projekt/assist/) team for the collection of Indian isolates.
Footnotes
Published ahead of print 13 August 2012
REFERENCES
- 1. Carapetis JR, Steer AC, Mulholland EK, Weber M. 2005. The global burden of group A streptococcal diseases. Lancet Infect. Dis. 5:685–694 [DOI] [PubMed] [Google Scholar]
- 2. Cunningham MW. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Devi U, Borah PK, Mahanta J. 2011. The prevalence and antimicrobial susceptibility patterns of beta-hemolytic streptococci colonizing the throats of schoolchildren in Assam, India. J. Infect. Dev. Ctries. 5:804–808 [DOI] [PubMed] [Google Scholar]
- 4. Ferretti JJ, et al. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. U. S. A. 98:4658–4663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Haggar A, et al. 2012. Clinical and microbiologic characteristics of invasive Streptococcus pyogenes infections in North and South India. J. Clin. Microbiol. 50:1626–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jain A, Shukla VK, Tiwari V, Kumar R. 2008. Antibiotic resistance pattern of group-a beta-hemolytic streptococci isolated from north Indian children. Indian J. Med. Sci. 62:392–396 [PubMed] [Google Scholar]
- 7. Sagar V, et al. 2004. Molecular heterogeneity among north Indian isolates of group A Streptococcus. Lett. Appl. Microbiol. 39:84–88 [DOI] [PubMed] [Google Scholar]
- 8. Sagar V, Kumar R, Ganguly NK, Chakraborti A. 2008. Comparative analysis of emm type pattern of group A Streptococcus throat and skin isolates from India and their association with closely related SIC, a streptococcal virulence factor. BMC Microbiol. 8:150. [DOI] [PMC free article] [PubMed] [Google Scholar]