Abstract
Seventy-four unrelated clinical isolates of Streptococcus pneumoniae harboring the tet(M) gene were studied. Seven strains with low tetracycline (Tc) MICs (0.25 to 0.5 μg/ml) were found to harbor truncated tet(M) alleles that were inactivated by different frameshift mutations. In contrast, five strains bore deletions in the tet(M) promoter region, among which four displayed increased Tc MICs (16 to 64 μg/ml). The same promoter mutations were detected in Tc-resistant mutants selected in vitro from various susceptible strains. Sequence analysis revealed that these deletions might impede the formation of the transcriptional attenuator located immediately upstream of tet(M). Expression in Enterococcus faecalis of a tet(M) reporter gene transcribed from these promoter mutants conferred a level of Tc resistance similar to that observed in the parental S. pneumoniae strains. These results show that different levels of Tc susceptibility found in clinical isolates of S. pneumoniae can be explained by frameshift mutations within tet(M) and by alterations of the upstream transcriptional attenuator.
INTRODUCTION
In European countries, tetracycline (Tc) is still used in significant amounts for human medicine, despite the availability of alternative antibiotics (10, 19), and it remains one of the most used antibiotics in veterinary medicine (1). For Streptococcus pneumoniae, resistance to Tc remains an important epidemiological trait (19). The major mechanism of Tc resistance is due to ribosome protection by the Tet(M) protein (3, 23), while protection by Tet(O) is less common (27). Tet(M) and probably Tet(O) promote the release of tetracycline from the 30S ribosomal subunit, in a GTP-dependent manner (7, 8).
The tet(M) genes are located mainly on conjugative transposons belonging to the Tn916-Tn1545 or Tn5253 family, largely accounting for the widespread distribution of these genes (5, 6, 9, 11, 21, 26). The tet(M) promoter region of Tn916 contains two major features (22) that control the transcription of the downstream gene: the −35 and −10 boxes, which define the binding site for RNA polymerase, and a short open reading frame corresponding to a 28-amino-acid peptide and containing an inverted repeat sequence located immediately upstream of tet(M) that acts as a transcriptional attenuator (22). Transcriptional analyses revealed that tet(M) expression results from an extension of a small transcript representing the upstream leader region into the resistance determinant (22). In the presence of a subinhibitory concentration of Tc, a significant increase in the amount of tet(M) transcription could be observed, resulting from increased transcriptional readthrough from the upstream transcript (2, 22). In France, 10% to 20% of S. pneumoniae clinical isolates are Tc resistant (Centre National de Référence des Pneumocoques [CNRP] [http://www.invs.sante.fr/Espace-professionnels/Centres-nationaux-de-reference/Rapports-d-activites-et-liens/Rapports-d-activite/Rapport-CNR-pneumocoque-2010]), and most strains harbor the tet(M) gene (data not shown). Interestingly, different studies made the point that silent tet(M) genes exist in S. pneumoniae and other streptococcal species (25). The aim of this work was to characterize the heterogeneity of tet(M) sequences in S. pneumoniae clinical isolates and to determine the impact of some relevant rearrangements on the level of expression of Tc resistance.
MATERIALS AND METHODS
Strains.
Five hundred invasive S. pneumoniae strains originating from 40 distinct French geographical regions were randomly selected from our CNRP collection and investigated for the presence of the tet(O) and tet(M) genes by PCR. Among these strains, tet(O) was never detected, and the 74 strains harboring tet(M) were studied further.
Susceptibility testing.
Tc MICs were determined on Mueller-Hinton (MH) agar supplemented with 5% horse blood. Spotting of 105 CFU was done with a Steers replicator device on media containing increasing Tc concentrations. MICs were read after 18 h of incubation at 37°C, and susceptibility was interpreted according to CLSI breakpoints (4). Independent measurements were done with a standardized Etest according to the manufacturer's instructions (bioMérieux, France).
Selection of resistant mutants.
In vitro Tc-resistant mutants were selected using an inoculum of 108 to 109 CFU/ml on MH agar supplemented with 5% horse blood and 6 μg/ml of Tc. Resistance was confirmed by MIC determination and Etest after isolation of single colonies.
DNA manipulations.
Chromosomal DNA was extracted as previously described (12). Detection of tet(M), tet(O), and the Tn916-Tn1545 family integrase gene int-Tn by PCR was performed using specific primers as previously described (16). All PCR amplifications were performed with the high-fidelity Pfu DNA polymerase (Promega, France) according to the manufacturer's instructions. PCR products [tet(M) promoter regions and full-length genes] were sequenced using a BigDye Terminator kit and an automated model 377 DNA sequencer (Applied Biosystems). Sequence analysis was performed using Bioedit.
The tet(M) promoter regions [from position −1 downstream of the tet(M) start codon to position −419] were amplified with a forward primer containing an EcoRI site (5′-GGGGGGAATTCGGTACTTGAAAAGAACGGGAG-3′) and a reverse primer containing a BamHI site (5′-CCCAAGGATCCCCATGTGATTTTCCTCCATTCA-3′). Using the same primers, selected S. pneumoniae tet(M) promoters were cloned between the corresponding sites of pTCV-lac (17) (Fig. 1). Recombinant vectors were introduced into Enterococcus faecalis BM4110 by electroporation (24), and the β-galactosidase activities were assayed as previously described (17). Electroporants were selected on erythromycin (5 μg/ml).
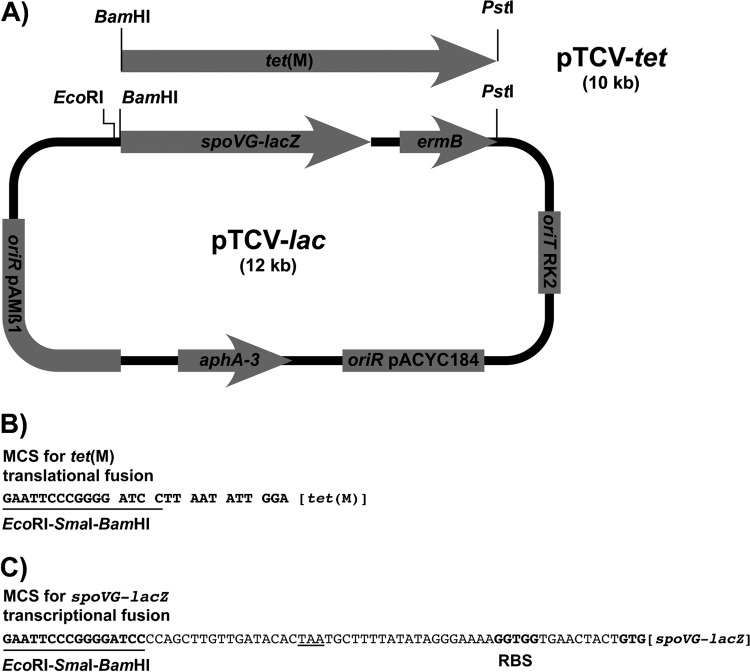
Fig 1.
Schematic representations of plasmids pTCV-lac and pTCV-tet. (A) The components of the parental vector pTCV-lac are (i) the spoVG-lacZ translational gene fusion; (ii) the genes aphA3 and ermB, conferring resistance to kanamycin and erythromycin, respectively; (iii) the replication origins (oriR) of plasmids pACYC184 and pAMβ1; and (iv) the transfer origin (oriT) of the IncP plasmid RK2. Plasmid pTCV-tet was constructed by replacing the BamHI-PstI spoVG-lacZ-ermB fragment of pTCV-lac with a BamHI-PstI fragment containing the tet(M) gene of Tn916 devoid of its translational signals, including the first codon. The plasmid components are not drawn to scale. (B) Expression of the tet(M) reporter gene of pTCV-tet was obtained following cloning of EcoRI-BamHI fragments containing various tet(M) promoter regions to provide appropriate transcriptional and translational signals. MCS, multiple cloning site. (C) Expression of the spoVG-lacZ reporter gene of pTCV-lac was obtained following cloning of the same EcoRI-BamHI fragments as those for panel B to provide appropriate transcriptional signals. Note that in all the spoVG-lacZ fusions analyzed in this study, tet(M) translation ends prematurely at the underlined TAA stop codon and hence should not interfere with that of the downstream reporter gene.
The vector pTCV-tet was constructed (Fig. 1) to test the contributions of different tet(M) promoter regions to the level of expression of Tc resistance. To this end, a 2,049-bp DNA fragment containing a promoterless functional pneumococcal tet(M) gene was amplified by PCR using a forward primer containing a BamHI site (underlined) (5′-GGGGGGGATCCTTAATATTGGAGTTTTAGCTCAT-3′) and a reverse primer containing a PstI site (underlined) (5′-GGGGGCTGCAGCCACTTGTAGTTTATAATAACTATCTC-3′). The corresponding BamHI-PstI PCR product was cloned in place of lacZ-ermB into pTCV-lac (17) to give pTCV-tet (Fig. 1). Selected S. pneumoniae tet(M) promoters were cloned into the EcoRI and BamHI sites of pTCV-tet, i.e., upstream of the tet(M) reporter gene, and the recombinant vectors were introduced into E. faecalis BM4110 by electroporation (17). Electroporants were selected on kanamycin (1,000 μg/ml).
RNA manipulations and real-time quantitative PCR (qRT-PCR) experiments.
RNA extractions were performed on exponentially growing S. pneumoniae cells (optical density at 650 nm [OD650] = 0.5) grown in brain heart infusion (BHI) medium at 37°C from a freshly isolated colony inoculated on Columbia agar supplemented with 5% horse blood. RNA was extracted using TRIzol Plus RNA purification reagent (Life Technologies, Grand Island, NY) as recommended by the manufacturer. Briefly, bacterial pellets were washed and resuspended in 25 mM Tris-10 mM EDTA solution supplemented with 20% glucose. RNA isolation was based on mechanical disruption with a FastPrep apparatus (Anachem, United Kingdom) followed by isolation of nucleic acids by use of TRIzol Plus and chloroform-isoamyl alcohol solutions. Nucleic acid precipitation was done with isopropanol, and residual DNA was removed using a Turbo DNA-free kit (Life Technologies, Grand Island, NY). Reverse transcription was done on 1 μg total RNA with an iScript cDNA synthesis kit (Bio-Rad, France), in parallel with a negative-control experiment where the reverse transcriptase enzyme was omitted to confirm the absence of residual DNA contamination.
qRT-PCR was performed using an SsoFast Evagreen Supermix kit in a MyiQ device (Bio-Rad) with tet(M)-specific primers (5′-ATGCTTGCTCCTATTGTATTGG and 5′-TGTATGCTCGTGAAAGATATTCC) and gyrA-specific primers (5′-CAATATGCTCGCTATCCAA and 5′-GACGAACAACCACTTCTT). Expression of tet(M) in the different S. pneumoniae strains was calculated by the ΔΔCT method, using the expression of the gyrA housekeeping gene as a reference. Each RNA quantification was done with biological duplicates (independent cultures and RNA extractions) in four technical replicates (qRT-PCRs).
RESULTS AND DISCUSSION
Sequence heterogeneity of the tet(M) gene and upstream promoter region in pneumococcal isolates expressing different levels of resistance to Tc.
Seventy-four unrelated S. pneumoniae clinical isolates collected by the CNRP during 2003 to 2009 and harboring tet(M) but exhibiting a wide range of susceptibilities to Tc (MICs of 0.25 to 64 μg/ml) were analyzed. Eighteen strains were Tc susceptible (MICs of ≤2 μg/ml), 5 were Tc intermediate (MIC = 4 μg/ml), and 51 were Tc resistant (MICs of ≥8 μg/ml). The presence of the integrase gene int-Tn in all isolates suggested that the tet(M) genes were carried by Tn916-related elements.
Pneumococcal strains with MICs of ≤1 μg/ml and harboring silent tet(M) genes were mentioned in previous studies (5, 25), but their tet(M) sequences were not characterized. We observed that among the 18 Tc-susceptible strains, the 7 displaying the lowest Tc MICs (0.25 to 0.5 μg/ml) harbored frameshifts within tet(M), resulting in a 10-bp deletion at positions 620 to 630 (n = 4), a 2-bp deletion at positions 1014 and 1015 (n = 2), or a T insertion at position 379 (n = 1). To characterize the sequence heterogeneity of this resistance determinant in our panel of strains, 30 randomly chosen strains, including 5 Tc-susceptible strains (MICs of 1 to 2 μg/ml), 2 strains with intermediate susceptibility (MIC of 4 μg/ml), and 23 resistant strains (MICs of 8 to 64 μg/ml), were similarly sequenced. No frameshift in tet(M) was observed for the five Tc-susceptible strains (data not shown). The Tet(M) amino acid sequences of 11 strains were identical to that encoded by Tn916 or Tn1545 (15), whereas those of the remaining 19 strains differed by only one of the following amino acid substitutions: K38R, V70M, V70I, Q108R, N553T, H556K, T573I, P613L, and V624A. However, the Tc MICs of all Tet(M) variants except for strain 52 (MIC of 1 μg/ml; see below) were ≥8 μg/ml, suggesting that none of these mutations could be associated with low-level Tc MICs.
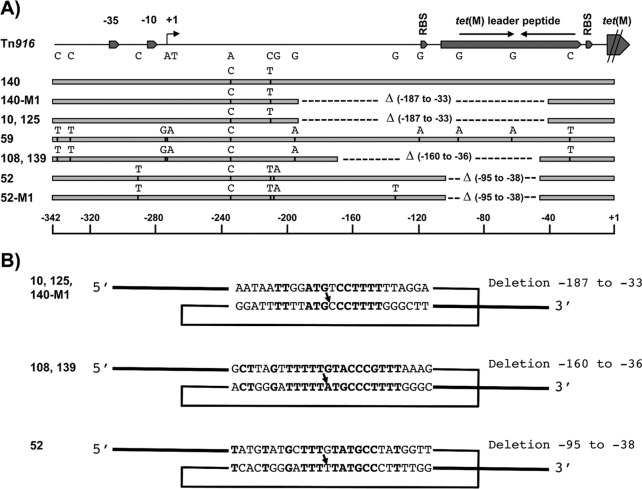
Since expression of tet(M) is inducible and regulated by transcriptional attenuation (2, 22), we used PCR analysis of the length polymorphism of the tet(M) promoter region in the 74 pneumococcal strains displaying variable Tc MICs to investigate if insertions or deletions could be detected. Four strains (10, 108, 125, and 139) (Fig. 2A) with Tc MICs ranging from 16 to 64 μg/ml, as well as strain 52, with an MIC of 1 μg/ml, were found to harbor shorter promoter regions than those in Tn916. Sequence analysis of these promoter regions [nucleotides (nt) −339 to +1; numbering throughout the text starts from the first base of the tet(M) initiation codon] revealed that strains 10, 108, 125, and 139 displayed a large deletion upstream of tet(M) that drastically altered the stem-loop forming the transcriptional attenuator (Fig. 2A; Table 1) (22). The deleted segment extended from positions −187 to −33 in strains 10 and 125 and from positions −160 to −36 in strains 108 and 139. Strain 52 contained a smaller deletion, extending from positions −95 to −38 (Fig. 2A), that is analyzed in detail below. We also sequenced the tet(M) promoter regions of two strains with a Tc MIC of 2 μg/ml (strains 140 and 59) whose PCR products displayed the expected size. Point mutations were detected in these promoter regions compared to those in Tn916, but none could be associated with any obvious functional alterations that could account for the observed low-level resistance.
Fig 2.
tet(M) promoter regions in pneumococcal isolates displaying different levels of resistance to tetracycline. (A) Schematic comparison of the 339-bp sequence upstream of the first base of the tet(M) initiation codon of Tn916 from E. faecalis DS16 (GenBank accession no. U09422) with those of the corresponding regions in WT isolates 140, 10, 59, 108, 139, and 52 and in the relevant mutants 140-M1 and 52-M1. The positions of the transcriptional signals (−35 and −10 boxes and +1 start site), the leader peptide (containing an inverted repeat acting as a transcriptional terminator), and the tet(M) translational signals are depicted in the Tn916 sequence. The positions of the mutations and the extents of the deletions (△) observed in pneumococcal sequences are indicated by small vertical bars and horizontal dashed lines, respectively. (B) Proposed homologous recombination events accounting for the deletions observed in strains 10, 125, and 140-M1 (positions −187 to −33), strains 108 and 139 (positions −160 to −36), and strains 52 and 52-M1 (positions −95 to −38). The short repeated recombining sequences containing mismatches with Tn916 sequences are boxed, and the crossover points are indicated by arrows. The resulting deletion sequences are shown with bold letters. Numbering of the deletions in panels A and B was determined relative to the first base of the tet(M) translation codon in the Tn916 sequence.
Table 1.
Analysis of tet(M) expression as assayed by determination of MICs, qRT-PCR analysis, and β-galactosidase activity assay
| S. pneumoniae isolate | MIC (μg/ml) | qRT-PCR resulta |
E. faecalis transformant datab |
|||
|---|---|---|---|---|---|---|
| MIC (μg/ml) | β-Gal activity (Miller units)c |
|||||
| Noninduced cells | Induced cells | Noninduced cells | Induced cells | |||
| 140 | 2 | 18 | 114 | 2 | 15 | 168 |
| 140-M1 | 64 | 650 | 551 | 64 | 669 | 1,188 |
| 10 | 64 | 308 | 470 | 64 | 627 | 1,197 |
| 59 | 2 | 7 | 41 | 1.5 | 14 | 76 |
| 108 | 16 | 302 | 379 | 24 | 288 | 420 |
| 52 | 1 | 776 | 880 | 1 | 252 | 213 |
| 52-M1 | 32 | 890 | 904 | 12 | — | — |
Relative levels of tet(M) transcripts were measured by qRT-PCR analysis of RNAs extracted from exponentially growing S. pneumoniae cells cultivated without or with a subinhibitory concentration of Tc (0.2 μg/ml). Expression levels were normalized by using gyrA as an internal standard and are indicated as threshold cycle ratios. Values are means for four technical replicates and are representative of two independent experiments showing less than 10% variation.
E. faecalis BM4110 was transformed with the reporter plasmid pTCV-tet(M) or with pTCV-lac containing the tet(M) promoter regions of the corresponding pneumococcal strains. —, not tested.
β-Galactosidase was assayed on exponentially growing E. faecalis cells cultivated without or with a subinhibitory concentration of Tc (0.2 μg/ml). Values are representative of two independent experiments showing less than 10% variation.
Convergent in vitro and in vivo selection of tet(M) mutants displaying increased levels of resistance to Tc.
It is worth noting that the deletions characterized in clinical isolates 10 and 108 might have been generated in vivo through homologous recombination between short repeated sequences containing mismatches (Fig. 2B), a recombination mechanism already described for S. pneumoniae (18). To demonstrate that these recombination events associated with increased Tc resistance could occur in vitro under laboratory conditions, Tc-susceptible S. pneumoniae strains 59 and 140 (MIC of 2 μg/ml), which possessed a tet(M) promoter region almost identical to that in Tn916, were plated on agar containing 6 μg/ml of Tc. Tc-resistant mutants were selected at a frequency of about 10−7, and all mutants selected from these two strains exhibited either the previous large deletion extending from positions −187 to −33 seen in strains 10 and 125 (MICs of 32 to 64 μg/ml) or that from positions −160 to −36 seen in strains 108 and 139 (MICs of 16 to 32 μg/ml) (Fig. 2A; Table 1).
Higher Tc resistance levels in WT pneumococcal isolates and mutant strains are due to increased tet(M) transcription.
The level of tet(M) transcription was analyzed by qRT-PCR on total RNAs extracted from wild-type (WT) clinical isolates 10, 59, 108, and 140 and the mutant derivative 140-M1, cultivated without and with a subinhibitory concentration of Tc. As expected, the results obtained revealed that tet(M) in strains 140 and 59 (Tc MIC of 2 μg/ml), which possessed Tn916-like promoter regions, was transcribed at a relatively low level that was induced 6-fold in the presence of Tc. In contrast, in WT strains 10 and 108 and the mutant strain 140-M1 (Tc MICs of 16 to 64 μg/ml), which possessed a truncated promoter region, tet(M) transcription was intrinsically higher (>15-fold) and not significantly upregulated in the presence of Tc.
These promoter regions were cloned into the low-copy-number expression vector pTCV-tet (Fig. 1), which enabled tet(M) expression from the cloned transcriptional and translational signals, and the functional impact of these deletions on Tc resistance was further characterized in a single heterologous host, E. faecalis BM4110 (Table 1). The Tc MIC of the recipient strain was 0.5 μg/ml and increased to 2 μg/ml when the Tn916-like tet(M) regulatory regions of S. pneumoniae 140 and 59 were cloned. Introduction of the regulatory region with the large deletion (nt −187 to −33) from strains 10 and 140-M1 resulted in a 32-fold increase in the Tc MIC compared to that of strain 140 (Table 1). Similarly, the deletion extending from positions −160 to −36 (strain 108) was associated with a 16-fold increase in the Tc MIC compared to that of strains 59 and 140. As shown in Table 1, the Tc MICs determined for E. faecalis harboring the tet(M) fusions perfectly reflected those of the parental pneumococcal strains.
The promoter regions were also cloned into the low-copy-number expression vector pTCV-lac, which enabled spoVG-lacZ expression from the cloned transcriptional signals. The corresponding β-galactosidase activities reflected the promoter strengths of the cloned fragments in E. faecalis cells cultivated under uninduced or induced conditions. As shown in Table 1, the promoters originating from Tc-susceptible pneumococcal isolates 140 and 59, possessing a functional transcription terminator, were associated with a low level of β-galactosidase activity under uninduced conditions that was increased 5- to 10-fold in the presence of Tc. As expected, the mutated promoters from strains 10, 108, and 140-M1, which were resistant to Tc (MICs of 24 to 64 μg/ml) and devoid of a transcription terminator, were 20- to 40-fold more active under uninduced conditions and were not upregulated by Tc (Table 1).
Evidence for translational control of tet(M) expression in S. pneumoniae strain 52.
Surprisingly, strain 52, with an MIC of 1 μg/ml, contained a smaller deletion extending from positions −95 to −38 that also removed the transcriptional terminator in its entirety (Fig. 2A; Table 1). Accordingly, qRT-PCR analysis revealed that the level of tet(M) mRNA was similar to those observed in strains 10 and 108, which were resistant to Tc, and did not increase in cells cultivated with Tc. These results were confirmed by expression analysis experiments carried out in E. faecalis which showed that the corresponding promoter region did not confer resistance to Tc (1 μg/ml), although it directed a level of β-galactosidase synthesis that was >15-fold higher than those obtained with the low-level resistant strains 140 and 59. We therefore concluded that the low level of Tc resistance in strain 52 was not due to a low level of transcription of tet(M), and we carried out a mutational analysis to decipher the molecular basis of this intriguing expression phenotype. A resistant S. pneumoniae strain 52 mutant (MIC of 32 μg/ml) was selected at a frequency of 10−7 on agar plates containing 6 μg/ml of Tc. This resistant mutant, 52-M1, possessed a G130T substitution, i.e., a mutation in a region never reported to be involved in the control of tet(M) expression (Fig. 2A; Table 1). We confirmed by qRT-PCR analysis that tet(M) expression was similar in strains 52 and 52-M1 and that the corresponding mutated promoter also conferred high-level resistance to Tc on E. faecalis.
To understand why strain 52, with a high level of constitutive tet(M) transcription, displayed susceptibility to Tc, we analyzed the secondary structure of tet(M) mRNA from positions −145 to +1. A putative stem-loop structure having a free energy of formation of −15.6 kcal/mol (28) and pairing with a 10-nt segment which is located immediately downstream of the tet(M) ribosome binding site (RBS) and includes the first four coding bases (positions −136 to −127) was predicted (Fig. 3). This suggests a control of tet(M) expression by translational attenuation. Consistent with this suggestion, the mutation in the 52-M1 promoter region associated with high-level Tc resistance is located in the middle of this segment (Fig. 3) and should impair the formation of the stem-loop (ΔG = −6.6 kcal/mol), thus enabling more efficient tet(M) translation with an increased MIC (Table 1). To substantiate this hypothesis, we independently selected three additional mutants of strain 52 that were highly resistant to Tc (16 to 32 μg/ml) and sequenced the corresponding promoter regions. This analysis revealed the presence of single point mutations located in the same 10-bp segment—G132T and G132C transversions and a G130A transition—which all similarly affect its pairing (Fig. 3). Based on these results, we conclude that this hairpin structure acts as a translational attenuator that inhibits tet(M) expression in isolate 52 (13, 14, 20). We postulate that the formation of this translational attenuator is impeded by that of the large transcriptional attenuator in fully functional Tn916-like tet(M) promoters (strains 140 and 59) and by the large deletions that remove the 5′ arm of the 10-bp stem in mutants 10, 125, and 140-M1 (from positions −187 to −33) or strains 108 and 139 (from positions −160 to −36).
Fig 3.
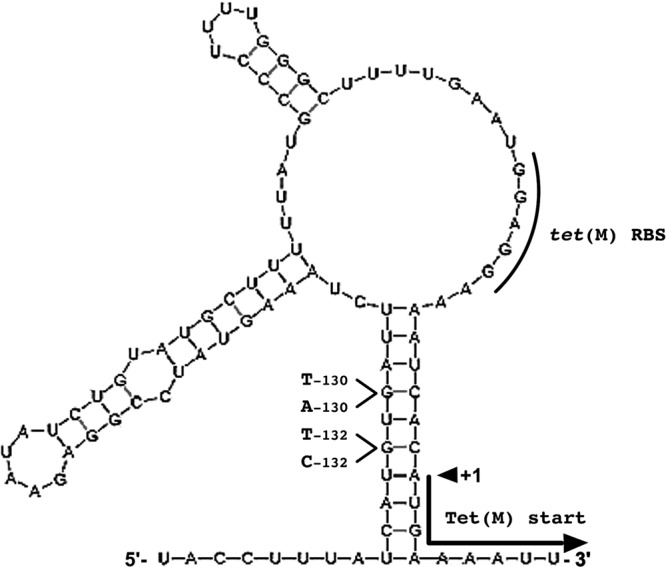
Secondary structure of tet(M) mRNA from strain 52. Compared to that in Tn916, the tet(M) promoter region of this strain exhibits a deletion extending from positions −95 to −38 (see Fig. 2A). The mRNA structure [positions −145 to +1; numbering was done relative to the first base of the tet(M) translation codon in the Tn916 sequence] was computed using the Mfold program (28). The tet(M) RBS and +1 translational start site are indicated. The mutations associated with increased levels of resistance to tetracycline (G130T in mutant 52-M1, G132T and G132C transversions, and G130A transition) and affecting the pairing of the stem that sequesters the tet(M) translational start site are shown. The free energy of interaction (ΔG) of the most stable structure of the tet(M) mRNA of strain 52 was −28.5 kcal/mol (−15.6 kcal/mol for the corresponding 10-nt stem) and increased to −23.5 kcal/mol (−6.6 kcal/mol for the corresponding 10-nt stem) in the resistant mutants.
In conclusion, we showed in this study that pneumococcal isolates containing tet(M) sequences but remaining susceptible to Tc possess pseudogenes resulting from frameshift mutations and encoding inactive truncated Tet(M) proteins. On the other hand, we also showed that pneumococcal strains highly resistant to Tc displayed rearrangements in the tet(M) promoter region that removed the transcriptional terminator controlling tet(M) expression and could readily be obtained in vitro under laboratory conditions. Importantly, we provide evidence that high-level tet(M) transcription could be associated with a low level of Tc resistance due to a deletion in the promoter region that favors the formation of a new stem-loop interfering with tet(M) translation. However, it is worth mentioning that some Tc-susceptible strains (e.g., 140 and 59) possess an apparently fully functional tet(M) transcriptional unit, which suggests that still-uncharacterized factors control the phenotypic expression of Tc resistance in S. pneumoniae isolates.
ACKNOWLEDGMENTS
This study was supported by a subvention of the Institut de Veille Sanitaire to the CNRP and by an INSERM grant.
Footnotes
Published ahead of print 16 July 2012
REFERENCES
- 1. Cars O, Mölstad S, Melander A. 2001. Variation in antibiotic use in the European Union. Lancet 357:1851–1853 [DOI] [PubMed] [Google Scholar]
- 2. Celli J, Trieu-Cuot P. 1998. Circularization of Tn916 is required for expression of the transposon-encoded transfer functions: characterization of long tetracycline-inducible transcripts reading through the attachment site. Mol. Microbiol. 28:103–117 [DOI] [PubMed] [Google Scholar]
- 3. Chopra I, Roberts M. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Clinical and Laboratory Standards Institute 2009. Performance standards for antimicrobial susceptibility testing. Approved standard M100-S19. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 5. Cochetti I, et al. 2007. New Tn916-related elements causing erm(B)-mediated erythromycin resistance in tetracycline-susceptible pneumococci. J. Antimicrob. Chemother. 60:127–131 [DOI] [PubMed] [Google Scholar]
- 6. Cochetti I, Tili E, Mingoia M, Varaldo PE, Montanari MP. 2008. erm(B)-carrying elements in tetracycline-resistant pneumococci and correspondence between Tn1545 and Tn6003. Antimicrob. Agents Chemother. 52:1285–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Connell SR, et al. 2002. The tetracycline resistance protein Tet(O) perturbs the conformation of the ribosomal decoding centre. Mol. Microbiol. 45:1463–1472 [DOI] [PubMed] [Google Scholar]
- 8. Connell SR, et al. 2003. Mechanism of Tet(O)-mediated tetracycline resistance. EMBO J. 22:945–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Del Grosso M, Scotto d'Abusco A, Iannelli F, Pozzi G, Pantosti A. 2004. Tn2009, a Tn916-like element containing mef(E) in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 48:2037–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Goossens H, et al. 2005. Outpatient antibiotic use in Europe and association with resistance: a cross-national database study. Lancet 365:579–587 [DOI] [PubMed] [Google Scholar]
- 11. Izdebski R, et al. 2007. Clonal diversity and resistance mechanisms in tetracycline-nonsusceptible Streptococcus pneumoniae isolates in Poland. Antimicrob. Agents Chemother. 51:1155–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Janoir C, Varon E, Kitzis MD, Gutmann L. 2001. New mutation in parE in a pneumococcal in vitro mutant resistant to fluoroquinolones. Antimicrob. Agents Chemother. 45:952–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kozak M. 2005. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene 361:13–37 [DOI] [PubMed] [Google Scholar]
- 14. Kudla G, Murray AW, Tollervey D, Plotkin JB. 2009. Coding sequence determinants of gene expression in Escherichia coli. Science 324:255–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Oggioni MR, Dowson CG, Smith JM, Provvedi R, Pozzi G. 1996. The tetracycline resistance gene tet(M) exhibits mosaic structure. Plasmid 35:156–163 [DOI] [PubMed] [Google Scholar]
- 16. Poyart C, Jardy L, Quesne G, Berche P, Trieu-Cuot P. 2003. Genetic basis of antibiotic resistance in Streptococcus agalactiae strains isolated in a French hospital. Antimicrob. Agents Chemother. 47:794–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Poyart C, Trieu-Cuot P. 1997. A broad-host-range mobilizable shuttle vector for construction of transcriptional fusions to beta-galactosidase in gram-positive bacteria. FEMS Microbiol. Lett. 156:193–198 [DOI] [PubMed] [Google Scholar]
- 18. Prudhomme M, Libante V, Claverys JP. 2002. Homologous recombination at the border: insertion-deletions and the trapping of foreign DNA in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. U. S. A. 99:2100–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Riedel S, et al. 2007. Antimicrobial use in Europe and antimicrobial resistance in Streptococcus pneumoniae. Eur. J. Clin. Microbiol. Infect. Dis. 26:485–490 [DOI] [PubMed] [Google Scholar]
- 20. Seo SW, Yang J, Jung GY. 2009. Quantitative correlation between mRNA secondary structure around the region downstream of the initiation codon and translational efficiency in Escherichia coli. Biotechnol. Bioeng. 104:611–616 [DOI] [PubMed] [Google Scholar]
- 21. Stapleton P, et al. 2004. Characterisation of viridans group streptococci with different levels of Tet(M)-mediated tetracycline resistance. Int. J. Antimicrob. Agents 24:439–443 [DOI] [PubMed] [Google Scholar]
- 22. Su YA, He P, Clewell DB. 1992. Characterization of the tet(M) determinant of Tn916: evidence for regulation by transcription attenuation. Antimicrob. Agents Chemother. 36:769–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Taylor DE, Chau A. 1996. Tetracycline resistance mediated by ribosomal protection. Antimicrob. Agents Chemother. 40:1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Trieu-Cuot P, Carlier C, Courvalin P. 1988. Conjugative plasmid transfer from Enterococcus faecalis to Escherichia coli. J. Bacteriol. 170:4388–4391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Varaldo PE, Montanari MP, Giovanetti E. 2009. Genetic elements responsible for erythromycin resistance in streptococci. Antimicrob. Agents Chemother. 53:343–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Villedieu A, et al. 2003. Prevalence of tetracycline resistance genes in oral bacteria. Antimicrob. Agents Chemother. 47:878–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Widdowson CA, Klugman KP, Hanslo D. 1996. Identification of the tetracycline resistance gene, tet(O), in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 40:2891–2893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]