Abstract
New Delhi metallo-β-lactamase 1 (NDM-1) is a key enzyme that the pathogen Klebsiella pneumonia uses to hydrolyze almost all β-lactam antibiotics. It is currently unclear why NDM-1 has a broad spectrum of activity. Docking of the representatives of the β-lactam families into the active site of NDM-1 is reported here. All the β-lactams naturally fit the NDM-1 pocket, implying that NDM-1 can accommodate the substrates without dramatic conformation changes. The docking reveals two major binding modes of the β-lactams, which we tentatively name the S (substrate) and I (inhibitor) conformers. In the S conformers of all the β-lactams, the amide oxygen and the carboxylic group conservatively interact with two zinc ions, while the substitutions on the fused rings show dramatic differences in their conformations and positions. Since the bridging hydroxide ion/water in the S conformer is at the position for the nucleophilic attack, the S conformation may simulate the true binding of a substrate to NDM-1. The I conformer either blocks or displaces the bridging hydroxide ion/water, such as in the case of aztreonam, and is thus inhibitory. The docking also suggests that substitutions on the β-lactam ring are required for β-lactams to bind in the S conformation, and therefore, small β-lactams such as clavulanic acid would be inhibitors of NDM-1. Finally, our docking shows that moxalactam uses its tyrosyl-carboxylic group to compete with the S conformer and would thus be a poor substrate of NDM-1.
INTRODUCTION
The recent outbreak of colistin- and carbapenem-resistant Klebsiella pneumoniae isolates in Detroit, MI (18, 20) emphasizes that multidrug resistance is a major global threat to human health. Although the mechanisms of multidrug resistance are unknown, hydrolysis of β-lactam antibiotics by β-lactamases is the most common cause of resistance in clinically important Gram-negative bacteria (3, 4). The number of members of the β-lactamases exceeded 890 by late 2009, and β-lactamases are categorized into four classes, A, B, C, and D, on the basis of their sequence similarity (4). Enzymes in classes A, C, and D are serine β-lactamases that contain no divalent metal ions and employ an active-site serine to hydrolyze their substrates by forming an acyl intermediate (1). Class B consists of metallo-β-lactamases (MBLs), most of which typically contain two zinc ions at the active site to facilitate the β-lactam hydrolysis. Class B enzymes can be further categorized into three subclasses of B1, B2, and B3 (10) and are regarded as a therapeutic challenge because the vast majority of the MBL enzymes show a broad spectrum of activity on hydrolysis of therapeutic β-lactams (1).
Recently, a new β-lactamase, called New Delhi metallo-β-lactamase 1 (NDM-1), has been identified in the pathogenic bacteria of patients. NDM-1 was originally found in India (8) and has spread worldwide to countries including Australia (23), Belgium (2), Canada (16), China (6), Germany (22), Italy (17), Japan (37), Norway (26), Spain (27), Switzerland (24), the United Kingdom (15), and the United States (5). NDM-1 has a fold common to the other MBLs and shows the ability to hydrolyze most β-lactams (3).
Hundreds of crystal structures of the β-lactamases have been deposited in the Protein Data Bank (PDB), including the recent structures of NDM-1 (11, 13, 14, 39). However, no structures of MBLs in complex with real substrates have been reported. This situation not only leaves the hydrolysis mechanisms of MBLs elusive but also slows the discovery of effective MBL inhibitors. Although academic and industrial researchers have worked extensively on the discovery of MBL inhibitors for several decades, the current compounds cannot be used as therapeutics to suppress the hydrolysis of β-lactams by the superbugs (1, 32). Most MBL inhibitors, including the compounds that simulate the intermediates of the reactions, such as mercaptocarboxylate for VIM-2 (35), show affinity only at the micro- or submicromolar level (32). This paper explores the bound conformations of the representatives of the β-lactam subfamilies (Fig. 1) by docking simulation. Our docking reveals a common pattern for binding of the β-lactams, which may simulate the bound conformation of the genuine substrates. The docking also shows that the widely open pocket of NDM-1, as revealed by the crystal structures (11, 13, 14, 39), can naturally adopt most β-lactams, thus providing valuable hints for the design of NDM-1 inhibitors.
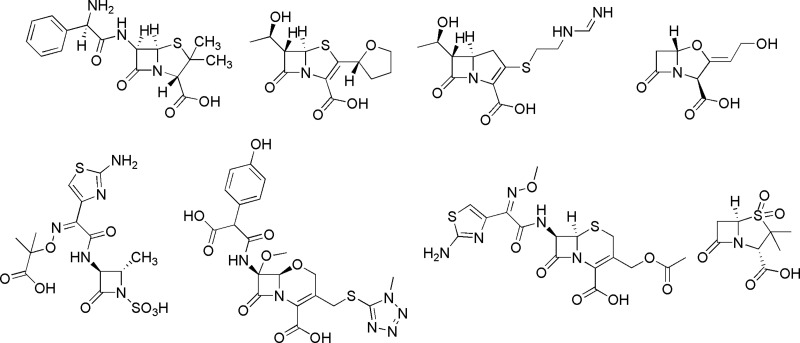
Fig 1.
Chemical structures of β-lactams.
MATERIALS AND METHODS
The following representatives of the β-lactam subtypes were manually built in a graphic terminal by using the hydrolyzed product of ampicillin as the starting model (PDB accession number 3Q6X) (39): ampicillin and sulbactam (penam subfamily), cefotaxime (cephem), moxalactam (oxacephem), clavulanic acid (clavam), aztreonam (monobactam), meropenem (carbapenem), and faropenem (penem) (Fig. 1). In an assumption of no changes on the enantiomeric configurations when the β-lactam substrates are converted to their products, the enantiomeric conformation of the hydrolyzed ampicillin was taken as the template to build the models of ampicillin and other β-lactams since they share two chiral centers on the β-lactam rings (Fig. 1).
The models of the β-lactams were docked by the program AutoDock (version 4.2) (12, 19) into the active site of a monomeric NDM-1 that was taken from the dimer in the crystal structure (PDB accession number 3Q6X) (39). The dihedral angles of the β-lactam models were optimized by the program to best fit into the pocket of NDM-1. The docking of ampicillin was performed against the NDM-1 structures with rigid or flexible side chains of the active-site residues (Ile35, Leu65, Met67, Val73, Try93, Asn220), except for the metal binding residues. These two methods of docking produced overall comparable results. However, the flexible docking output indicated only 73% poses that bind to the active site of NDM-1, while 27% poses bind somewhere else. In comparison, the rigid docking showed 100% poses bound to the active site of NDM-1. Therefore, the rigid monomeric NDM-1 structure was used for docking of all other β-lactams. A typical box with 60 by 60 by 60 grid points equally spaced at 0.375 Å per grid was generated using AutoGrid (19). The following parameters and Lamarckian genetic algorithm were used for the docking: random initial orientation and position, population size of 150, maximum number of energy evaluations of 25 million, maximum number of generations of 27,000, mutation rate of 0.02, crossover rate of 0.8, and 100 docking runs to output 100 docked conformations.
RESULTS
Structure of NDM-1.
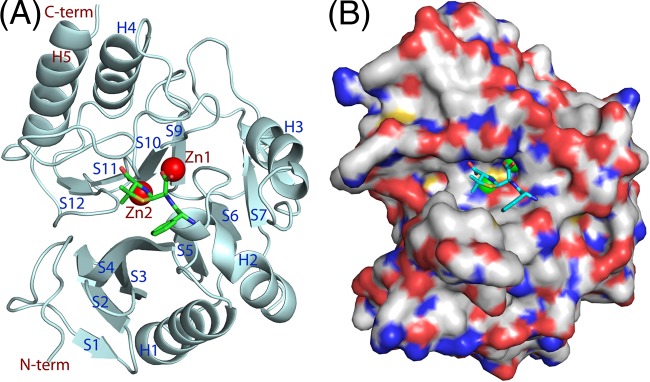
As shown by the crystal structure (PDB accession number 3Q6X), NDM-1 shares a common folding architecture with other MBLs and contains two zinc ions as its catalytic metals that chelate with His120/His122/His189 and Asp124/Cys208/His250, respectively (11, 13, 14, 39). The monomer of NDM-1 contains five major helices and 12 β strands that are marked H1 to H5 and S1 to S12 in Fig. 2, following the class B β-lactamase (BBL) numbering scheme (9). However, NDM-1 contains a short helix before H2 and a short strand after H3 and also has a helix in the place of S8 of other MBLs (9). The active-site pocket of NDM-1 in the crystal structures (11, 13, 14, 39) is widely open, with two zinc ions sitting in the bottom of the cavity.
Fig 2.
Structure of NDM-1 (drawn from PDB accession number 3Q6X). (A) Ribbon diagram of NDM-1. The α helices and β stands are marked H1 to H5 and S1 to S12, respectively, following the BBL numbering scheme (9). However, NDM-1 has a helix to correspond to S8 of the BBL scheme and contains a short helix before H2 and a short β strand after H3. (B) Surface presentation of NDM-1. Red, oxygen; blue, nitrogen; white, carbon.
Docked conformations of the β-lactams.
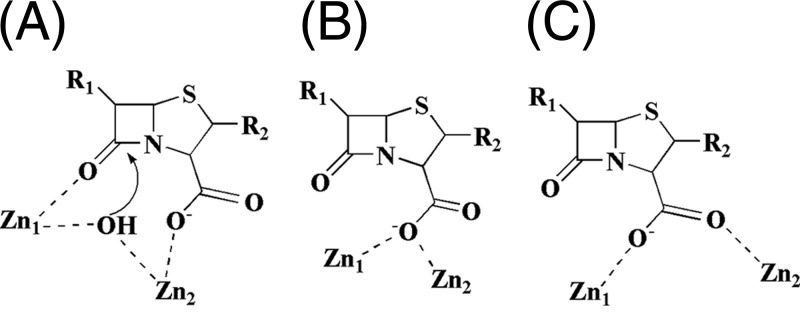
The program AutoDock (version 4.2) (12) automatically groups the 100 output poses into clusters when their overall root mean square deviations (RMSDs) are less than 2 Å. However, this classification does not well present the key feature of the β-lactam binding. We alternatively reclustered the outputs on the basis of the interactions of the carboxylic and carbonyl groups of β-lactams, which are two key functional groups for the catalysis and are shared by all β-lactams, with the zinc ions of NDM-1 (Fig. 1). Thus, all the docked poses of the β-lactams can be categorized into two binding modes. In the first mode, the carbonyl oxygen of the amide of β-lactams coordinates with zinc ion 1 (Zn1) (Fig. 3A), while the carboxylic oxygen of the fused rings chelates with zinc ion 2 (Zn2). Since the binding geometry in this mode is ready for the nucleophilic attack by the bridging hydroxide ion/water, which is a widely accepted mechanism for β-lactam hydrolysis found by people in the field of MBLs (7, 13, 28, 33, 39), we tentatively name this binding mode the S (substrate) conformer. In the second mode, the carboxylic group of β-lactams either displaces the bridging hydroxide ion/water (Fig. 3B) or simultaneously coordinates with both zinc ions to switch the amide group away from the metal ions (Fig. 3C). As the result, the hydrolysis of β-lactams is abolished in the second binding mode, and thus, we tentatively name it the I (inhibitor) conformer.
Fig 3.
Potential interactions of β-lactams with the zinc ions of NDM-1. (A) Proposed active form of the β-lactam interaction with the zinc ions. The bridging OH− would perform the nucleophilic attack. R1 and R2, substitutions of β-lactams. (B and C) Two types of inhibitory interactions of β-lactams with the zinc ions.
In the S conformers, the distance between negatively charged carboxylic oxygen of β-lactams and positively charged Zn2 of NDM-1 is 1.6 to 1.7 Å, while the distance between the carbonyl of β-lactam amide and Zn1 varies over a range of from 1.7 to 2.8 Å for the docked poses with the lowest energy (Table 1). The carbonyl and carboxylic oxygens of the β-lactams form distorted tetrahedrons, respectively, with Zn1 and Zn2 in all the S conformers, in spite of a slight variation in the positions of the fused β-lactam rings in each docked conformation. This binding geometry implies that zinc ions play an essential role in the substrate binding and the catalysis and is consistent with the early proposal (25). The interactions of the S or I conformers with the zinc ions are conserved in all the docked β-lactams and are thus understood in the following description of the binding of individual β-lactams.
Table 1.
Distances between zinc ions and carbonyl/carboxylic oxygens of β-lactams
| β-Lactam | Distance (Å) |
|
|---|---|---|
| Zn1 to carbonyl oxygen of β-lactam | Zn2 to carboxylic oxygen of β-lactam | |
| Ampicillin | 1.9 | 1.6 |
| Penicillin G | 2.0 | 1.6 |
| Piperacillin | 1.9 | 1.6 |
| Cefepime | 1.8 | 1.6 |
| Cefotaxime | 1.8 | 1.6 |
| Ceftazidime | 2.8 | 1.7 |
| Cefuroxime | 1.8 | 1.6 |
| Faropenem | 2.2 | 1.6 |
| Imipenem | 2.0 | 1.6 |
| Meropenem | 1.7 | 1.6 |
Ampicillin.
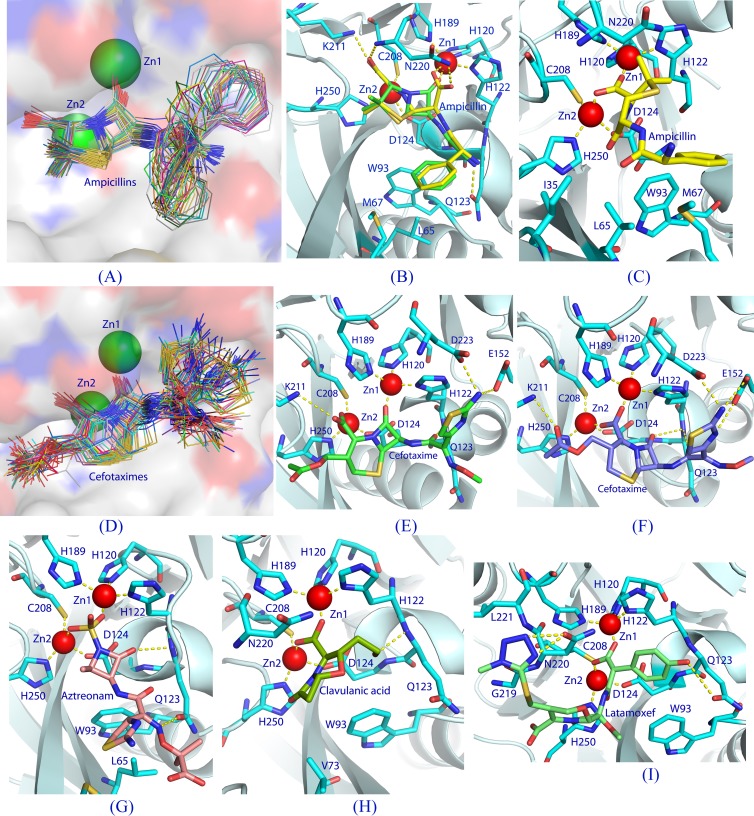
The 100 docked conformations of ampicillin were categorized into five clusters by AutoDock (version 4.2) and regrouped into the S and I conformers, which count for populations of 89% and 11%, respectively (Table 2). The lowest binding energies of the S and I conformers were −12.07 and −11.74 kcal/mol, respectively, corresponding to the estimated Ki values of 1.4 and 2.5 nM, respectively. A plot of all the 89 S conformers together shows that the positions of the β-lactam ring and the interactions of the carboxylic and carbonyl groups with the zinc ions are conserved, with a root mean standard deviation of <1 Å (Fig. 4A). However, the conformation and position of the phenyl group of ampicillin vary dramatically, forming a pattern like the open tail of a peacock (Fig. 4A). It is unknown which phenyl conformation of ampicillin would simulate the substrate binding, although the S conformer with the lowest binding energy is well superimposed over the hydrolyzed ampicillin in the crystal structure of NDM-1 (Fig. 4B). The S conformer with the lowest binding energy interacts with NDM-1 in a pattern similar to that for the reaction product in the crystal structure of NDM-1 (PDB accession number 3Q6X) (39), except for the hydrogen bond between Asn220 and the hydrolyzed ampicillin (Fig. 4B). The S conformer with the lowest binding energy forms three hydrogen bonds with Nz of Lys211, OE1 of Gln123, and the backbone nitrogen of Asp124, in addition to the van der Waals interactions with Ile35 Leu65, Met67, Val73, Trp93, and Asn220 (Fig. 4B). The I conformer with the lowest energy forms no hydrogen bonds but has van der Waals contacts with Ile35, Leu65, Met67, Val73, Trp93, and Asn220 (Fig. 4C).
Table 2.
Summary of β-lactam docking to NDM-1a
| β-Lactam | Experimentally measuredb |
S conformer |
I conformer |
|||||
|---|---|---|---|---|---|---|---|---|
| Km (μM) | kcat/Km (s−1 μM−1) | Lowest binding energy (kcal/mol) | Estimated Ki (nM) | Population (%) | Lowest binding energy (kcal/mol) | Estimated Ki (nM) | Population (%) | |
| Ampicillin | 22 | 2.2 | −12.07 | 1.4 | 89 | −11.74 | 2.5 | 11 |
| Penicillin G | 16 | 3.0 | −12.35 | 0.9 | 79 | −12.53 | 0.7 | 21 |
| Piperacillin | 12 | −12.70 | 0.5 | 97 | −11.56 | 3.4 | 3 | |
| Cefepime | 77 | −11.29 | 5.3 | 83 | −12.18 | 1.2 | 17 | |
| Cefotaxime | 10 | −12.04 | 1.5 | 71 | −14.23 | 0.04 | 29 | |
| Cefoxitin | 49 | −10.61 | 16.7 | 51 | −12.32 | 0.9 | 26 | |
| −12.58c | 0.6c | 4c | −11.10d | 7.3d | 11d | |||
| −13.46e | 0.1e | 8e | ||||||
| Ceftazidime | 181 | −11.02 | 8.4 | 8 | −12.7 | 0.5 | 86 | |
| −11.25d | 5.7d | 6d | ||||||
| Cefuroxime | 8 | −12.06 | 1.4 | 45 | −12.57 | 0.6 | 55 | |
| Cephalothin | 10 | −11.90 | 1.9 | 35 | −14.30 | 0.03 | 62 | |
| −12.02d | 1.5d | 3d | ||||||
| Faropenem | −12.11 | 1.3 | 14 | −12.76 | 0.4 | 74 | ||
| −11.96d | 1.7d | 12d | ||||||
| Imipenem | 94 | 4.3 | −10.39 | 24.2 | 62 | −11.31 | 5.2 | 17 |
| −10.18c | 34.4c | 7c | −10.79d | 12.2d | 14d | |||
| Meropenem | 49 | 2.6 | −10.64 | 15.8 | 84 | −11.65 | 2.9 | 5 |
| −11.59c | 3.2c | 11c | ||||||
| Clavulanic acid | Not detected | −11.44 | 4.1 | 100 | ||||
| Aztreonam | Not detected | −12.84 | 0.4 | 93 | ||||
| −12.38e | 0.8e | 7e | ||||||
| Sulbactam | −10.79 | 4.7 | 17 | −11.61 | 3.1 | 57 | ||
| −11.55d | 3.4d | 26d | ||||||
| Moxalactam | −10.97 | 9.1 | 1 | |||||
| −15.12f | 0.008f | 99f | ||||||
In the S conformer, the amide oxygen of the β-lactams interacts with Zn1, while the carboxylic group coordinates with Zn2. The I conformer has the carboxylic or sulfate oxygens interacting with Zn1 and Zn2.
The Km and kcat/Km values are taken from the papers by Yong et al. (38) and by Thomas et al. (28), respectively.
The S2 conformer has switched interactions with the zinc ions from the S conformer.
The I2 conformer has the orientation of the carboxylic group opposite that of I1.
In the I3 conformer, the bridging hydroxide/water is displaced by the carboxylic or sulfate oxygen.
In the I4 conformer, the tyrosyl-carboxylic group chelates with the zinc ions.
Fig 4.
Docking of β-lactams into the NDM-1 binding pocket. (A) A plot of all S conformers of docked ampicillin (total, 89 sticks) into the NDM-1 active site (surface presentation). Positional variation of the phenyl groups forms a pattern like the open tail of a peacock. (B) Interactions between NDM-1 and the S conformer (green sticks) of ampicillin with the lowest binding energy. The dotted lines represent hydrogen bonds or coordination with the metals. The hydrolyzed ampicillin from the crystal structure of the NDM-1-product complex (PDB accession number 3Q6X) is plotted with yellow sticks. (C) Interactions of the I conformer of ampicillin with NDM-1. (D) Binding of all the S conformers (71 total) of cefotaxime to the NDM-1 pocket. (E) Interactions of the S conformer of cefotaxime with NDM-1. (F) Interactions of the I conformer of cefotaxime with NDM-1. (G) An inhibitory conformation of aztreonam. (H) A representative binding of clavulanic acid. (I) An inhibitory binding of moxalactam (latamoxef) to NDM-1 in which the tyrosyl-carboxylic group of moxalactam interacts with both zinc ions.
Cefotaxime.
The five clusters of cefotaxime output from the program AutoDock (version 4.2) were reclassified into the S and I conformers, which have populations of 71% and 29%, respectively, and estimated Ki values of 1.5 and 0.04 nM, respectively (Table 2). The superposition of all the S conformers shows that the positions of the β-lactam ring and the interactions of the carboxylic and carbonyl groups with the zinc ions are well comparable (Fig. 4D). However, two substitutions on the β-lactam ring dramatically change their position and conformation (Fig. 4D). The S conformer with the lowest binding energy forms four hydrogen bonds with Gln123, Glu152, Lys211, and Asp223 (Fig. 4E), while the I conformer forms nine hydrogen bonds with Gln123, Asp124, Glu152, Lys211 Asn220, and Asp223 (Fig. 4F). Both S and I conformers show van der Waals interactions with Ile35, Val73, Trp93, and Gln123.
Faropenem.
AutoDock (version 4.2) outputs only three clusters for faropenem, which correspond to 14% S conformers (the best Ki, 1.3 nM) and two I conformers, I1 and I2 (74% population and Ki of 0.4 nM for I1 and 12% population and Ki of 1.7 nM for I2; Table 2). The I1 and I2 conformers have the opposite orientations of the β-lactam ring. The S conformer with the lowest energy forms five hydrogen bonds with Gln123, Asp124, Lys211, and Asn220, in addition to the van der Waals interactions with Ile35, Val73, and Trp93. The I conformer with the lowest energy forms two hydrogen bonds with Asn220 and His250, in addition to the van der Waal contacts with Ile35, Val73, Trp93, Gln123, Glu152, and Asp223.
Imipenem.
Imipenem had a wide distribution of the docked conformations, as shown by the output of nine clusters from AutoDock (version 4.2). The S conformers count for 69% of the population and can be further grouped into two conformations (62% and 7%; Table 2) in which the β-lactam ring has the opposite orientations and thus switched interactions of the carbonyl and carboxylic groups with Zn1 and Zn2. The I conformers of imipenem can also be divided into two groups, I1 and I2, in which I1 (17%) and I2 (14%) have the opposite orientations of their β-lactam rings. The S conformer with the lowest energy forms two hydrogen bonds with the side chain nitrogen of Lys211 and backbone carbonyl of Leu218, in addition to van der Waal contacts with Ile35, Val73, Trp93, and Asn220. The I conformer with the lowest energy forms two hydrogen bonds with Glu152 and Asp223, in addition to van der Waals interactions with Leu65, Met67, Val73, Trp93, and Asn220.
Aztreonam.
Aztreonam is the only β-lactam that contains no fused ring (Fig. 1). The 100 poses have a wide distribution of conformations and were grouped into 14 clusters. However, all the conformers have the sulfate group of aztreonam either blocking (93%) or displacing (7%) the bridging hydroxide/water and are thus inhibitory (Table 2). The I conformer with the lowest binding energy has a sulfate group coordinating with both zinc ions and forms two hydrogen bonds with Gln123 (Fig. 4G), in addition to van der Waals interactions with Leu65, Met67, and Trp93.
Clavulanic acid.
The 100 poses of clavulanic acid unanimously have the I conformation (Table 2). The I conformer with the lowest binding energy forms two hydrogen bonds with the backbone nitrogen atoms of Gln123 and Asp124, in addition to the van der Waals interactions with Val73, Trp93, Gln123, and Asn220 (Fig. 4H).
DISCUSSION
Evaluation of docking results.
Since the docked pose with the lowest binding energy does not always represent the true binding of a ligand in a biological system (34), an auxiliary standard would be required to identify the true conformation of ligand binding. We feel that the population of clusters from the docking might be a helpful hint for judgment of the correct conformation simulating ligand binding in a biological system. The β-lactams with a large population of the S conformer, such as ampicillin, would be expected to be substrates of NDM-1, while the large population of the I conformer would indicate a poor substrate. This prediction for NDM-1 is in general consistent with the experimental results (Table 2). However, the population of clusters would not have a quantitative meaning for analysis of docking results, and the population percentile that defines whether a β-lactam behaves as a substrate or an inhibitor would vary in individual cases. Besides, the estimated Ki values of the β-lactams from AutoDock (version 4.2) are about 3 to 4 orders of magnitude better than the experimental data (Table 2) and possibly result from the parameter settings of the program. Therefore, the docking may qualitatively identify a substrate or an inhibitor and would be a relative means for comparison of β-lactams.
Potential bound conformations of β-lactam substrates.
Since no crystal structures of any MBLs in complex with a substrate have been reported, the bound conformations of the β-lactam substrates remain veiled. Our docking experiments reveal that the β-lactams bound to the active site of NDM-1 in either the S or I conformation (Table 2; Fig. 4). In the S conformation, the interactions between the zinc ions of NDM-1 and the amide and carboxylic oxygens of the β-lactams are comparable for all the β-lactam substrates that have been docked, while the substitutions on the β-lactam rings dramatically change their conformations and positions (Fig. 4). Therefore, we hypothesize that the conformations and interactions of the β-lactam rings in the S conformers must simulate the true binding of the β-lactam substrates to NDM-1. This hypothesis is supported by several lines of evidence. First, the interactions of the docked S conformation of ampicillin with the lowest binding energy are similar to those of the hydrolyzed product, as shown by the structure superposition in Fig. 4B, except for the hydrolyzed ring of ampicillin. Second, in the S conformers, the bridging hydroxide/water is located at a position about 4 Å away from the amide carbon of the β-lactams and may thus be ready for nucleophilic attack. This pathway is consistent with the hydrolysis mechanism that has been unanimously proposed by several research groups (7, 21, 28–30, 33, 39). Third, the chelation of the amide oxygen of the β-lactams with Zn1, which was proposed to be an essential configuration of the catalysis on the basis of the crystal structures of IND-7 (36) and NDM-1 (39), is similar to that in the S conformers. Last and most importantly, our docking results are, in general, consistent with the enzymatic property of the β-lactam substrates. All β-lactam substrates that can be hydrolyzed by NDM-1 (28, 38) can be docked with a significant percentage of the S conformers, while the β-lactams that are resistant to NDM-1 hydrolysis, such as aztreonam and clavulanic acid, were docked in only I conformers (Table 2).
Insight into the broad spectrum of activity of NDM-1.
It has been puzzling how NDM-1 is capable of hydrolyzing almost all antibiotics containing the β-lactam scaffold (31). The docking of the various β-lactams described in this paper provides a hint to this puzzle. Since the docking experiments were to fit variable conformations of the β-lactams into the rigid NDM-1 pocket, the good accommodation of all the β-lactams to the active site of NDM-1 must imply no global or allosteric conformation changes upon the binding of β-lactams. Instead, the widely open binding pocket of NDM-1 in the crystal structure (Fig. 2B) may adopt β-lactams with a variety of substitutions, although minor changes of local conformation around the binding pocket of NDM-1 might be possible, too. If so, the minor conformational changes would likely be associated with the N-terminal fragment around S3 (Fig. 2A) that has significant variation in sequence and three-dimensional conformation across MBL families, but this is yet to be confirmed by the crystal structure of NDM-1 in complex with substrates.
Hints on design of NDM-1 inhibitors.
Discovery of clinically usable inhibitors of any MBLs has not been successful (1, 32). Our docking provides some hints on the design of NDM-1 inhibitors. The docking suggests that the high electrophilicity of the NDM-1 binding pocket with two zinc ions on its bottom may be the major binding force and thus prefers to bind negatively charged inhibitors. This hypothesis is consistent with the early argument that zinc ion plays critical roles both in the binding of substrates and in the hydrolysis of β-lactams (25). The neighboring of the amide oxygen and carboxylic group of the fused rings in most β-lactams matches well with the geometry of the zinc pocket (Fig. 3 and 4) and may thus explain why most MBL enzymes have a wide spectrum of activity. On the basis of this observation, we expected that a β-lactam containing an additional negatively charged group would compete with binding of the S conformer of a substrate and thus inhibit the enzyme activity. For example, 99 out of 100 docked conformations of moxalactam have its tyrosyl-carboxylic group instead of the carboxylic group on the fused ring of β-lactams interacting with the zinc ions (Fig. 1 and 4I). Thus, moxalactam would be expected to be a pool substrate or even an inhibitor of NDM-1, although this possibility needs to be confirmed by enzymatic assay. Another example is aztreonam. The negatively charged sulfate group of aztreonam either blocks or replaces the bridging hydroxide/water in all docked conformations (Fig. 4G), suggesting that aztreonam would behave as an inhibitor. This prediction is consistent with the enzymatic assay, in that no hydrolysis activity of NDM-1 was detected (38).
In addition to the electrophilic interactions with the zinc ions, the interactions of the remaining pockets of NDM-1 would also be important to determine whether a β-lactam works as a substrate or an inhibitor. Since the negatively charged carboxylic group has a stronger tendency to chelate with two positively charged zinc ions and thus to inhibit the NDM-1 activity, a helper is necessary to orient the β-lactam ring in a correct position to chelate with the zinc ions in the S conformation. This may be achieved by a relatively large substitution that links to either side of the lactam ring. Thus, the β-lactams without large substitutions would behave as an inhibitor, as observed in the case of clavulanic acid, in which the 100% poses in the I conformation from our docking (Fig. 1 and 4H) agreed well with no detection of hydrolysis by NDM-1 (38). In addition, the majority of a small β-lactam, sulbactam (Fig. 1), was docked in the I conformation (83%; Table 2), and sulbactam would thus be expected to be a poor substrate.
Conclusions.
(i) The docked S conformation, in which the amide oxygen and carboxylic acid of β-lactams interact, respectively, with Zn1 and Zn2, may simulate the binding of a true substrate to NDM-1. (ii) The natural fit of various β-lactams into the rigid pocket of NDM-1, as revealed by the docking, suggests that no dramatic conformation changes are required for NDM-1 to recognize β-lactam substrates. (iii) Substitutions on the fused β-lactam ring are essential for β-lactams to bind in the S conformation, and thus, small β-lactams such as clavulanic acid may behave as inhibitors. (iv) β-Lactams with an additional negatively charged group, such as moxalactam, may disturb the substrate binding in the S conformation and may thus be a poor substrate or even an inhibitor.
ACKNOWLEDGMENTS
This work was supported in part by NIH grant GM59791 to H.K.
We thank Pei-Ying Wu and Wenjun Cui for their proofreading.
Footnotes
Published ahead of print 23 July 2012
REFERENCES
- 1. Bebrone C. 2007. Metallo-beta-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. Biochem. Pharmacol. 74:1686–1701 [DOI] [PubMed] [Google Scholar]
- 2. Bogaerts P, et al. 2011. Emergence of NDM-1-producing Enterobacteriaceae in Belgium. Antimicrob. Agents Chemother. 55:3036–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bush K. 2010. Alarming β-lactamase-mediated resistance in multidrug-resistant Enterobacteriaceae. Curr. Opin. Microbiol. 13:558–564 [DOI] [PubMed] [Google Scholar]
- 4. Bush K, Jacoby GA. 2010. Updated functional classification of beta-lactamases. Antimicrob. Agents Chemother. 54:969–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Centers for Disease Control and Prevention 2010. Update: detection of a Verona integron-encoded metallo-beta-lactamase in Klebsiella pneumoniae—United States. MMWR Morb. Mortal. Wkly. Rep. 59:1212. [PubMed] [Google Scholar]
- 6. Chen Y, Zhou Z, Jiang Y, Yu Y. 2011. Emergence of NDM-1-producing Acinetobacter baumannii in China. J. Antimicrob. Chemother. 66:1255–1259 [DOI] [PubMed] [Google Scholar]
- 7. Crisp JR, et al. 2007. Structural basis for the role of Asp-120 in metallo-beta-lactamases. Biochemistry 46:10664–10674 [DOI] [PubMed] [Google Scholar]
- 8. Deshpande PC, et al. 2010. New Delhi metallo-beta lactamase (NDM-1) in Enterobacteriaceae: treatment options with carbapenems compromised. J. Assoc. Physicians India 58:147–149 [PubMed] [Google Scholar]
- 9. Galleni M, et al. 2001. Standard numbering scheme for class B β-lactamases. Antimicrob. Agents Chemother. 45:660–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Garau GI, et al. 2004. Update of the standard numbering scheme for class B beta-lactamases. Antimicrob. Agents Chemother. 48:2347–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guo YJ, et al. 2011. A structural view of the antibiotic degradation enzyme NDM-1 from a superbug. Protein Cell 2:384–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huey R, Morris GM, Olson AJ, Goodsell DS. 2007. A semiempirical free energy force field with charge-based desolvation. J. Comput. Chem. 28:1145–1152 [DOI] [PubMed] [Google Scholar]
- 13. Kim YC, et al. 2011. Structure of apo- and monometalated forms of NDM-1—a highly potent carbapenem-hydrolyzing metallo-β-lactamase. PLoS One 6:e24621 doi:10.1371/journal.pone.0024621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. King D, Strynadka N. 2011. Crystal structure of New Delhi metallo-β-lactamase reveals molecular basis for antibiotic resistance. Protein Sci. 20:1484–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kumarasamy KK, et al. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10:597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kus JV, et al. 2011. New Delhi metallo-{beta}-lactamase-1: local acquisition in Ontario, Canada, and challenges in detection. CMAJ 183:1257–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mammina C, et al. 2010. Multiclonal emergence of carbapenem-resistant Klebsiella pneumoniae in Tuscany, Italy. Int. J. Antimicrob. Agents 36:576–578 [DOI] [PubMed] [Google Scholar]
- 18. Marchaim D, et al. 2011. Outbreak of colistin-resistant, carbapenem-resistant Klebsiella pneumoniae in metropolitan Detroit, Michigan. Antimicrob. Agents Chemother. 55:593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Morris GM, et al. 1998. Automated docking using a Lamarckian genetic algorithm and empirical binding free energy function. J. Comput. Chem. 19:1639–1662 [Google Scholar]
- 20. Neuner EA, et al. 2011. Treatment and outcomes in carbapenem-resistant Klebsiella pneumoniae bloodstream infections. Diagn. Microbiol. Infect. Dis. 69:357–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Page MI, Badarau A. 2008. The mechanisms of catalysis by metallo beta-lactamases. Bioinorg. Chem. Appl. 2008:576297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pfeifer Y, et al. 2011. Molecular characterization of blaNDM-1 in an Acinetobacter baumannii strain isolated in Germany in 2007. J. Antimicrob. Chemother. 66:1998–2001 [DOI] [PubMed] [Google Scholar]
- 23. Poirel L, Lagrutta E, Taylor P, Pham J, Nordmann P. 2010. Emergence of metallo-β-lactamase NDM-1-producing multidrug resistant Escherichia coli in Australia. Antimicrob. Agents Chemother. 54:4914–4916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Poirel L, et al. 2011. Molecular analysis of NDM-1-producing enterobacterial isolates from Geneva, Switzerland. J. Antimicrob. Chemother. 66:1730–1733 [DOI] [PubMed] [Google Scholar]
- 25. Rasia RM, Vila AJ. 2004. Structural determinants of substrate binding to Bacillus cereus metallo-β-lactamase. J. Biol. Chem. 279:26046–26051 [DOI] [PubMed] [Google Scholar]
- 26. Samuelsen Ø, et al. 2011. Identification of NDM-1-producing Enterobacteriaceae in Norway. J. Antimicrob. Chemother. 66:670–672 [DOI] [PubMed] [Google Scholar]
- 27. Solé M, et al. 2011. First description of an Escherichia coli strain producing NDM-1 carbapenemase in Spain. Antimicrob. Agents Chemother. 55:4402–4404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Thomas PW, et al. 2011. Characterization of purified New Delhi metallo-β-lactamase-1 (NDM-1). Biochemistry 50:10102–10113 [DOI] [PubMed] [Google Scholar]
- 29. Tioni MF, et al. 2008. Trapping and characterization of a reaction intermediate in carbapenem hydrolysis by B. cereus metallo-beta-lactamase. J. Am. Chem. Soc. 130:15852–15863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tomatis PE, et al. 2008. Adaptive protein evolution grants organismal fitness by improving catalysis and flexibility. Proc. Natl. Acad. Sci. U. S. A. 105:20605–20610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Walsh TR, Toleman MA. 2011. The new medical challenge: why NDM-1? Why Indian? Expert Rev. Anti Infect. Ther. 9:137–141 [DOI] [PubMed] [Google Scholar]
- 32. Walsh TR, Toleman MA, Poirel L, Nordmann P. 2005. Metallo-beta-lactamases: the quiet before the storm. Clin. Microbiol. Rev. 18:306–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang Z, Fast W, Valentine AM, Benkovic SJ. 1999. Metallo-beta-lactamase: structure and mechanism. Curr. Opin. Chem. Biol. 3:614–622 [DOI] [PubMed] [Google Scholar]
- 34. Warren GL, et al. 2006. A critical assessment of docking programs and scoring functions. J. Med. Chem. 49:5912–5931 [DOI] [PubMed] [Google Scholar]
- 35. Yamaguchi Y, et al. 2007. Crystallographic investigation of the inhibition mode of a VIM-2 metallo-beta-lactamase from Pseudomonas aeruginosa by a mercaptocarboxylate inhibitor. J. Med. Chem. 50:6647–6653 [DOI] [PubMed] [Google Scholar]
- 36. Yamaguchi Y, et al. 2010. Structure of metallo-beta-lactamase IND-7 from a Chryseobacterium indologenes clinical isolate at 1.65-Å resolution. J. Biochem. 147:905–915 [DOI] [PubMed] [Google Scholar]
- 37. Yamamoto T, Takano T, Iwao Y, Hishinuma A. 2011. Emergence of NDM-1-positive capsulated Escherichia coli with high resistance to serum killing in Japan. J. Infect. Chemother. 17:435–439 [DOI] [PubMed] [Google Scholar]
- 38. Yong D, et al. 2009. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53:5046–5054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang H, Hao Q. 2011. Crystal structure of NDM-1 reveals a common {beta}-lactam hydrolysis mechanism. FASEB J. 25:2574–2582 [DOI] [PubMed] [Google Scholar]