Abstract
Historically, the incidence of gentamicin resistance in Campylobacter has been very low, but recent studies reported a high prevalence of gentamicin-resistant Campylobacter isolated from food-producing animals in China. The reason for the high prevalence was unknown and was addressed in this study. PCR screening identified aminoglycoside resistance genes aphA-3 and aphA-7 and the aadE–sat4–aphA-3 cluster among 41 Campylobacter isolates from broiler chickens. Importantly, a novel genomic island carrying multiple aminoglycoside resistance genes was identified in 26 aminoglycoside resistant Campylobacter coli strains. Sequence analysis revealed that the genomic island was inserted between cadF and COO1582 on the C. coli chromosome and consists of 14 open reading frames (ORFs), including 6 genes (the aadE–sat4–aphA-3 cluster, aacA-aphD, aac, and aadE) encoding aminoglycoside-modifying enzymes. Analysis by pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing indicated that the C. coli isolates carrying this unique genomic island were clonal, and the clone of PFGE subtype III and sequence type (ST) 1625 was particularly predominant among the C. coli isolates examined, suggesting that clonal expansion may be involved in dissemination of this resistance island. Additionally, we were able to transfer this genomic island from C. coli to a Campylobacter jejuni strain using natural transformation under laboratory conditions, and the transfer resulted in a drastic increase in aminoglycoside resistance in the recipient strain. These findings identify a previously undescribed genomic island that confers resistance to multiple aminoglycoside antibiotics. Since aminoglycoside antibiotics are used for treating occasional systemic infections caused by Campylobacter, the emergence and spread of this antibiotic resistance genomic island represent a potential concern for public health.
INTRODUCTION
Campylobacter is one of the most common food-borne pathogens worldwide. Two thermophilic species, Campylobacter jejuni and Campylobacter coli, are responsible for most Campylobacter infections in humans. Both C. jejuni and C. coli are prevalent in food-producing animals, and the extensive use of antimicrobials in food animal production has led to an increasing prevalence of antibiotic-resistant Campylobacter strains, which can be transmitted to humans via the food chain (1, 3).
Clinical therapy of Campylobacter-induced gastroenteritis, when needed, is often conducted with macrolide and fluoroquinolone antibiotics, but severe systemic infections, which occur sporadically, requires the use of an aminoglycoside antibiotic, such as gentamicin (2). Aminoglycosides are highly potent, broad-spectrum, bactericidal antibiotics and are commonly used in the treatment of infections caused by both Gram-negative and Gram-positive organisms in humans (25). Additionally, several aminoglycoside agents, such as neomycin and amikacin, are also used in conventional broiler chicken and swine production in some countries, including China. Among the known mechanisms of aminoglycoside resistance, enzymatic modification is the most important and prevalent one in several bacterial species, including Campylobacter spp. (17, 25). Based on the reactions they catalyze, aminoglycoside-modifying enzymes are divided into three groups: aminoglycoside acetyltransferases (AACs), aminoglycoside nucleotidyltranferases (ANTs), and aminoglycoside phosphotransferases (APHs) (17). In Campylobacter, each group of aminoglycoside-modifying enzymes has been detected. The 3′,9-aminoglycoside adenyltransferase gene (aadA) and 6-aminoglycoside adenyltransferase gene (aadE) confer resistance to streptomycin; the aacA4 gene, encoding an AAC, confers resistance to gentamicin and tobramycin (11, 15), and 3′-APHs, which are the most frequently reported, are responsible for resistance to kanamycin and neomycin (30).
The aminoglycoside resistance genes in Campylobacter have been detected on multidrug-resistant plasmids, integrons, and transposons (9, 13). In addition, the aminoglycoside resistance gene cluster, aadE–sat4–aphA-3, has been detected on transmissible plasmids in C. jejuni but has not been identified in C. coli (9, 13). Although kanamycin resistance has been reported in multiple studies, the prevalence of gentamicin-resistant Campylobacter is generally low (<1%) in studies from various countries (3, 12, 27). However, two recent studies conducted in China indicated that the frequency of aminoglycoside resistance, especially gentamicin resistance, is high (>20%) in C. coli isolated from both broiler chickens and swine (4, 16). The resistance mechanisms and the reason for the high prevalence of gentamicin-resistant Campylobacter in China are not known. To answer these questions, we analyzed aminoglycoside-resistant Campylobacter isolates derived from chickens and identified a unique genetic structure containing multiple aminoglycoside resistance genes, including aadE–sat4–aphA-3 and aacA-aphD, on the chromosome of C. coli. This novel chromosomal aminoglycoside resistance gene cluster has not been reported previously and is associated with clonal isolates of C. coli, suggesting the possibility of clonal expansion of aminoglycoside-resistant C. coli in broiler chickens in China. Furthermore, we demonstrated the potential dissemination of this unique genetic structure from C. coli to C. jejuni under laboratory conditions. These findings provide new insights into the dissemination of aminoglycoside resistance in Campylobacter.
MATERIALS AND METHODS
Campylobacter strains and susceptibility testing.
In total, 138 Campylobacter isolates (C. jejuni, n = 90; C. coli, n = 48), derived from two broiler chicken slaughterhouses located in eastern (Penglai) and western (Shenxian) regions of Shandong province, were used in this study. Detailed information on their susceptibilities to various antimicrobials (including gentamicin) is provided in a previous publication (4). In this study, the isolates were further examined for their susceptibility to kanamycin, neomycin, and amikacin using the agar dilution method, according to the guideline M31-A3 of the Clinical and Laboratory Standards Institute (CLSI) (5). The reference strain C. jejuni ATCC 33560 was used as a quality control strain. The breakpoint for kanamycin is 64 μg/ml according to the CLSI (5). Since there are no CLSI breakpoints for neomycin and amikacin in Campylobacter, we used the breakpoints of the two agents for Enterobacteriaceae, which are 32 μg/ml and 64 μg/ml, respectively (5, 26).
Detection of aminoglycoside resistance genes and molecular analysis.
All kanamycin-resistant strains were selected to investigate the known aminoglycoside resistance genes aphA-1, aphA-3, and aphA-7 and the aadE-sat4-aphA-3 gene cluster, which were previously found in Campylobacter (9, 14, 22). The primers used to detect the aphA-3 gene and aadE-sat4-aphA-3 gene cluster were described previously (9), and the primers designed for the amplification of aphA-1 (aphA-1-F, 5′-CGTATTTCGTCTCGCTCAG-3′; aphA-1-R, 5′-CCGACTCGTCCAACATCA-3′) and aphA-7 (aphA-7-F, 5′-ATCCGATAAACTGAAAGTAC-3′; aphA-7-R, 5′-ATAATCCGGTTCAAGTCCC-3′) were based on previously published sequences (14, 22). Genomic DNA was isolated from the strains using a Wizard genomic DNA purification kit (Promega, Madison, WI). Plasmid DNA was extracted using a Qiagen plasmid extraction midikit (Qiagen, Hilden, Germany). Southern blotting was conducted to determine the location of the aadE–sat4–aphA-3 gene cluster; the primers specific for the gene cluster (CF, 5′-GGATGGATTCCTATGAAAACAT-3′; CR, 5′-GGCTTTGTTCATCTTCATACTCT-3′) were designed according to previously published sequences (9), and the PCR product was used as the probe for hybridization. The probe was nonradioactively labeled with a DIG High Prime I DNA labeling and detection starter kit (Roche Diagnostics, Mannheim, Germany). Hybridization was performed at 42°C for 14 h. Membranes were washed twice at room temperature (22 to 25°C) in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.5% SDS for 5 min and twice at 68°C in 0.1× SSC, 0.5% SDS for 15 min. DIG was detected with specific antibodies using a DIG High Prime I DNA labeling and detection starter kit (Roche Diagnostics, Mannheim, Germany) according to the recommendations of the supplier. In addition, the flanking regions of the gene cluster were sequenced by a modified random primer sequencing walking strategy as described previously (29).
Pulsed-field gel electrophoresis (PFGE).
PFGE was performed using a CHEF-DR III apparatus (Bio-Rad Laboratories, Hercules, CA), according to the rapid protocol for Campylobacter (18). Salmonella H9812 was used as the reference marker (digested with XbaI), while all Campylobacter isolates were digested with SmaI. The running conditions were as follows: 0.5× Tris-borate-EDTA–1% SeaKem Gold agarose (FMC BioProducts, Rockland, ME) at 14°C and 6 V/cm for 18 h with switch times ranging from 6.75 s to 38.35 s and an included angle of 120°. The dendrograms were constructed from the PFGE data by UPGMA (unweighted pair group method with arithmetic average) with the Dice coefficient using the InfoQuest FP software version 4.5 (Bio-Rad Laboratories).
Multilocus sequence typing (MLST).
MLST of the seven housekeeping genes (aspA, glnA, gltA, glyA, pgm, tkt, and uncA) for C. coli was performed following the method described previously (7). Briefly, all the housekeeping genes were amplified, and the PCR products were run on an agarose gel to confirm the correct amplicon size and then purified using a Tiangen purification midikit following the manufacturer's instructions (Tiangen, Beijing, China). All purified products were sent to the Beijing Genomic Institute (BGI) for sequencing using the forward and reverse primers of these genes. The allelic profiles and the sequence types were generated by carrying out a BLAST search with the Campylobacter sequences in the MLST database (http://www.pubmlst.org/campylobacter).
Natural transformation.
A transformation assay using Mueller-Hinton agar was performed according to the method described by Wang and Taylor with minor modifications (28). Genomic DNAs purified from aminoglycoside-resistant C. coli strains served as the donors, while aminoglycoside-sensitive C. jejuni NCTC11168 was used as the recipient. Briefly, fresh recipient cells (20- to 24-h growth on Mueller-Hinton [MH] agar, grown microaerobically at 42°C) were spread on MH agar at about 5 × 107 cells per plate (in triplicate) and incubated for 6 h (microaerobically at 42°C). Subsequently, 1 μg of genomic DNA of the donor strain was spotted directly onto the inoculated agar, without additional spreading, and incubation was continued for 5 h at 42°C under microaerobic conditions. The cells were harvested from the MH agar and plated on a selective MH agar plate containing kanamycin (60 μg/ml) (KMHA), which was further incubated for 48 to 72 h at 42°C under microaerobic conditions. The recipient (without donor DNA) inoculated on the same MH agar plate served as a negative control. Transformants on the KMHA plates were selected and confirmed by PCR. The stability of aminoglycoside resistance in the transformants was investigated using the method reported by Kim et al. (10).
Nucleotide sequence accession number.
The sequence of the novel gene cluster described in this paper has been submitted to GenBank under accession number JQ655275.
RESULTS
Characterization of aminoglycoside resistance in Campylobacter.
Among the 138 Campylobacter isolates tested (C. jejuni, n = 90; C. coli, n = 48), 41 (29.7%, including 5 C. jejuni and 36 C. coli) were resistant to both kanamycin (MIC ≥ 256 μg/ml) and neomycin (MIC ≥ 32 μg/ml), and 36 (26%, including 2 C. jejuni and 34 C. coli) were resistant to amikacin (MIC ≥ 64 μg/ml). All 36 amikacin-resistant isolates were also resistant to both kanamycin and neomycin. The prevalence of resistance to the tested aminoglycosides was significantly higher in C. coli (>71%) than in C. jejuni (<5.6%) (P < 0.01). Over half of the kanamycin-resistant isolates (65.9%, 26/41) harbored the aadE–sat4–aphA-3 cluster, and all of them were C. coli. In addition, all these aadE–sat4–aphA-3-carrying C. coli isolates were resistant to gentamicin. The other 15 kanamycin-resistant isolates harbored only either aphA-7 or aphA-3 and were not resistant to gentamicin. The aphA-7 gene was present only in the Campylobacter isolates from Shenxian, including 2 C. jejuni and 10 C. coli isolates. The single aphA-3 gene was observed only in three C. jejuni strains, one from Shenxian and two from Penglai. No aphA-1 gene was detected among the kanamycin-resistant isolates. The distribution of kanamycin resistance genes in Campylobacter is summarized in Table 1.
Table 1.
Distribution of kanamycin resistance determinants in Kmr isolates detected by PCR
| Kanamycin resistance gene(s) | Species | No. of Kmr isolates: |
|
|---|---|---|---|
| SX (n = 18) | MH (n = 23) | ||
| aphA-1 | C. jejuni | 0 | 0 |
| C. coli | 0 | 0 | |
| aphA-3a | C. jejuni | 1 | 2 |
| C. coli | 0 | 0 | |
| aphA-7 | C. jejuni | 2 | 0 |
| C. coli | 10 | 0 | |
| aadE–sat4–aphA-3 | C. jejuni | 0 | 0 |
| C. coli | 5 | 21 | |
The isolates harbored only aphA-3 (strains harboring aphA-3 as part of aadE-sat4-aphA3 were not included in this category).
Genotyping.
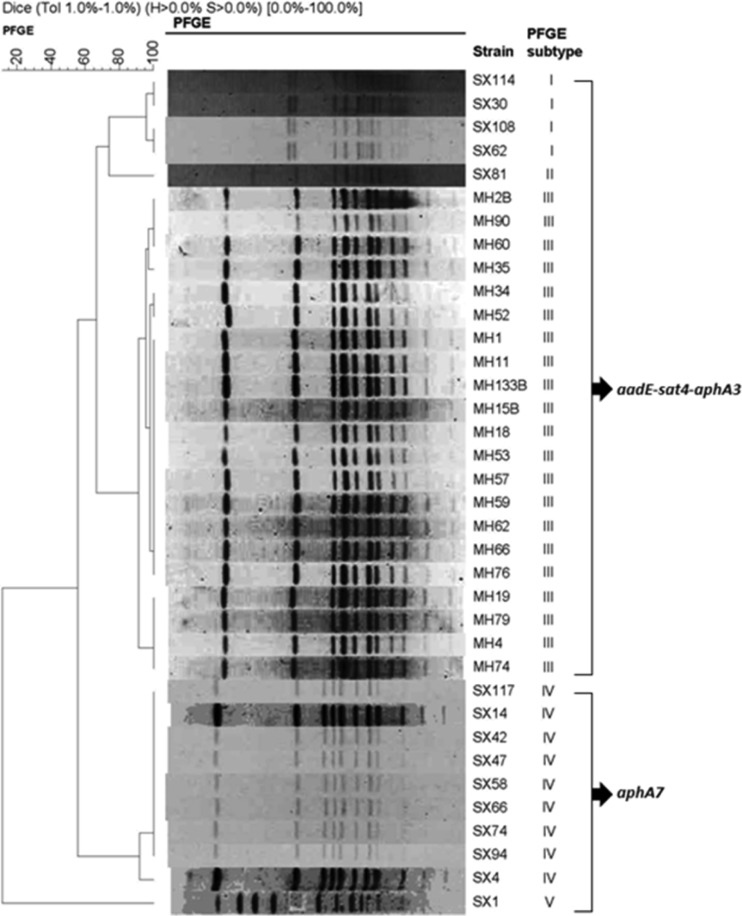
To facilitate understanding of the transmission of the aminoglycoside resistance genes, the genetic diversity of the 36 aminoglycoside-resistant C. coli isolates that harbored aphA7 or the aadE–sat4–aphA-3 cluster was determined by PFGE using SmaI digestion (Fig. 1). Using a cutoff of 90% genetic similarity, the isolates were clustered into 5 PFGE subtypes, with subtype III accounting for the majority (21/36) of the isolates. Subtypes I, II, and III harbored the aadE–sat4–aphA-3 cluster (n = 26), while subtypes IV and V carried the aphA7 gene. The high genetic similarity (>95%) of the 21 isolates (subtype III) (Fig. 1) that all originated from Penglai (MH strains) suggested that these isolates might have originated from a single clone. However, the isolates from Shenxian (SX strains) exhibited four different PFGE subtypes (I, II, IV, and V).
Fig 1.
PFGE-based dendrogram for 36 kanamycin-resistant C. coli strains harboring the aphA-7 gene or the aadE–sat4–aphA-3 gene cluster.
Twelve aminoglycoside-resistant isolates representative of all the PFGE subtypes and four aminoglycoside-susceptible isolates (SX71, SX73, MH16, and MH25) were selected for MLST typing. The results identified six sequence types (STs) (Table 2). Four STs were identified for the aminoglycoside resistant isolates: ST 5604 (SX30, SX108 and SX114 in PFGE subtype I), ST 1586 (SX81 in PFGE subtype II), ST 1625 (MH4, MH34, MH57 and MH62 in PFGE subtype III), and ST 5510 (SX94, SX117, and SX58 in PFGE subtype IV; SX1 in PFGE subtype V). ST 5604 represents a newly identified ST and is first reported in this study. In general, each PFGE subtype corresponds to a single ST, indicating the consistency of the two typing methods. Notably, all of the typed aminoglycoside resistant isolates belong to clonal complex 828 (Table 2). Two STs were found for the typed aminoglycoside-susceptible isolates, including ST 4248 (SX71 and SX73) and ST 4268 (MH16 and MH25), whose clonal complexes have not been assigned.
Table 2.
Aminoglycoside susceptibility, ST, clonal complex, and PFGE pattern of C. coli strains from broiler chickens
| Aminoglycoside susceptibility | Strains | ST | Clonal complex | PFGE pattern |
|---|---|---|---|---|
| Resistant | SX114 | 5604 | 828 | I |
| SX30 | 5604 | 828 | I | |
| SX108 | 5604 | 828 | I | |
| SX81 | 1586 | 828 | II | |
| MH57 | 1625 | 828 | III | |
| MH62 | 1625 | 828 | III | |
| MH34 | 1625 | 828 | III | |
| MH4 | 1625 | 828 | III | |
| SX94 | 5510 | 828 | IV | |
| SX117 | 5510 | 828 | IV | |
| SX58 | 5510 | 828 | IV | |
| SX1 | 5510 | 828 | IV | |
| Susceptible | SX71 | 4248 | —a | NDb |
| SX73 | 4248 | — | ND | |
| MH16 | 4268 | — | ND | |
| MH25 | 4268 | — | ND |
—, not assigned.
ND, not determined.
Identification of a novel aminoglycoside resistance gene island.
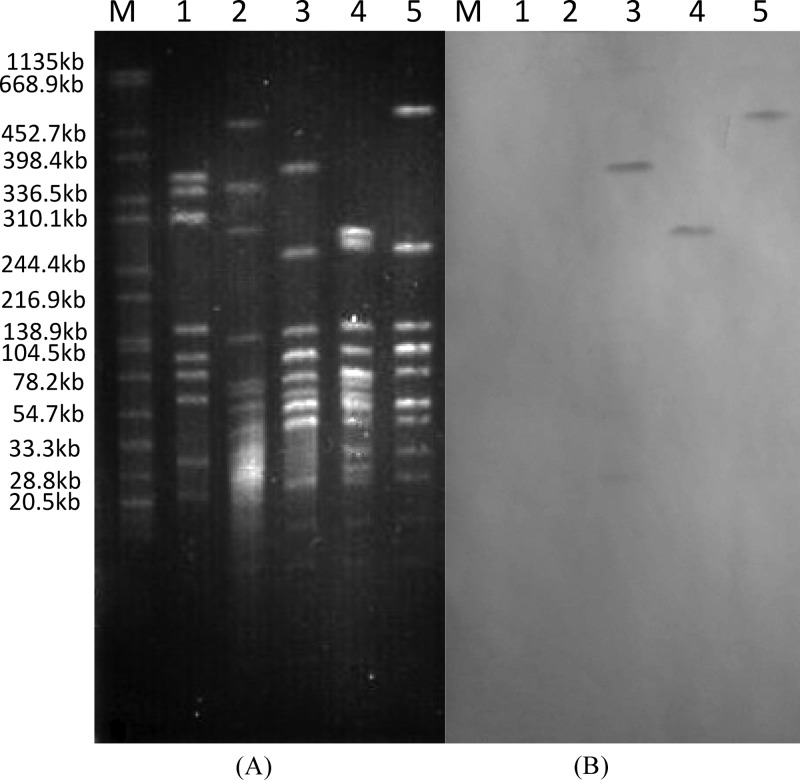
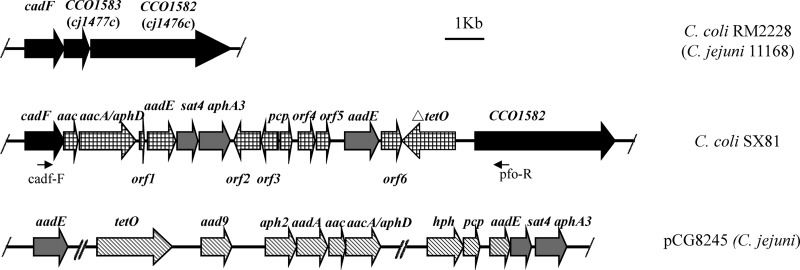
Three isolates harboring the aadE–sat4–aphA-3 gene cluster (SX81, MH57, and SX114), which belonged to three different PFGE subtypes (Fig. 1) and three different ST types (Table 2), were selected for further genetic analysis. No plasmids were obtained from these isolates by extraction using the Qiagen plasmid extraction midikit and the Wizard Plus SV miniprep purification system (Promega), suggesting that the resistance genes were not carried by plasmids. Southern blotting of PFGE-separated DNA of all tested strains indicated that the aadE–sat4–aphA-3 cluster was located on large chromosomal DNA fragments digested by SmaI (Fig. 2). The surrounding genetic environment of the aadE–sat4–aphA-3 cluster in SX81 was sequenced by a modified random primer walking strategy, starting from the aadE gene and the aphA-3 gene. A 15.3-kb segment of the chromosome was sequenced and was found to contain 16 open reading frames (ORFs), including the aadE–sat4–aphA-3 cluster (Fig. 3; Table 3). A unique segment of 14 ORFs, including several antimicrobial resistance genes [six genes encoding aminoglycoside-modifying enzymes and a truncated tet(O) gene], was inserted between the cadF gene and the CCO1582 gene in the chromosomal DNA of C. coli SX81. In the sequenced genome of C. coli RM2228 (7), the CCO1583 gene is located between cadF and CCO1582. A similar genetic organization was also observed in C. jejuni (Fig. 3). However, the gene CCO1583 (or cj1477c in C. jejuni) was replaced by the 14-ORF segment in C. coli SX81. Additionally, the G+C content for this 14-gene cluster is 37.3%, which is significantly higher than that (31.4%) of the entire genome of C. coli RM2228. These results indicate that C. coli SX81 has acquired a genomic island of 14 ORFs inserted between cadF and COO1582 on the chromosome.
Fig 2.
(A) PFGE plugs of kanamycin-resistant C. coli SX81, SX114, and MH57 digested with SmaI; (B) Southern hybridization of the aadE–sat4–aphA-3 cluster to genomic DNA of these isolates. Lane 1, NCTC11168; lane 2, ATCC 33559; lane 3, SX81; lane 4, SX114; lane 5, MH57; lane M, DNA standards.
Fig 3.
Genomic organization of the aminoglycoside resistance island in C. coli SX81 in comparison with plasmid pCG8245 and chromosomal genes of C. coli RM2228. Arrows indicate the positions and directions of transcription of the genes. Homologous genes in the three fragments are displayed with the same types of shading. The locations of primers cadf-F and pfo-R used to detect the unique genomic island are indicated.
Table 3.
Annotation of genes in the genomic island flanked by cadF and cco1582 in C. coli SX81
| Gene or ORF | Coding region (bp) | G+C content (%) | Length (aa)a | Length (aa)b | % Identity/% positive (no. of aa) | Annotation and/or accession no. |
|---|---|---|---|---|---|---|
| cadF | 121–1119 | 34.5 | 332 | 332 | 99/99 (332) | Fibronectin-binding protein, Campylobacter coli JV20; ZP_07401432 |
| aac | 1133–1498 | 24.9 | 121 | 121 | 100/100 (121) | Acetyltransferase, Enterococcus faecalis T2; ZP_05426921 |
| aacA-aphD | 1499–2938 | 24.7 | 479 | 479 | 100/100 (479) | Bifunctional aminoglycoside modifying enzyme AacA-AphD, Staphylococcus aureus; NP_863643 |
| orf1 | 2996–3112 | 19.7 | 38 | Unknown (no significant match in database) | ||
| aadE | 3204–3902 | 37.4 | 233 | 225 | 97/97 (225) | Streptomycin aminoglycoside 6-adenyltransferase, Enterococcus faecalis D6; EEU82268 |
| sat4 | 3914–4444 | 37.9 | 176 | 176 | 100/100 (176) | Streptothricinacetyltransferase, Enterococcus faecium; ZP_05674679.1 |
| aphA-3 | 4537–5331 | 44.9 | 264 | 264 | 100/100 (264) | Aminoglycoside phosphotransferase type III, Enterococcus faecalis; YP_783930 |
| orf2 | 5439–6105 | 44.4 | 224 | 199 | 80/82(199) | Hypothetical protein BSEG_01714, Bacteroides dorei 5_1_36, D4; EEO45573 |
| orf3 | 6127–6550 | 41.3 | 150 | 141 | 99/99 (141) | Hypothetical protein BSEG_01715, Bacteroides dorei5_1_36, D4; EEO45574 |
| pcp | 6620–6913 | 51.2 | 139 | 97 | 100/100 (97) | Pyrrolidone carboxylate peptidase, Bacteroides sp. 9_1_42FAA; ZP_04541315 |
| orf4 | 7067–7495 | 31.7 | 132 | 120 | 95/98 (120) | Hypothetical protein, Streptococcus suis; CBZ42064 |
| orf5 | 7527–7871 | 42.0 | 181 | 104 | 98/100 (104) | Unknown, Campylobacter jejuni; AAW34157 |
| aadE | 8249–9115 | 35.4 | 288 | 288 | 99/99 (288) | Putative adenyltransferase, Pediococcus acidilactici; YP_001965484 |
| orf6 | 9203–9720 | 58.9 | 196 | 172 | 88/89(172) | Hypothetical protein CLONEX_02761, Clostridium nexile DSM 1787; EEA81276 |
| Δtet(O) | 9735–11096 | 39.2 | 639 | 449 | 100/100 (449) | Tetracycline resistance protein, Campylobacter coli RM2228; ZP_00371083 |
| CCO1582 | 11568–15128 | 35.3 | 1186 | 1186 | 99/99 (1186) | Pyruvate ferredoxin/flavodoxin oxidoreductase family protein, Campylobacter coli RM2228; ZP_00368185 |
Size of the ortholog protein. aa, amino acids.
Length of the SX81 peptide.
The annotation of the 14-gene island, including the flanking genes cadF and CCO1582, is shown in Table 3. Among the six genes encoding aminoglycoside-modifying enzymes in this cluster, sat4 and aphA-3 in the aadE–sat4–aphA-3 cluster show 100% nucleotide identity with their homologs in pCG8245 (13), a multidrug-resistant plasmid identified in C. jejuni. The aadE gene in the aadE–sat4–aphA-3 cluster of SX81 is 702 bp, encoding a full-length protein that is 97% identical (amino acids) to the homolog in Enterococcus faecalis. However, the aadE homolog in pCG8245 was truncated (only 516 bp long) and was not functional (13). The aac gene located immediately downstream of cadF in SX81 showed only 47.2% nucleotide identity to the aac gene in pCG8245, although they have the same length (13). The aac gene in SX81 encodes an acetyltransferase enzyme that shows 100% amino acids identity to its homolog in E. faecalis T2. The aacA-aphD gene encodes a protein of 479 amino acids that has 100% identity to the bifunctional aminoglycoside-modifying enzyme AacA-AphD in Staphylococcus aureus. However, the aacA (aphD) gene in SX81 exhibits only 47.7% nucleotide identity to the homolog in pCG8245. Furthermore, the aacA-aphD gene in pCG8245 was truncated and encodes only 281 amino acids (13).
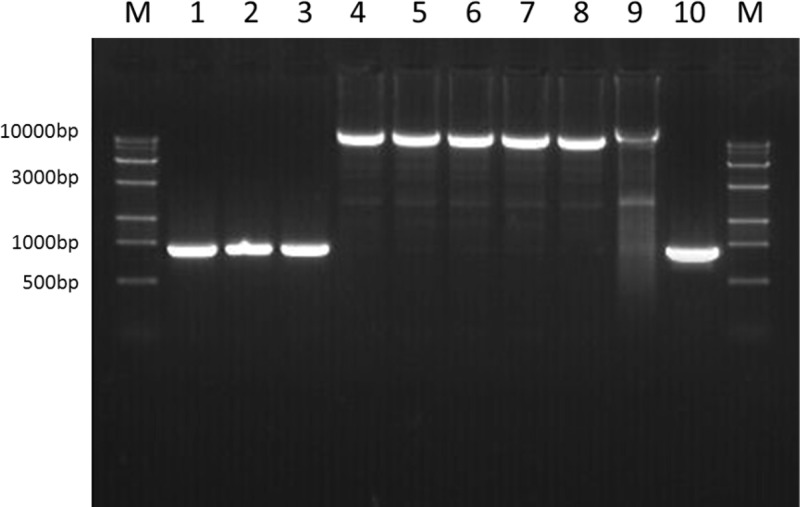
To determine whether this unique genomic island exists in the other two aadE–sat4–aphA-3 cluster-containing C. coli strains (SX114 and MH57), a long-range PCR assay was conducted using a Premix LA Taq version 2.0 kit (TaKaRa, Dalian, China) and primers cadf-F (5′-GCTCAAGCAATGACACTAAAG-3′) and pfo-R (5′-GGAGAACTAGGTGTAATAGGAT-3′), which are located in the conserved regions of the cadF and COO1582 genes, respectively. An amplicon of 10.6 kb was obtained from both SX114 and MH57 (Fig. 4), and sequencing results showed that it was identical to that in SX81 (data not shown). Furthermore, this unique structure was also detected in the other 23 aadE–sat4–aphA-3 cluster-harboring C. coli isolates using the above-mentioned PCR assays (data not shown). In contrast, the examined aminoglycoside susceptible isolates SX71, SX73, and MH16 did not yield this 10.6-kb fragment but showed a 908-bp fragment (Fig. 4), which was determined to be the CCO1583 gene by sequencing. Additionally, PCR testing of the kanamycin-resistant isolates that harbored only aphA-7 or aphA-3 did not identify the 14-gene cluster.
Fig 4.
PCR detection of the region between cadF and COO1582 in various strains. Lanes 1 to 3, PCR products from kanamycin-susceptible isolates SX71, SX73, and MH16, respectively; lanes 4 to 6, PCR products from aminoglycoside-resistant strains SX81, SX114, and MH57, respectively; lanes 7 to 9, PCR products from the C. jejuni transformants of SX81, SX114, and MH57, respectively; lane 10, PCR products from C. jejuni NCTC11168.
Transfer of aminoglycoside resistance via natural transformation.
Campylobacter is naturally transformable by taking up DNA from the environment. As the unique aminoglycoside-resistant genomic island is inserted into the chromosome of C. coli and is not associated with a plasmid or transposable element, we examined if the gene cluster can be transferred to C. jejuni by natural transformation under laboratory conditions. The genomic DNAs of the three C. coli strains (SX81, MH57, and SX114) were used as the donor DNA, and C. jejuni NCTC11168 was used as the recipient strain. With each donor DNA, aminoglycoside resistant transformants were obtained. A 10.6-kb fragment was amplified using the primers cadf-F and pfo-R from the transformants (Fig. 4), and DNA sequencing confirmed that the PCR products contained the unique 14-gene cluster, indicating that this genomic island is transferable to C. jejuni 11168 by natural transformation. The MICs of streptomycin, kanamycin, gentamicin, neomycin, and amikacin in the recipient, donors, and transformants are summarized in Table 4. Compared to the recipient strain, the transformants showed drastically increased MICs of the five tested aminoglycosides, which were comparable to the MICs in the donor strains. This result suggests that once transferred to C. jejuni, the 14-gene cluster confer a high-level resistance to aminoglycoside antibiotics. Additionally, passage of the transformants in antibiotic-free medium indicated that the acquired resistance and the 14-gene cluster remained stable after 14 passages (data not shown).
Table 4.
MICs of aminoglycosides in the recipient and donor strains as well as transformantsa
| Antimicrobial agent | MIC (mg/liter) for: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NCTC11168 | SX81 | NT81 | NT1481 | SX114 | NT114 | NT14114 | MH57 | NT57 | NT1457 | |
| Streptomycin | 8 | >512 | >512 | >512 | >512 | >512 | >512 | >512 | >512 | >512 |
| Kanamycin | 8 | >512 | >512 | >512 | 512 | 512 | 512 | >512 | >512 | >512 |
| Gentamicin | 1 | 128 | 256 | 256 | 256 | 256 | 256 | 256 | 256 | 256 |
| Neomycin | 2 | 32 | 32 | 32 | 64 | 64 | 64 | 32 | 32 | 32 |
| Amikacin | 4 | 32 | 32 | 32 | 32 | 32 | 32 | 64 | 64 | 64 |
NT strains are transformants of C. jejuni 11168 with donor DNA from SX81 (NT81), SX114 (NT114), and MH57 (NT57). NT14 strains are transformants passaged in MH broth in the absence of kanamycin.
DISCUSSION
In the present study, a previously undescribed genomic island containing 14 ORFs and conferring aminoglycoside resistance was identified in C. coli isolates derived from broiler chickens. This island is inserted between cadF and COO1582 on the C. coli chromosome and has a G+C content significantly higher than that of the entire C. coli genome. The conclusion that the aminoglycoside resistance island is located on chromosome is based on the following findings. First, no plasmids were isolated from the examined C. coli isolates. Second, the resistance island was found to be inserted between chromosomally located genes cadF and CCO1582, which are highly conserved on C. coli and C. jejuni genomes and have not been found on any plasmids. Third, the aadE–sat4–aphA-3 cluster-specific probe hybridized to large SmaI-digested genomic fragments in SX81, SX114, and MH57 (∼280 kb, ∼370 kb, and ∼500 kb, respectively) (Fig. 2). Additionally, this resistance island can be readily transferred to C. jejuni via natural transformation, and the genomic location of the transferred resistance genes was further confirmed by long-range PCR (Fig. 4) and sequence analysis. As transfer of large plasmids to C. jejuni by natural transformation is quite difficult, its ease of transfer by natural transformation also suggests a chromosomal location of the resistance island. Together, these data indicate that this aminoglycoside resistance gene cluster represents a genomic island inserted into the chromosome of C. coli.
This genomic island is unique in several respects. First, it contains genes encoding all known aminoglycoside-inactivating enzymes (AACs, ANTs, and APHs) and thus confers resistance to multiple aminoglycoside antibiotics. Second, it is inserted into the chromosome and is not associated with any known mobile elements or carried by a plasmid, suggesting that transmission is likely mediated by homologous recombination. Third, the genomic island was found in C. coli isolates that belong to three clones as defined by PFGE and MLST analysis but was associated with the high prevalence of aminoglycoside-resistant C. coli in chickens in China, suggesting that this aminoglycoside resistance island might have undergone spread by both horizontal gene transfer and clonal expansion in the poultry production system in Shandong province of China.
Previously, the gene cluster aadE–sat4–aphA-3 was detected on plasmids such as pCG8245 in Campylobacter (9, 13). In the genomic island identified in this study, all of the aminoglycoside resistance genes encode constitute ORFs and thus are all presumably functional. In contrast, the aadE gene in the aadE–sat4–aphA-3 cluster and the aacA-aphD gene in pCG8245 were truncated and not functional (13). aacA-aphD is the only identified gene encoding a bifunctional aminoglycoside-modifying enzyme responsible for both gentamicin and kanamycin resistance in Campylobacter (13, 19). Identification of this gene in the genomic island explains why the kanamycin-resistant C. coli isolates were also resistant to gentamicin. To the best of our knowledge, this is the first report of a chromosomal location of the gene aacA-aphD and the gene cluster aadE–sat4–aphA-3 in Campylobacter. In pCG8245, aadE–sat4–aphA-3 was next to a hybrid of two H. pylori transposons, ISHp608 and IS606 (13); however, in the genomic island identified in this study, no transposon-like elements were found between cadF and COOl582. Based on these observations, we speculate that the entire structure of the aminoglycoside resistance genomic island might be derived from a pCG8245-like plasmid that lost its ability to replicate and was subsequently integrated into the chromosome of C. coli by insertion.
Interestingly, all six genes encoding aminoglycoside-modifying enzymes identified in this study appear to have originated from Gram-positive bacteria, including Enterococcus, Staphylococcus, and Pediococcus acidilactici. The aac, aacA-aphD, sat4, and aphA3 genes showed 100% identity to their corresponding homologs, and the two aadE genes also showed more than 97% identity to their Gram-positive homologs (Table 3). This observation is consistent with previous findings by other investigators (6, 21) and further suggests the transfer of antibiotic resistance genes between Gram-positive bacteria and Campylobacter. Additionally, several hypothetical ORFs in the genomic island share high homologies with ORFs in Bacteroides dorei (Table 3), a Gram-negative organism. Together, these findings suggest that the genes in the genomic island originated from divergent sources.
The C. coli isolates harboring the genomic island were from two slaughterhouses located in two geographically separated areas (SX and MH), suggesting the dissemination of C. coli carrying this aminoglycoside resistance island in different regions of Shandong province in China. The antibiotic usage records in the two regions indicated that aminoglycoside agents, such as neomycin, amikacin, and gentamicin, were commonly used for curing disease in broiler chickens. This suggests that the emergence and spread of aminoglycoside resistance in Campylobacter are likely driven by the selection pressure from antibiotic usage. Based on the genotyping results from PFGE and MLST (Table 2 and Fig. 1), the C. coli isolates carrying the antibiotic resistance island can be classified into three different clones. In Campylobacter, PFGE and MLST are often used in combination to determine genetic relationship, and isolates of the same PFGE type and ST are regarded as clonal. Clone 1 isolates are PFGE subtype III and ST 1625, while clones 2 and 3 are PFGE subtype I and ST 5604 and PFGE subtype II and ST 1586, respectively. Clone 1 is predominant among the aminoglycoside-resistant isolates examined in this study, suggesting endemic spread of this clone in the broiler production system in Shandong province of China. Furthermore, all three clones belong to clonal complex 828, which was previously identified in C. coli isolates from both humans and food-producing animals (23, 24).
The aminoglycoside resistance island identified in C. coli in this study is no longer associated with mobile elements, but it might be disseminated via natural transformation mediated homologous recombination. This possibility was demonstrated under laboratory conditions, in which the genes of the genomic island were transferred from C. coli to C. jejuni and drastically increased aminoglycoside resistance in the recipient C. jejuni strain (Table 4). This finding suggests that spread of this aminoglycoside resistance island between C. coli and C. jejuni could occur in nature, as Campylobacter species are naturally transformable. If this occurs, it will pose a concern for public health, as C. jejuni is more frequently associated with human campylobacteriosis than C. coli. Aminoglycoside antimicrobials, such as gentamicin, are used for treating systemic infections in humans caused by Campylobacter (2). Although cases of systemic campylobacteriosis are rare, the clinical consequences can be severe, as the infection may lead to bacteremia, abortion, or even neonatal death (20). Thus, transmission of aminoglycoside-resistant Campylobacter represents a concern for food safety and public health.
In conclusion, this study provides the first report of a C. coli genomic island that harbors genes conferring resistance to multiple aminoglycoside antibiotics. This unique genomic island consists of 14 ORFs and is disseminated among C. coli isolates derived from broiler chickens in China. Horizontal transfer and clonal expansion of the aminoglycoside resistance island were observed, which highlights the need for enhanced efforts to monitor the spread of this genomic island in different Campylobacter species.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (U1031004), Special Fund for Agro-scientific Research in the Public Interest (201203040), Key Projects in the National Science & Technology Pillar Program during the Twelfth Five-year Plan Period (2012BAK01B02), and Specialized Research Fund for the Doctoral Program of Higher Education (SEFDP, 20100008120001).
Footnotes
Published ahead of print 6 August 2012
REFERENCES
- 1. Alfredson DA, Korolik V. 2007. Antibiotic resistance and resistance mechanisms in Campylobacter jejuni and Campylobacter coli. FEMS Microbiol. Lett. 277:123–132 [DOI] [PubMed] [Google Scholar]
- 2. Blaser MJ, Engberg J. 2008. Clinical aspects of Campylobacter jejuni and Campylobacter coli infections, p 99–121 In Nachamkin I, Szymanski CM, Blaser MJ. (ed), Campylobacter, 3rd ed ASM Press, Washington, DC [Google Scholar]
- 3. Bywater R, et al. 2004. A European survey of antimicrobial susceptibility among zoonotic and commensal bacteria isolated from food-producing animals. J. Antimicrob. Chemother. 54:744–754 [DOI] [PubMed] [Google Scholar]
- 4. Chen X, et al. 2010. Prevalence and antimicrobial resistance of Campylobacter isolates in broilers from China. Vet. Microbiol. 144:133–139 [DOI] [PubMed] [Google Scholar]
- 5. Clinical and Laboratory Standards Institute 2008. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals; informational supplement. CLSI document M31–A3. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 6. Courvalin P. 1994. Transfer of antibiotic resistance genes between gram-positive and gram-negative bacteria. Antimicrob. Agents Chemother. 38:1447–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dingle KE, Colles FM, Falush D, Maiden MCJ. 2005. Sequence typing and comparison of population biology of Campylobacter coli and Campylobacter jejuni. J. Clin. Microbiol. 43:340–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Reference deleted.
- 9. Gibreel A, Skold O, Taylor DE. 2004. Characterization of plasmid-mediated aphA-3 kanamycin resistance in Campylobacter jejuni. Microb. Drug Resist. 10:98–105 [DOI] [PubMed] [Google Scholar]
- 10. Kim JS, Carver DK, Kathariou S. 2006. Natural transformation-mediated transfer of erythromycin resistance in Campylobacter coli strains from turkeys and swine. Appl. Environ. Microbiol. 72:1316–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee MD, et al. 2002. Class 1 integron-associated tobramycin-gentamicin resistance in Campylobacter jejuni isolated from the broiler chicken house environment. Antimicrob. Agents Chemother. 46:3660–3664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Luangtongkum T, et al. 2006. Effect of conventional and organic production practices on the prevalence and antimicrobial resistance of Campylobacter spp. in poultry. Appl. Environ. Microbiol. 72:3600–3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nirdnoy W, Mason CJ, Guerry P. 2005. Mosaic structure of a multiple-drug-resistant, conjugative plasmid from Campylobacter jejuni. Antimicrob. Agents Chemother. 49:2454–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ouellette M, Gerbaud G, Lambert T, Courvalin P. 1987. Acquisition by a Campylobacter-like strain of aphA-1, a kanamycin resistance determinant from members of the family Enterobacteriaceae. Antimicrob. Agents Chemother. 31:1021–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pinto-Alphandary H, Mabilat C, Courvalin P. 1990. Emergence of aminoglycoside resistance genes aadA and aadE in the genus Campylobacter. Antimicrob. Agents Chemother. 34:1294–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Qin SS, et al. 2011. Antimicrobial resistance in Campylobacter coli isolated from pigs in two provinces of China. Int. J. Food Microbiol. 146:94–98 [DOI] [PubMed] [Google Scholar]
- 17. Ramirez MS, Tolmasky ME. 2010. Aminoglycoside modifying enzymes. Drug Resist. Updat. 13:151–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ribot EM, Fitzgerald C, Kubota K, Swaminathan B, Barrett TJ. 2001. Rapid pulsed-field gel electrophoresis protocol for subtyping of Campylobacter jejuni. J. Clin. Microbiol. 39:1889–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rouch DA, Byrne ME, Kong YC, Skurray RA. 1987. The aacA-aphD gentamicin and kanamycin resistance determinant of Tn4001 from Staphylococcus aureus: expression and nucleotide sequence analysis. J. Gen. Microbiol. 133:3039–3052 [DOI] [PubMed] [Google Scholar]
- 20. Smith JL. 2002. Campylobacter jejuni infection during pregnancy: long-term consequences of associated bacteremia, Guillain-Barré syndrome, and reactive arthritis. J. Food Prot. 65:696–708 [DOI] [PubMed] [Google Scholar]
- 21. Taylor DE, Courvalin P. 1988. Mechanisms of antibiotic resistance in Campylobacter species. Antimicrob. Agents Chemother. 32:1107–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tenover FC, Elvrum PM. 1988. Detection of two different kanamycin resistance genes in naturally occurring isolates of Campylobacter jejuni and Campylobacter coli. Antimicrob. Agents Chemother. 32:1170–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thakur S, Morrow WEM, Funk JA, Bahnson PB, Gebreyes WA. 2006. Molecular epidemiologic investigation of Campylobacter coli in swine production systems, using multilocus sequence typing. Appl. Environ. Microbiol. 72:5666–5669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thakur S, et al. 2009. Genotyping of Campylobacter coli isolated from humans and retail meats using multilocus sequence typing and pulsed-field gel electrophoresis. J. Appl. Microbiol. 106:1722–1733 [DOI] [PubMed] [Google Scholar]
- 25. Vakulenko SB, Mobashery S. 2003. Versatility of aminoglycosides and prospects for their future. Clin. Microbiol. Rev. 16:430–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vali L, et al. 2007. Antibiotic resistance and molecular epidemiology of Escherichia coli O26, O103 and O145 shed by two cohorts of Scottish beef cattle. J. Antimicrob. Chemother. 59:403–410 [DOI] [PubMed] [Google Scholar]
- 27. Varela NP, Friendship R, Dewey C. 2007. Prevalence of resistance to 11 antimicrobials among Campylobacter coli isolated from pigs on 80 grower-finisher farms in Ontario. Can. J. Vet. Res. 71:189–194 [PMC free article] [PubMed] [Google Scholar]
- 28. Wang Y, Taylor DE. 1990. Natural transformation in Campylobacter species. J. Bacteriol. 172:949–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang K, McClure JA, Elsayed S, Conly JM. 2009. Novel staphylococcal cassette chromosome mec type, tentatively designated type VIII, harboring class A mec and type 4 ccr gene complexes in a Canadian epidemic strain of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 53:531–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang Q, Plummer P. 2008. Mechanisms of antibiotic resistance in Campylobacter, p 263–276 In Nachamkin I, Szymanski CM, Blaser MJ. (ed), Campylobacter, 3rd ed ASM Press, Washington, DC [Google Scholar]