Abstract
Asunaprevir (ASV; BMS-650032) is a hepatitis C virus (HCV) NS3 protease inhibitor that has demonstrated efficacy in patients chronically infected with HCV genotype 1 when combined with alfa interferon and/or the NS5A replication complex inhibitor daclatasvir. ASV competitively binds to the NS3/4A protease complex, with Ki values of 0.4 and 0.24 nM against recombinant enzymes representing genotypes 1a (H77) and 1b (J4L6S), respectively. Selectivity was demonstrated by the absence of any significant activity against the closely related GB virus-B NS3 protease and a panel of human serine or cysteine proteases. In cell culture, ASV inhibited replication of HCV replicons representing genotypes 1 and 4, with 50% effective concentrations (EC50s) ranging from 1 to 4 nM, and had weaker activity against genotypes 2 and 3 (EC50, 67 to 1,162 nM). Selectivity was again demonstrated by the absence of activity (EC50, >12 μM) against a panel of other RNA viruses. ASV exhibited additive or synergistic activity in combination studies with alfa interferon, ribavirin, and/or inhibitors specifically targeting NS5A or NS5B. Plasma and tissue exposures in vivo in several animal species indicated that ASV displayed a hepatotropic disposition (liver-to-plasma ratios ranging from 40- to 359-fold across species). Twenty-four hours postdose, liver exposures across all species tested were ≥110-fold above the inhibitor EC50s observed with HCV genotype-1 replicons. Based on these virologic and exposure properties, ASV holds promise for future utility in a combination with other anti-HCV agents in the treatment of HCV-infected patients.
INTRODUCTION
Hepatitis C virus (HCV) infects an estimated 170 million individuals worldwide (52). Primary infection is asymptomatic; however, most HCV infections progress to a chronic state that can gradually evolve into cirrhosis, liver failure, and/or hepatocellular carcinoma. In the United States, these clinical manifestations are the leading indication for liver transplantation (20).
Seven major genotypes and several subtypes exist based on the sequence heterogeneity of the HCV genome; their distribution, transmission, and disease progression differ substantially (25). While the distribution of genotypes 1 and 2 is global, genotype 3 predominates in Southeast Asia, genotype 4 in Africa and Egypt, genotype 5 in South Africa, and genotype 6 primarily in Hong Kong and Vietnam. In the United States, genotype 1 accounts for more than 70% of all HCV infections (24). Although genotype does not predict the outcome of infection, different genotypes are associated with variable responses to interferon-based treatment options (17, 20, 29). Recently, the direct-acting antivirals (DAAs) boceprevir and telaprevir were approved for use in combination regimens with pegylated alfa interferon and ribavirin, resulting in enhanced sustained virologic response (SVR) rates (66 to 75%) in genotype 1 treatment-naïve patients (22, 39). Although these regimens are more efficacious in patients infected with genotype 1, complicated three-times-daily dosing schedules and significant side effects, such as anemia, add further limitations to the well-documented tolerability issues associated with interferon-based regimens and can lead to higher discontinuation rates or dose reductions (20, 32, 50). This highlights the need for more effective, better-tolerated therapeutic agents for HCV infection.
HCV is a member of the genus Hepacivirus in the family Flaviviridae (43), which comprises small, enveloped, single-stranded, positive-sense RNA viruses (33). The 9.6-kb HCV RNA genome consists of one large open reading frame flanked on the 5′ and 3′ ends by untranslated regions important for efficient replication and translation of the polyprotein. The HCV polyprotein is processed into proteins that are essential for the virus replication cycle (2). The bifunctional NS3 protein consists of an N-terminal catalytic subunit that, when complexed with its activating cofactor NS4A, is required for cleavage of the polyprotein and subsequent viral replication (3, 41). Inhibition of proteolytic cleavage has been an effective approach for anti-HCV therapy since its identification (2). Efforts were aided early on by the availability of a crystal structure of the enzyme's NS3/4A protease active site (1). Using the crystal structure in conjunction with computer-aided structure design, peptido-mimetic compounds were designed to occupy the enzyme active site, resulting in highly active, selective inhibitors of the HCV NS3/4A serine protease complex (7, 9, 27).
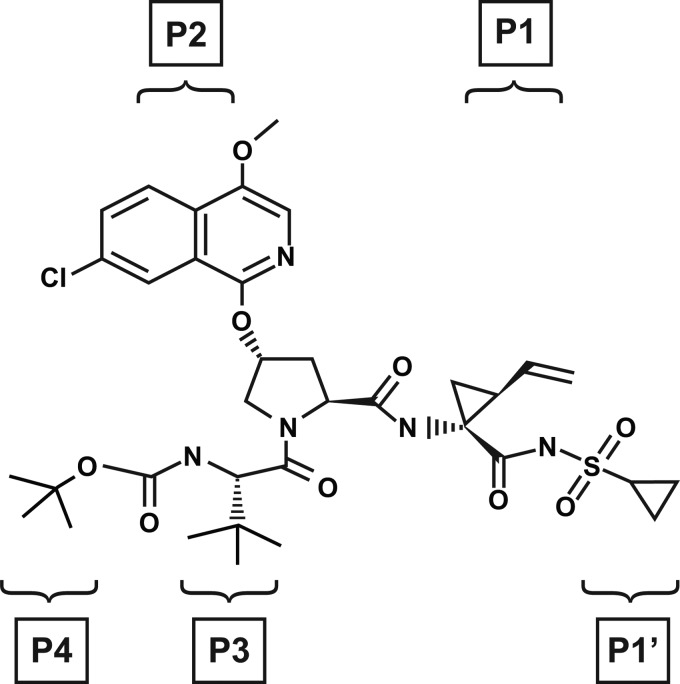
A structure-based drug discovery approach used computer-generated models of the tripeptide core of BILN-2061 (ciluprevir) (26) bound to the HCV NS3/4A protease complex (31). When the carboxylate moiety of a ciluprevir derivative was replaced with a proprietary cyclopropyl acylsulfonamide moiety, a significant enhancement in both intrinsic and cell activity was observed compared with the equivalent carboxylate derivative (31). Lead optimization subsequently led to the identification of the selective NS3 protease inhibitor (PI) asunaprevir (ASV; BMS-650032) (Fig. 1) (45). Here we report the preclinical profile that supported the selection of ASV for subsequent clinical development.
Fig 1.
Chemical structure of ASV (BMS-650032): N-(tert-butoxycarbonyl)-3-methyl-l-valyl-(4R)-N-((1R,2S)-1{[(cyclopropylsulfonyl)amino]carbonyl}-2-vinylcyclopropyl) −4-[(4-methoxy-7-chloroisoquinolin-1-yl)oxy]-l-prolinamide (45). P and P′ labeling indicates the ASV moieties that correspond to the natural HCV substrate positions occupying the NS3 protease active site.
MATERIALS AND METHODS
Compounds, cells, and viruses.
ASV (N-(tert-butoxycarbonyl)-3-methyl-l-valyl-(4R)-N-((1R,2S)-1{[(cyclopropylsulfonyl)amino]carbonyl}-2-vinylcyclopropyl)-4-[(4-methoxy-7-chloroisoquinolin-1-yl)oxy]-l-prolinamide), daclatasvir, and the HCV NS5B polymerase inhibitor BMS-791325 were synthesized by Bristol-Myers Squibb (BMS) (19, 23, 45). The NS3 PI telaprevir (VX-950 is used to denote the compound synthesized at BMS), the pyrimidine analog 2′-C-methylcytidine, the non-nucleoside inhibitor HCV-796, and NM-107, the active component of the prodrug valopicitabine (12), were synthesized at BMS (6). Recombinant human alpha interferon 2b (rIFN-α; Intron A) was from Myoderm Medical Supply (Norristown, PA). Ribavirin was from Sigma-Aldrich Co. (St. Louis, MO). HCV serum samples of strains (H77c, J4L6S, HC-J6, HC-J8, SA52, ED43, SA13, and HK-6A) representing six major HCV genotypes and GB virus-B (GBV-B) were a gift from Jens Bukh. Cell lines included the human hepatoma cells HuH-7 (Ralf Bartenschlager, University of Heidelberg, Germany), HuH-7.5 (APATH, Brooklyn, NY), MT2 (NIH AIDS Research and Reference Reagent Program), HepG2, HeLa, HEK293, and MRC5 (American Type Culture Collection [ATCC]). The cell lines HuH-7, HeLa, HEK293, and MRC5 were propagated in Dulbecco's modified Eagle's medium (DMEM) containing 10% heat-inactivated fetal bovine serum (FBS) and 2 mM l-glutamine. Huh-7 cells that stably maintain HCV replicons were propagated as subconfluent monolayers in DMEM containing 10% FBS, 2 mM l-glutamine, and 0.5 mg/ml Geneticin (G418; Invitrogen Corp., Carlsbad, CA) (13, 45). MT-2 and HepG2 cells were propagated in RPMI 1640 containing 10% heat-inactivated FBS and 2 mM l-glutamine. Human rhinovirus serotype 2 (HRV2) and human coronavirus strain OC43 (HCoV OC43) were from ATCC. HRV2 was propagated in HeLa cells; HCoV OC43 was propagated in MRC5 cells. The proviral DNA clone of HIV NL4-3 was obtained from the NIH AIDS Research and Reference Reagent Program.
HCV NS3/4A protease genotype assays.
Published methods for the construction, expression, and purification of recombinant full-length NS3/4A complexes representing HCV genotypes 1a (H77c) and 1b (J4L6S) (46) were used to generate homogeneous full-length NS3/4A protease complexes representing the six major HCV genotypes (HC-J6, HC-J8, S52, ED43, SA13, and HK-6A). The susceptibility of purified recombinant NS3/4A protease complexes was assessed using fluorescence resonance energy transfer (FRET) assays (47). The 50% inhibitory concentration (IC50) was generated as described previously (14).
Enzyme-based selectivity assays.
To assess in vitro selectivity, compounds were counterscreened against GBV-B NS3/4A protease, human neutrophil elastase (Sigma), porcine pancreatic elastase (Sigma), human α-chymotrypsin (Sigma), human cathepsin A (R&D Systems, Minneapolis, MN), and human liver cathepsin B (Calbiochem, San Diego, CA). Compounds were diluted in assay buffer in 10% dimethyl sulfoxide (DMSO). GBV-B NS3/4A protease (0.3 nM) replaced HCV-NS3/4A protease in the FRET-based assay, as described above. All other assays were performed in 96-well plates with 1% DMSO and measured continuous substrate hydrolysis at 405 nm on a Spectramax (Molecular Devices, Sunnyvale, CA) plate spectrophotometer.
Human neutrophil elastase, porcine pancreatic elastase, and human pancreatic α-chymotrypsin reaction mixtures contained 50 mM Tris-HCl (pH 8.0), 50 mM NaCl, 0.1 mM EDTA, 0.05% Tween 20, and 20 nM human elastase, 25 nM porcine elastase, or 1 nM chymotrypsin, respectively. Reactions were initiated by addition of substrate (elastases, 5 μM succinyl-AAPV-amino-4-methylcoumarin (AMC) [Sigma]; chymotrypsin, 5 μM LLVY-AMC [Sigma]) and monitored fluorimetrically at an excitation wavelength (Ex) of 360 nm and an emission wavelength (Em) of 480 nm. Cathepsin A assays were performed per the manufacturer's instructions with (7-methoxycoumarin-4-yl)acetyl-RPPGFSAFK(dinitrophenyl)-OH (R&D Systems) as the substrate; reactions were monitored at Ex/Em of 565/580 nm. Cathepsin B assay mixtures contained 50 mM sodium acetate (NaOAc) (pH 5.5), 1 mM tris(2-carboxyethyl)phosphine, 5 nM cathepsin B, and 2 μM Z-FR-AMC (Calbiochem) as the substrate, and reactions were monitored at Ex/Em of 360/460 nM. Factor XA assay mixtures contained 100 mM NaPO4 (pH 7.5), 0.5% polyethylene glycol (PEG) 8000, 200 mM NaCl, 1 nM human factor XA (Enzyme Research Laboratories, South Bend, IN), and 400 μM S-2222 (Diapharma, West Chester, OH) as the substrate. Factor XIA assay mixtures contained 50 mM HEPES (pH 7.5), 0.1% PEG-8000, 5 mM KCl, 145 mM NaCl, 0.14 nM human factor XIA (Hematologic Technologies, Essex Junction, VT), and 250 μM S-2366 (Diapharma) as the substrate. Factor VIIA assay mixtures contained 50 mM HEPES (pH 7.5), 150 mM NaCl, 5 mM CaCl2, 0.1% PEG-8000, 1.8 nM factor VIIA (Novo Screen), and 1 mM S-2288 (Diapharma) as the substrate. Kallikrein assay mixtures contained 50 mM Tris (pH 7.5), 100 mM NaCl, 0.5% PEG-8000, 0.2 nM kallikrein (Enzyme Research Laboratories), and 400 μM S-2302 (Diapharma) as the substrate. Thrombin assay mixtures contained 100 mM sodium phosphate (pH 7.5), 200 mM NaCl, 0.5% PEG-8000, 0.2 nM thrombin (Enzyme Research Laboratories), and 200 μM S-2366 (Diapharma) as the substrate. Trypsin assay mixtures contained 100 mM NaPO4 (pH 7.5), 200 mM NaCl, 0.5% PEG-8000, 0.332 nM trypsin (Sigma), and 200 μM S-2222 (Diapharma) as the substrate.
Cell lines and viral constructs.
The genotype 1b (Con1) replicon cell line was constructed as described previously (16). A replicon cell line representing genotype 1a (H77) was generated as described previously (14), except that mutations were introduced into the genes encoding the NS3 helicase domain (proline to leucine at position 1496) and NS5A (serine to isoleucine at position 2204) to improve replication in cell culture (21). A genotype 2a (JFH-1) cell line was generated (15) by introducing the JFH-1 sequence from NS3 to NS5B (51) into the genotype 1b (Con1) backbone. Recombinant HCV hybrid replicon clones, in which the NS3 protease domain (amino acids 1 to 181) from Con1 or JFH-1 replicons was replaced with NS3 protease sequences from HCV genotypes 2b (HC-J8) and 3a (S52), were generated as described previously (11). PCR primers for amplifying and inserting the 2b (HC-J8) NS3 protease sequence contained anchoring sequences for PCR-mediated restrictionless cloning into the 2a (JFH-1) replicon (forward oligonucleotide, 5′ GCT CCC ATT ACT GCT TAC ACT CAG CAA AC 3′; reverse oligonucleotide, 5′ CAC AGC TGG CGG CGT ACT GTT G 3′). Primers for inserting 3a (S52) NS3 sequences into a 1b (Con1) replicon carrying two cell culture-adaptive mutations (A1289G and S1560G) in the NS3 helicase were designed similarly (forward oligonucleotide, 5′ CCT TTG AAA AAC ACG ATA ATA CCA TGG CGC CGA TCA CAG CAT ACG CCC AGC 3′; reverse oligonucleotide, 5′ GGG ACG AGT TGT CCG TGA AGA CCG GAG ACC TAG CCT GTG TGC TAA GGG 3′). To construct plasmids harboring consensus 2b (HC-J8) and 3a (S52) NS3/4A regions, first-strand cDNA was synthesized using random hexamers and a Superscript II first-strand synthesis system for RT-PCR (Life Technologies, Grand Island, NY). The HCV genotype 2b NS3/4A gene was PCR amplified using primers 5′-GTG AAG CTT ATG GCT CCC ATT ACT GCT TAC ACT CAG-3′ and 5′-GAT CCT GCA GTT AGC ATT CTT CCA TCT CAT CAA AG-3′, cloned into pCR2.1 (Life Technologies), and sequenced. A consensus NS3/4A sequence was assembled, and a HindIII-EcoRI fragment containing the NS3/4A sequence was cloned into pcDNA3.1P-Zeo (Life Technologies), creating construct p50A-1. The HCV genotype 3a NS3/4A gene was PCR amplified using primers 5′-GCT CTA GAA GCT TAT GGC CCC GAT CAC AGC ATA CGC CCA-3′ and 5′-GAT CCT GCA GTT AGC ACT CCT CCA TCT CAT CGT ATT G-3′, cloned into pCR2.1, and sequenced. A consensus HCV genotype 3a NS3/4A clone was assembled to create p51-1. Plasmids p50A-1 and p51-1 were used as templates for “touchdown” PCR amplification (13) (Life Technologies) using Platinum Taq high-fidelity DNA polymerase. PCR products were purified using a DNA Clean and Concentrate kit (Zymo Research, Irvine, CA). Purified amplicon (2 μg) was used as a “megaprimer” in a QuikChange II XL mutagenesis reaction (Agilent Technologies, Santa Clara, CA) using recombinant plasmids containing 2a (JFH-1) or 1b (Con1) replicon sequences as the template. All recombinant NS3 hybrid replicon clones were confirmed by DNA sequencing.
HCV genotype 4a NS3 protease sequences (amino acids 1 to 181) from strain ED43 were PCR amplified and cloned into the 1b (Con1) replicon shuttle vector (46), which includes a Renilla luciferase (RLuc) reporter for quantifying HCV replication. Constructs were verified by sequence analysis, and alignments were performed using Sequencher software (Gene Codes, Ann Arbor, MI). The genotype 4a NS3 protease sequence was amplified, cloned into the shuttle vector, and transformed into STBL-2 cells. DNA template was prepared, and replicon RNA was transcribed from the T7 promoter for subsequent phenotyping in a transient HCV replicon transfection assay (46).
A recombinant HIV NL-RLuc virus, in which RLuc replaced a section of nef from NL4-3, was constructed by cotransfection of two plasmids: pNLRLuc and pVSVenv (W. Blair and T. Spicer, HIV-1 reporter viruses and their use in assaying anti-viral compounds, PCT international patent application WO 2001096610, December 2001). The pNLRLuc contains the NL-RLuc DNA cloned into pUC18 at the PvuII site, while pVSVenv contains the gene for VSV G protein linked to a long-terminal-repeat promoter. Transfections were performed at a 1:3 ratio of pNLRLuc to pVSVenv on 293T cells using Lipofectamine Plus (Invitrogen), and titers of the resulting pseudotype virus were determined in MT-2 cells. The bovine viral diarrhea virus (BVDV) luciferase replicon genome carrying an in-frame firefly luciferase reporter gene was described previously (36).
Cell culture virology assays.
For HCV replicons containing an RLuc reporter, compound antiviral activities (50% effective concentration [EC50]), were determined as described previously (46). For replicons lacking a reporter gene, activity was measured using a FRET-labeled peptide substrate (10 μM), as described previously (46). BVDV assays were performed as described previously (36), except that incubations were maintained for 4 days.
Antiviral susceptibility of HIV, HRV2, and HCoV was determined by incubation with serial dilutions of compound. For recombinant HIVs expressing RLuc, antiviral activity was evaluated by luciferase assay 5 days postinfection using a dual-luciferase kit (Promega). The diluted passive lysis solution was premixed with both the luciferase assay substrate and the Stop & Glo substrate (2:1:1 ratio). To each aspirated sample well, 40 μl of the mixture was added, and luciferase activity was measured immediately on a Wallac TriLux (PerkinElmer, Waltham, MA). For determination of activities against HRV2 and HCoV OC43, compound and virus (multiplicity of infection, 0.1) were incubated with cells for 96 h, and cell protection was used as a measure of virus production. For the cell protection assay, 17 μl of Cell-Titer Blue reagent (Promega) was added to each well. Plates were then incubated for 2 h at room temperature before fluorescence was measured using a Spectramax Gemini EM instrument (Molecular Devices) set to Ex/Em of 530/580 nm.
Cytotoxicity was determined by incubating cells (3,000 to 10,000 cells/well) with serially diluted test compounds or DMSO for 5 days (MT-2 cells) or 4 days (all other cell types). Cell viability was quantitated using an MTS assay for MT-2 or a Cell-Titer Blue reagent (Promega, Madison, WI) assay for HEK-293, HuH-7, HepG2, and MRC5 cells, and 50% cytotoxic concentrations (CC50s) were calculated. For the HCV and BVDV replicon assays, CC50s were determined from the same wells that were later used to determine EC50s.
Inhibitor combination studies.
For combination studies, inhibitors (rIFN-α and inhibitors of NS3, NS5A, and NS5B) were each tested at 11 concentrations. Stock solutions, at 200 times the final assay concentrations, were prepared by 2- or 3-fold dilution in DMSO prior to addition to cells or media. Compounds were tested alone and in two- or three-drug combinations with ASV. HCV genotype 1b (Con1) replicon cells were exposed to compounds for 4 days, and inhibition was determined using the FRET-labeled peptide assay (46). Cytotoxicity of combined agents was monitored in parallel by Cell-Titer Blue staining. Concentration-response curves were used to assess the degree of antagonism, additivity, or synergy at the 50, 75, or 90% effective level. Combination effects were analyzed using in-house software to determine combination indices and 95% confidence intervals, applying the definition of Loewe's synergy cited by Chou (10). The analysis starts by fitting concentration-response curves to normalized responses from each therapy. These curves are fit as a four-parameter logistic model with a common minimum and maximum across all therapies. Conceptually, this equation can be written as
where Ij is 1 if therapy = j and 0 otherwise, for a j value of 1, 2, 3, 4, or 5. In the above equation, response represents the normalized data. For this assay, A is the bottom plateau common to all curves, D is the common top plateau, Bj is the “slope” parameter for the jth therapy, and Cj is the concentration that produces an effect equal to the average of A and D for the jth therapy. The concentration response curves are then inverted and used to compute the combination index (CI):
In the above equation, for a given effective level, α, α[A], and α[B] are the concentrations of drugs that produce that specific level of effect individually, and α[A]B and α[B]A represent the concentrations of drugs in combination that would produce the same level of effect.
Combination studies with ribavirin were carried out essentially as described previously, using MacSynergy II software, developed by Prichard and Shipman (12). For each combination, six to eight replicate plates were analyzed per experiment. Antiviral activity was measured after 4 days in culture using an RLuc reporter expressed by the HCV genotype 1b replicon (16). A representative plate from each experiment was stained in parallel with Cell-Titer Blue reagents to assess cytotoxicity. Ribavirin concentrations that exhibited cytotoxic effects of >15% (typically, concentrations of 45 μM and 135 μM) were excluded from antiviral analysis. As recommended by guidelines in the MacSynergy II user manual, values of synergy or antagonism under 25 μM2% at 95% confidence were considered insignificant, values over 100 indicated strong synergy, and values less than 100 indicated strong antagonism. In three-dimensional plots, an overall response below the 0% plane was considered to indicate antagonism, a response above indicated synergy, and a flat surface indicated additivity.
In vitro protocols for predicting permeability, metabolic stability, and protein binding.
Compound permeability was assessed by the non-cell-based parallel artificial membrane permeability assay (PAMPA), as well as the cell-based Caco-2 cell model. PAMPAs were conducted in triplicate with 100 μM compound at room temperature for 4 h using pION lipid solution, at pH 7.4 and 5.5. Caco-2 cells (ATCC) were seeded at 80,000/cm2 on a 24-well polycarbonate filter membrane and incubated at 37°C with 5% CO2 and 95% relative humidity for approximately 21 days, with medium changes every 2 days. Culture medium consisted of DMEM with 10% FBS, 1% nonessential amino acids, 1% l-glutamine, 100 U/ml penicillin G, and 100 μg/ml streptomycin. The assay was performed at pH 7.4 using Hanks' balanced salt solution containing 10 mM HEPES as the transport medium. Studies were initiated by adding a buffer containing compound to either the apical (A-to-B transport) or basolateral (B-to-A transport) side of the monolayer and incubating for 2 h at 37°C. Compound transport was determined by liquid scintillation counting. Permeability coefficients (Pc) were determined as dA/(dt·S·C0), where dA/dt is the flux of test compound across the monolayer (nmol/s), S is the surface area of the cell monolayer (0.33 cm2), and C0 is the initial concentration (μM) in the donor compartment.
In vitro metabolic stability and clearance were determined from duplicate 96-well reaction mixtures containing liver microsomes isolated from rats (Sprague-Dawley; BD Biosciences, Woburn, MA), dogs (beagle; BD Biosciences), humans (pooled samples; BD Biosciences), and monkeys (cynomolgus; BD Biosciences). Assay mixtures contained 0.5 μM compound, 1% acetonitrile, 1 mg/ml protein, 100 mM potassium phosphate buffer (pH 7.4), and 6.7 mM magnesium chloride. Reactions were initiated by addition of 1 mM NADPH, and the mixtures were incubated at 37°C for 0, 10, 20, 30, 40, or 60 min. The reactions were quenched by the addition of two volumes of acetonitrile. Mixtures were centrifuged at 3,000 × g in an Allegra X-12 centrifuge with an SX4750 rotor (Beckman Coulter Inc., Fullerton, CA) at 10°C for 10 min; supernatants were analyzed using liquid chromatography-tandem mass spectrometry (LC-MS/MS). Several compounds were run in parallel as quality controls for low, medium, and high clearances. Compound disappearance rates were determined by regression analysis of concentration time profiles.
Protein binding was determined in fresh rat, dog, cynomolgus monkey, and human sera (pooled samples from Bioreclamation Inc., Hicksville, NY). Protein binding was also determined in the replicon assay medium (DMEM containing l-glutamine and 4% FBS), with and without 40% human serum, using equilibrium dialysis (Dianorm equilibrium dialyzer; Dianorm, Munich, Germany) in 0.1 M sodium chloride, 4 mM potassium chloride, 1 mM magnesium sulfate, and 30 mM sodium phosphate (pH 7.4) and dialysis membranes (HTDialysis, LLC, Gales Ferry, CT) with a 12,000- to 14,000 molecular mass cutoff. Dialysis cells (HTDialysis, LCC, Gales Ferry, CT) were rotated at 20 rpm on an orbital shaker at 37°C with 10% CO2 and 85% humidity. Protein binding was determined at a nominal compound concentration of 10 μM (n = 3). At 7 h, buffer samples (100 μl) were collected in a 96-well plate that contained 100 μl of species-specific serum. Serum samples (100 μl) were collected in a 96-well plate that contained protein binding buffer. A mixed-matrix solution contained 50% protein binding buffer and 50% blank serum of the species under investigation and was used to prepare standard curves. Samples were analyzed using LC-MS/MS, and percent protein binding was calculated from the concentration in buffer relative to that in serum.
In vivo exposure studies.
The amorphous free acid form of ASV was used in all studies. Plasma and tissue exposures were characterized in male FVB mice (20 to 25 g; Harlan Breeding Laboratories, Indianapolis, IN), male Sprague-Dawley rats (300 to 350 g; Hilltop Lab Animals, Scottsdale, PA) with indwelling cannulas implanted in the jugular vein or common bile duct, male beagles (ca. 9 to 12 kg; Marshall Farms, North Rose, NY) bearing vascular access ports, and male cynomolgus monkeys bearing vascular access ports (3.1 to 5.7 kg; Charles River Biomedical Research Foundation, Houston, TX). All animal procedures were performed under protocols approved by the Institutional Animal Care and Use Committee of the test facility. In all studies, coagulated blood samples taken after dosing were centrifuged at 4°C (1,500 to 2,000 × g); plasma samples were stored at −20°C until analyzed. Tissue samples were rinsed, blotted dry, weighed, and stored frozen. ASV in plasma and tissue samples was analyzed by LC-MS/MS. The lower limit of quantification (LLOQ) of ASV was 1 nM in plasma samples from all species, 5 nM in mouse and rat tissues, and 50 nM in dog and monkey tissues.
Mice (n = 9 per group; overnight fast) received ASV by oral gavage (5 mg/kg; vehicle of PEG-400–ethanol, 9:1). Blood samples (∼0.2 ml) were obtained by retro-orbital bleeding at 0.25, 0.5, 1, 3, 6, 8, and 24 h after dosing. Within each group, three animals were bled at 0.25, 3, and 24 h, three at 0.5 and 6 h, and three at 1 and 8 h, resulting in a composite pharmacokinetic profile. Livers and brains were also removed from mice at the terminal sampling points.
Rats (n = 3 per group; overnight fast) received ASV (amorphous free acid) by oral gavage (3, 5, 10, and 15 mg/kg) in PEG-400–ethanol (9:1). Serial blood samples (∼0.3 ml) were obtained from the jugular vein predosing (0 h) and at 0.25, 0.5, 0.75, 1, 2, 4, 6, 8, 24, and 48 h postdosing. To assess tissue exposure, rats were orally administered ASV (5 or 15 mg/kg, same vehicle as above), and blood, liver, and heart samples from two rats/group were obtained at 0.17, 0.5, 1, 2, 4, 6, 8, 24, 48, and 72 h after dosing.
Dogs (n = 3; overnight fast) were administered ASV by oral gavage at 3 or 6 mg/kg (3 mg/ml in 85% PEG-400–15% water). Serial blood samples were collected from vascular access ports at 0.08, 0.167, 0.25, 0.5, 0.75, 1, 2, 4, 6, 8, 24, and 48 h postdose. To assess tissue exposure, six male dogs (8.4 to 12.5 kg) were orally administered ASV (6 mg/kg), and blood, liver, and spleen samples were obtained from one dog each at 1, 3, 7, 24, 48, and 72 h postdosing.
Monkeys (n = 3 males; overnight fast) were administered ASV by oral gavage at 3 mg/kg (3 mg/ml in 85% PEG-400–15% water). Blood samples were collected at 0.25, 0.5, 0.75, 1, 2, 4, 6, 8, and 24 h postdose. To assess tissue exposure, three male and six female cynomolgus monkeys (2 to 5 kg) were orally administered ASV (10 mg/kg), and blood, liver, and spleen samples were obtained from one male each at 2, 8, and 24 h postdosing and from one female each at 0.5, 2, 4, 8, 24, and 30 h postdosing.
RESULTS
Proteolytic enzyme activity.
To assess the potential genotype spectrum coverage by ASV, enzyme IC50s against representatives of six major genotypes of HCV NS3/4A protease were determined (Table 1). These in vitro potencies were benchmarked against the NS3 PI VX-950 (Table 1), recently approved for use in a combination regimen with pegylated alfa interferon and ribavirin in patients infected with HCV genotype 1. ASV inhibited the NS3 proteolytic activity of genotype 1a (H77 strain) and genotype 1b (J4L6S strain), with IC50s of 0.7 and 0.3 nM, respectively. Against NS3/4A protease complexes representing genotypes 2 through 6, IC50s ranged from 0.9 to 320 nM. The in vitro potencies of VX-950 ranged from 12 to 537 nM against enzymes representing genotypes 1 through 6.
Table 1.
Potency of ASV and VX-950 against recombinant full-length HCV NS3/4A protease complexes representing different HCV genotypes
| Genotype (strain) | IC50 (nM)a |
|
|---|---|---|
| ASV | VX-950b | |
| 1a (H77) | 0.70 ± 0.06 | 12 ± 2 |
| 1b (J4L6S) | 0.30 ± 0.02 | 22 ± 6 |
| 2a (HC-J6) | 15 ± 1 | 95 ± 17 |
| 2b (HC-J8) | 78 ± 2 | 12 ± 1 |
| 3a (S52) | 320 ± 13 | 537 ± 4 |
| 4a (ED43) | 1.6 ± 0.1 | 12 ± 1 |
| 5a (SA13) | 1.7 ± 0.2 | 38 ± 3 |
| 6a (HK-6A) | 0.9 ± 0.1 | 86 ± 2 |
Data are means ± standard deviations from at least three independent experiments.
VX-950 was preincubated with each of the respective NS3/4A protease complexes for 30 min before the addition of substrate to obtain optimum potency. Preincubation of ASV and enzyme was not required to enhance potency.
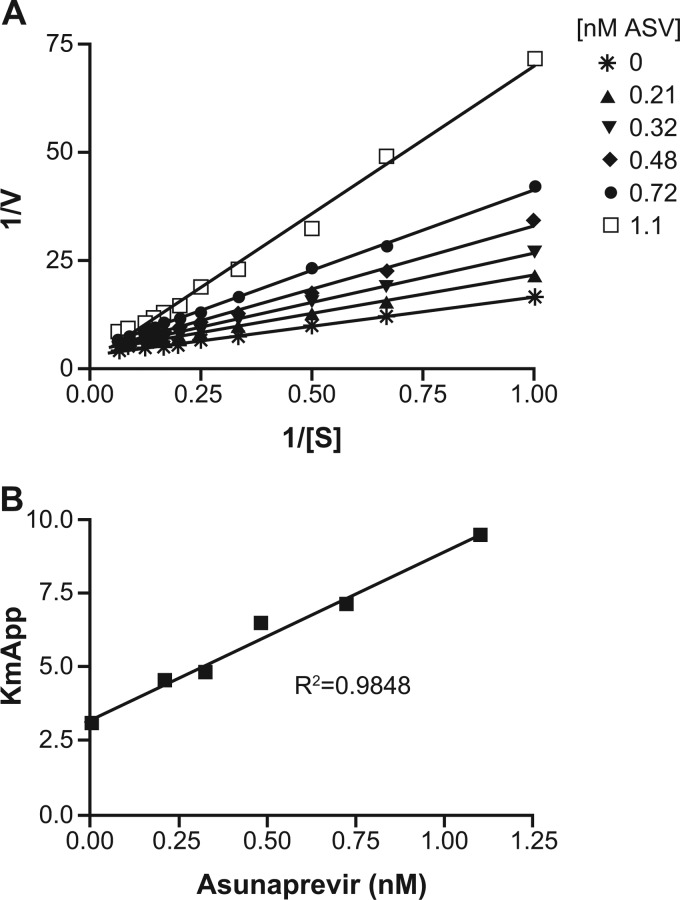
The mechanism of inhibition was established for ASV using purified recombinant full-length HCV NS3/4A protease complexes. The kinetic constants kcat/Km (rate of catalysis divided by Michaelis-Menten constant) of the HCV NS3/4A proteases representing genotype subtypes 1a and 1b, measured against the FRET-labeled peptide substrate loosely representing the HCV NS4A/NS4B cleavage site (47), were 0.9 ± 0.0 and 1.3 ± 0.5 μM−1 s−1, respectively. In order to determine the inhibition constant (Ki), apparent Km determinations for the FRET-labeled peptide substrate were calculated in the presence of increasing concentrations of ASV. Analysis of the data revealed that as concentrations increased, the apparent Km for the substrate increased while the maximal enzyme turnover rate (Vmax) remained constant, typical of a first-order competitive inhibitor. Slope replots from each of the resulting Lineweaver-Burk graphs were plotted against the compound concentration. The graph in Fig. 2 represents the data from a typical experiment. The average Ki values for ASV were approximately 0.4 nM and 0.2 nM for the HCV NS3/4A protease complexes representing genotype 1a and genotype 1b, respectively. This mechanism of inhibition differs from that reported for telaprevir. The acylsulfonamide of ASV interacts with the NS3 protease catalytic site in a noncovalent manner, whereas the α-ketoamide moiety of telaprevir covalently binds to the catalytic serine of NS3/4A protease and slowly reverses over time (38).
Fig 2.
Lineweaver-Burk plot and Ki determination for asunaprevir (ASV). (A) Lineweaver-Burk plot of 1/V (reciprocal of rate of change of fluorescence) versus 1/S (reciprocal of the substrate concentration). (B) Ki determination plot of the apparent Km value (slope obtained from the Lineweaver-Burk plot) versus ASV concentration.
The substrate specificity determinant of proteases is generally defined by the side chains flanking the cleavage site (35). The naturally occurring trans-cleavage substrates for HCV NS3/4A protease exhibit a strict requirement for a cysteine residue in the P1 position of the scissile bond. Owing to the large number of proteases present in the body, selectivity is an important feature for PIs designed as therapeutic agents. ASV selectivity was subsequently determined by evaluating its inhibitory effect on a panel of purified recombinant proteases (Table 2). Reference enzymes that are structurally and/or mechanistically similar to the target enzyme or of clinical significance were chosen for the in vitro selectivity studies. ASV exhibited an excellent selectivity index (>40,000) against all serine/cysteine proteases evaluated, which included human leukocyte elastase (human “sputum” elastase), porcine pancreatic elastase, and three members of the chymotrypsin family of serine proteases, compared with the IC50 against genotype 1a (H77c) NS3/4A protease. GBV-B belongs to the family Flaviviridae and is closely related to human HCV. Although GBV-B and HCV share only approximately 30% amino acid homology within the NS3 protease domain, the catalytic triad and the S1 specificity determinant residue, a phenylalanine at residue 154, are conserved. In addition, GBV-B and HCV NS3/4A proteases have overlapping substrate specificities. Against purified recombinant GBV-B NS3/4A protease, ASV had a selectivity index of approximately 30,000 for the HCV NS3/4A protease representing the genotype 1a H77c strain. These ASV selectivity indices compared favorably with those calculated for VX-950 against closely related human serine proteases and GBV-B versus its activity against the HCV genotype 1a NS3/4A protease (VX-950 selectivity index ≥ 27.5) (Table 2).
Table 2.
Potency of ASV and VX-950 against a panel of serine and cysteine proteases
| Protease | IC50 (μM)a |
|
|---|---|---|
| ASV | VX-950 | |
| HCV NS3/4A (1a [H77]) | 0.0007 ± 0.0001 | 0.012 ± 0.002 |
| GBV-B NS3/4A | 21 ± 6 | 7.7 ± 0.0 |
| Human leukocyte elastase | >50 | 3.2 ± 0.1 |
| PPE | >50 | 5.6 ± 0.3 |
| Human chymotrypsin | >50 | 45 ± 8 |
| Human cathepsin A | >5b | 0.33 ± 0.02 |
| Human cathepsin B | >50 | 1.7 ± 0.2 |
| Trypsin | >30 | ND |
| Thrombin | >30 | ND |
| Factor VIIa | >30 | ND |
| Factor Xa | >30 | ND |
| Factor XIa | >30 | ND |
| Kallikrein | >30 | ND |
Data are means ± standard deviations from at least three independent experiments. ND, not determined.
Assay interference was observed at 20 μM.
Antiviral activity in cell culture.
The activity of ASV against HCV RNA replication was measured using replicons representing the most clinically pertinent genotypes, 1a and 1b, as well as the genotype 2a JFH-1 strain (51). In addition, its effects against hybrid replicons encoding the NS3 protease domain representing genotype 2b, 3a, or 4a were also monitored. Inhibition of HCV RNA replication by ASV was monitored using a luciferase reporter assay; however, comparable inhibitory effects could be determined by directly measuring HCV RNA or protein expression (data not shown) (36). A summary of ASV EC50s generated using these assays is shown in Table 3. Against replicons encoding the NS3 protease domains representing genotypes 1a, 1b, and 4a, EC50s ranged from 1.2 to 4.0 nM. These compared favorably with the 427 to 1,137 nM values determined for VX-950. Against replicons encoding the NS3 protease domains of genotypes 2a and 3a, ASV exhibited significantly reduced activity (EC50s ranging from 67 to 1,162 nM), similar to results obtained for VX-950 (EC50s ranging from 108 to 2,731 nM) (Table 3).
Table 3.
Cell culture potency of ASV and VX-950 in HCV replicons
| Genotype (strain)a | Stable cell line or transient-transfection replicon assayb | EC50 (nM)c |
|
|---|---|---|---|
| ASV | VX-950 | ||
| 1a (H77c) | Stable cell line | 4.0 ± 0.3 | 995 ± 11 |
| 1b (Con1) | Stable cell line | 1.2 ± 0.3 | 427 ± 3 |
| 2a (JFH-1) | Stable cell line | 230 ± 74 | 229 ± 32 |
| 3a (S52) | Stable cell line | 1,162 ± 274 | 2,731 ± 188 |
| 2a (JFH-1) | Transient | 67 ± 23 | 108 ± 12 |
| 2b (HC-J8) | Transient | 480 ± 104 | 1,236 ± 96 |
| 1b (Con1) | Transient | 1.3 ± 0.1 | 266 ± 77 |
| 4a (ED43) | Transient | 1.8 ± 0.2 | 1,137 ± 223 |
Genotypes 2b, 3a, and 4a are NS3 protease hybrid-derived replicons.
Luciferase reporter used as readout.
Data are means ± standard deviations from at least three independent experiments.
ASV specificity was examined in different cell culture systems. ASV demonstrated no significant activity against BVDV, a pestivirus that is closely related to flaviviruses (48), or other RNA viruses at concentrations exceeding 10 μM (Table 4). Against a panel of cell lines representing liver, T lymphocytes, lung, cervix, and embryonic kidney, CC50s ranged from 11 to 38 μM (Table 4). The calculated therapeutic index for ASV was 6,250-fold using the genotype 1a replicon.
Table 4.
Selectivity of ASVa
| Virus | Cell type | EC50 (μM) | CC50 (μM)a |
|---|---|---|---|
| HCV-1ab | HuH-7 | 0.004 ± 0.0003 | 25 ± 5 |
| BVDVc | HuH-7 | 12 ± 2 | 32 ± 4 |
| HCoVOC43 | MRC5 | >32 | 32 ± 5 |
| HRV2 | MRC5 | >32 | 32 ± 5 |
| HIV | MT-2 | 13 ± 2 | 34 ± 3 |
| NA | HepG2 | NA | 38 ± 5 |
| NA | HeLa | NA | 19 ± 1 |
| NA | HEK293 | NA | 11 ± 2 |
Data are means ± standard deviations from at least three independent experiments. NA, not applicable.
Subgenomic replicon.
Full-length replicon.
Inhibitor combination studies in the HCV replicon.
The effects of combination therapy were explored using HCV genotype 1a and 1b replicon systems to determine whether drug combinations result in additive, antagonistic, or synergistic interactions. Six HCV antivirals, including rIFN-α, ribavirin, daclatasvir (HCV NS5A replication complex inhibitor), an NS5B inhibitor targeting the nucleoside-binding site (NM-107), and NS5B allosteric inhibitors targeting site I (BMS-791325) or site II (HCV-796) were tested in two-drug combinations with ASV. The NS3 PI and rIFN-α were combined at three different ratios. Concentration-response curves were fit to assess the antiviral effects of the drug treatment combinations, and the degree of antagonism, additivity, or synergy was determined at the 50, 75, and 90% effective levels. The combination indices were calculated using the definition of Loewe's synergy cited by Chou (10). The results showed a mixture of additive and synergistic effects for rIFN-α, as determined from the combination indices (Table 5). Additive and synergistic effects were also observed for daclatasvir (synergistic-additive) and NS5B inhibitors (additive or synergistic-additive) in two-drug combination studies (see Tables S1 to S3 in the supplemental material). Importantly, no drug antagonism was observed in any of the combinations evaluated. Effects of combining ASV in three-drug combinations were also explored. Combinations of the NS3 PI with rIFN-α and daclatasvir or rIFN-α and BMS-791325 yielded mixed synergy-additivity or additivity, respectively. As with the two-drug combinations, no antagonism was observed at any of the effective doses (see Tables S4 and S5 in the supplemental material). For combination studies between ASV and ribavirin, a technique similar to that used by Coelmont et al. (12) was used rather than the method described by Chou, which relies on obtaining a full EC50 curve in the absence of any overlapping cytotoxicity. In the genotype 1b system, ribavirin concentration-response curves resulted in EC50 and CC50s that were not well separated enough for analysis by the method of Chou (20 ± 5 μM and 68 ± 23 μM, respectively). Combination response data were analyzed using the MacSynergy II software developed by Prichard and Shipman (University of Michigan), with ASV demonstrating overall additivity (see Fig. S1 in the supplemental material).
Table 5.
Activity of ASV in combination with rIFNα
| Expt | ASV EC50 (nM) | rIFN-α EC50 (U/ml) | Ratio of rIFN-α to ASV | Combination index (confidence interval) |
Overall result | ||
|---|---|---|---|---|---|---|---|
| 50% effective | 75% effective | 90% effective | |||||
| 1 | 1.7 | 12.8 | 25:1 | 0.97 (0.89, 1.06) | 0.94 (0.83, 1.05) | 0.90 (0.74, 1.07) | Additivity |
| 10:1 | 0.77 (0.70, 0.84) | 0.81 0.71, 0.91) | 0.86 (0.70, 1.03) | Synergy/additivity | |||
| 4:1 | 0.74 (0.67, 0.80) | 0.82 (0.71, 0.92) | 0.91 (0.73, 1.09) | Synergy/additivity | |||
| 2 | 1.5 | 24.5 | 25:1 | 0.71 (0.66, 0.77) | 0.61 (0.54, 0.67) | 0.52 (0.43, 0.61) | Synergy |
| 10:1 | 0.95 (0.88, 1.01) | 0.77 (0.69, 0.85) | 0.63 (0.53, 0.73) | Synergy/additivity | |||
| 4:1 | 0.71 (0.65, 0.77) | 0.72 (0.64, 0.80) | 0.73 (0.60, 0.87) | Synergy | |||
In vitro permeability, metabolic stability, and serum protein binding.
The permeability of ASV was examined using a PAMPA model and the cell-based Caco-2 bidirectional permeability assay. The Pc for the NS3 PI in the PAMPA model were >473 nm/sec (pH 5.5) and >492 nm/sec (pH 7.4), similar to those of compounds that exhibit good (>80%) absorption in humans. In the Caco-2 cell bidirectional permeability assay, ASV permeability was dependent on the initial drug concentration; (B-A)/(A-B) efflux ratios greater than unity (∼31-fold at 5 μM and ∼3-fold at 25 μM) were calculated, suggesting that it is a substrate of efflux transporters such as P glycoprotein (Table 6).
Table 6.
Bidirectional permeability of ASV in Caco-2 cells as a function of concentration
| Compound | Concn (μM) | Pc (nm/s)a |
Efflux ratio | |
|---|---|---|---|---|
| A-B | B-A | |||
| ASV | 5 | 11 ± 2 | 341 ± 20 | 31 |
| ASV | 25 | 54 ± 3 | 174 ± 22 | 3 |
| Controlb | 5 | 9 ± 1 | 167 ± 25 | 19 |
Pc, permeability coefficient. Data are means ± standard deviations from three independent experiments.
P-glycoprotein substrate digoxin.
The rate of metabolism of ASV was evaluated in NADPH-fortified liver microsomes of various species. Using the rates of loss of parent compound at 0.5 μM, the predicted hepatic intrinsic clearance (CLh,int) values were 13.3, 13.3, 31.7, and 8.5 ml/min/kg for rats, dogs, monkeys, and humans, respectively. These values represent approximately 24, 43, 69, and 41% of hepatic blood flow in rats, dogs, monkeys, and humans and indicate that the NS3 PI may exhibit low to intermediate metabolic clearance. These results indicate that, as a fraction of hepatic blood flow, the predicted human CLh,int of ASV (41%) is more similar to that predicted for dogs (43%) than it is to that predicted for rats (24%) or monkeys (69%).
Serum protein binding can often attenuate the clinical efficacy of antiviral agents. The extent of ASV serum protein binding was assessed across species by equilibrium dialysis, while the direct effect of human serum binding on its antiviral activity was also examined in a replicon cell-based assay. In protein binding assays, ASV (10 μM) was 98.8% ± 0.5% bound to human serum protein and 97.2% to 98.8% bound to serum proteins in the animal species studied (rats, dogs, and cynomolgus monkeys). Additionally, protein binding of the compound (0.5, 3, and 10 μM) was determined in the replicon assay medium (containing 4% FBS) and found to be 77.6% ± 1.5%, 76.0% ± 4.8%, and 68.1% ± 1.1%, respectively. When the replicon buffer contained 40% human serum, ASV protein binding was increased to 97.4% ± 0.66%, 97.7% ± 0.31%, and 97.6% ± 0.54% at 0.5, 3, and 10 μM, respectively. Supplementation of 40% human serum to an HCV genotype 1b replicon assay resulted in a loss in ASV potency of approximately 6.5-fold. This loss in EC50 compared with a 6.8-fold loss observed for VX-950 (data not shown).
In vivo plasma and tissue exposures across species.
Plasma and liver concentrations, as well as that in other organs, were assessed in mice, rats, dogs, and monkeys (Table 7). FVB mice were dosed orally (PO) with ASV, and drug exposure was monitored over 24 h. Liver concentrations in all animals were highly elevated over those in plasma (Table 7), with liver-to-plasma ratios of ≥40. ASV was not detected in brain, indicating minimal penetration into that tissue. Similar studies were carried out in rats; the time courses of tissue (liver and heart) and plasma concentrations of ASV were monitored through 48 or 72 h after PO dosing. Analysis of samples revealed that liver contained the highest concentrations of ASV, with the elevated liver-to-plasma area under the curve (AUC) ratios (Table 7) similar to those observed in mice using a more limited data set. Compound exposure in the hearts of these rats was minimal when detected. In dogs, the time courses of tissue (liver and spleen) and plasma concentrations were determined through 72 h after PO dosing. Liver contained the highest concentrations of compound, and the liver-to-plasma ratios at each of the sampling times were variable, ranging from 10 to approximately 410, with a ratio of 40 obtained using AUC values. The other tissues (spleen) contained lower levels of ASV than liver, with tissue-to-plasma ratios at the various sampling times ranging from 0.5 to 1. In monkeys, the time courses of tissue (liver and spleen) and plasma concentrations of ASV were determined through 72 h after PO dosing. Over the 72-h study duration, liver tissue again contained the highest concentration of compound, while spleen contained considerably less, with concentrations in most samples below the LLOQ as determined by LC-MS/MS. While the liver-to-plasma ratios of ASV at each of the sampling times were variable, ranging from 39 to approximately 320, the estimate based on the AUC values was 151. The observed liver-to-plasma ratio after oral dosing to monkeys (∼150) was in between the ratios observed in mice (>80) or rats (∼360) and higher than that for dogs (∼40). When dose prediction analyses were considered (our unpublished results), it was assumed that the high liver levels observed across preclinical species would also translate to humans.
Table 7.
Tissue and plasma concentrations of ASV after PO dosing across speciesa
| Species (PO dose, mg/kg)b | Sample analyzed | C24 (nM) | T/P AUC ratio | Fold GT1a EC50c |
|---|---|---|---|---|
| Mouse (5) | Serum | <LLOQ | ||
| Liver | 714 ± 330d | >80e | 179 (28) | |
| Rat (15) | Plasma | 38 | 9.5 (1.5) | |
| Liver | 11,986 | 359 | 2,997 (461) | |
| Dog (6) | Plasma | 6 | 1.5 (0.2) | |
| Liver | 2,541 | 40 | 635 (98) | |
| Spleen | <LLOQ | 0.5 | ||
| Monkey (10) | Plasma | <LLOQ | ND | |
| Liver | 441 | 151 | 110 (17) | |
| Spleen | <LLOQ | ND |
C24, concentration at 24 h postdose; GT, genotype; L, liver; LLOQ, lower limit of quantification; ND, not determined; T, tissue; P, plasma.
Mice (n = 3 per time point) were dosed at 5 mg/kg and monitored for 24 h, rats (n = 2 per time point) were dosed at 15 mg/kg PO and monitored for up to 72 h postdose, dogs (n = 1 per time point) were dosed at 6 mg/kg PO and monitored for up to 72 h, and monkeys (n = 1 or 2 per time point) were dosed at 10 mg/kg PO and monitored for up to 30 h postdose. Average values are shown for rats and monkeys, whereas only single values were obtained for dogs.
Fold EC50 multiples of ASV were calculated by dividing the C24 value by the EC50 for ASV in HCV genotype 1a replicons (EC50 ≈ 4 nM). Values in parentheses represent a similar calculation using the ASV EC50 in 40% human serum (EC50 ≈ 26 nM).
Average ± standard deviation.
Liver-to-serum ratio was determined.
DISCUSSION
ASV is a linear peptide mimetic HCV NS3 PI discovered using a structure-based drug design approach to replace the carboxylate moiety of ciluprevir derivatives with a proprietary P1′ acylsulfonamide cyclopropyl moiety, which was chosen with the aim of significantly enhancing potency (Fig. 1) (7). Comprehensive profiling has demonstrated that ASV is a highly selective inhibitor of HCV NS3/4A protease complex enzymatic activity, effectively inhibiting four major HCV genotype classes (Table 1). It is most active against proteases representing genotype 1, the genotype responsible for the highest unmet medical need, while isolates representing genotypes 2 and 3 are the least susceptible to inhibition. The reduced activity against genotype 3 was anticipated, since the reported salt bridge that forms between the charged NS3 amino acid residues R155 and D168 in genotypes 1, 4, 5, and 6 (49), stabilizing the interaction of the bulky P2 moiety of ASV with the enzyme, would not form. This is a consequence of amino acid 168 in NS3 being a glutamine in genotype 3a. Replacement of the charged D168 by neutral residues, predicted to weaken or destroy the 155:168 salt bridge, results in reduced susceptibility to ASV (30). For the genotype 2 subtype, NS3 residue 122 is either a lysine (genotype 2a) or arginine (genotype 2b). This residue is close to charged residues D168 and R123, and replacement of the neutral serine by arginine could lead to conformational changes affecting the binding of ASV and NS3 PIs with similar bulky P2 moieties (49).
Kinetic characterization of ASV against HCV genotype 1a and 1b NS3/4A protease complexes in homogeneous in vitro assays indicated that it is a classic competitive inhibitor with Ki values in the picomolar range. The classic competitive inhibition curve, as calculated from a Lineweaver-Burk plot (Fig. 2), confirmed that ASV is a noncovalent, single-step competitive inhibitor rather than a covalent (telaprevir and boceprevir) or noncovalent (danoprevir), two-step slowly reversible inhibitor (40). In theory, NS3 PIs exhibiting slower off-rates from the enzyme could have greater efficacy in patients and result in lower doses and less frequent dosing. However, the off-rate would need to be longer than the drug exposure half-life to have an impact in patients. Since telaprevir and boceprevir are administered three times daily at high doses and danoprevir appears likely to require boosting with ritonavir to achieve twice-daily administration (18), it is unclear whether the observed in vitro slow off-rates confer any advantage over ASV. However, single- and multiple-ascending-dose monotherapy clinical studies with ASV indicated rapid robust responses at lower doses than reported for telaprevir, boceprevir, or danoprevir (37).
ASV was shown to be highly specific for the HCV NS3 serine protease, demonstrating micromolar activity against the closely related GBV-B NS3 protease (Table 2). Furthermore, it exhibited little or no activity against a panel of human serine or cysteine proteases, including elastase, the most homologous of these proteases. The selectivity index calculated for ASV was significantly greater than that calculated for VX-950 (telaprevir), which demonstrated submicromolar to low-micromolar activities against a number of human serine and cysteine proteases. The reported exposure of telaprevir administered to HCV-infected patients (maximum concentration [Cmax], 3.5 μg/ml; minimum concentration [Cmin], 2 μg/ml) is higher than the observed intrinsic potencies against some of these human proteases. This may have potential implications with respect to off-target side effects, such as the skin orders (rash and pruritus) reported with telaprevir (50). The reduced selectivity of VX-950 (cathepsin A IC50, ∼0.3 μM) may also limit the potential of combining this drug with HCV nucleoside and nucleotide prodrugs. Cathepsin A is believed to be involved in the initial activation of the prodrug moiety of the NS5B polymerase nucleotide PSI-7977 via cleavage of the amino acid ester moiety (34). Taken together, these results suggest that the selectivity profile of ASV has the potential to result in improved tolerability over telaprevir in vivo.
HCV genotypes 1a, 1b, and 4 account for the majority of HCV infections. Patients infected with these genotypes represent some of the more difficult-to-treat populations using the currently available alfa interferon-based therapies; therefore, a requirement for DAAs with promising activity against these genotypes still exists. HCV replicon cell systems representing NS3 protease genotype subtypes 1a, 1b, and 4a were susceptible to inhibition by ASV, as demonstrated by EC50s in the low-nanomolar range (Table 3). This in vitro antiviral activity correlates with the high rates of early viral suppression observed among genotype 1 and 4 patients treated with ASV in combination with pegylated alfa interferon and RBV in phase 2 studies (5). Cellular proteases are unlikely to be affected, given the calculated in vitro therapeutic index of ≥2,750 for ASV. ASV was also shown to exert minimal or no activity against other RNA viruses, including BVDV, HRV, HCoV, and HIV, further confirming its specificity for HCV. BVDV is the only virus encoding a serine protease, albeit of low sequence homology (52%) (33), and these selectivity results potentially indicate that or ASV exerts minimal off-target activities. In fact, in early clinical studies (37), rash and neutropenia did not appear to be associated with the administration of ASV.
Resistance to direct-acting NS3 PIs has been shown to emerge rapidly during monotherapy (44). To increase the barrier to resistance, clinical studies have shown that combination therapies can achieve improved sustained viral responses (22, 39). For an investigational agent to be combined with another HCV agent in patients, it is important to first demonstrate that no antagonism is observed in in vitro combination studies (34). Under the experimental conditions outlined here, additive or synergistic effects were obtained when ASV was examined in combination with HCV inhibitors, representing distinct mechanisms of action. Importantly, no enhanced cytotoxicity was observed with any of the combinations tested.
Assessment of the in vitro permeability properties of ASV indicated that the compound should be readily absorbed. However, this absorption may be influenced by efflux transporters such as P glycoprotein (Table 6) and lead to variable drug exposure. ASV was predicted to exhibit low to intermediate metabolic clearance, with calculated human hepatic clearance being more similar to that predicted for dogs than that for rats or monkeys. While a high proportion of ASV was shown to be bound to serum proteins, its intrinsic potency was not attenuated in an NS3/4A protease enzyme assay supplemented with albumin to a concentration equivalent to that in human serum (data not shown). However, modest attenuation of anti-NS3 protease activity was observed in an HCV genotype 1b replicon assay supplemented with 40% human serum. This loss in potency is comparable to values obtained for other NS3 PIs.
The exposure profile of ASV across four different animal species (mouse, rat, dog, and monkey) revealed its hepatotropic disposition in all tested species, as indicated by a high exposure in liver versus plasma (Table 7). Compound exposure in other tissues examined, such as brain and spleen, was comparable to or less than that observed in plasma. Subsequent studies indicated that there was active uptake of ASV into the hepatocyte and elimination via the bile across species (our unpublished results), suggesting that the compound was enriched in hepatocytes. This is considered a desirable property given that HCV is a disease of the liver, with viral replication occurring in hepatocytes. Moreover, limited systemic exposure of the compound would reduce the potential for nonspecific side effects. Using the animal data to provide insight into potential doses and exposures in humans suggests that clearance in humans was closer to that in dogs. After dogs were administered an oral dose of 6 mg/kg of ASV, the Cmin in the liver was ∼100-fold the EC50 against genotype 1a replicons supplemented with 40% human serum. Using a simple body surface area conversion (42), a dose of approximately 200 mg was predicted to provide sufficient efficacy in humans (data not shown). This ASV dose administered twice daily provided rapid and robust antiviral activity in monotherapy studies and when combined with pegylated interferon and ribavirin (5, 37), and this was subsequently chosen for phase 2b studies.
Based on its overall preclinical profile, ASV was considered a suitable investigational agent for evaluation in clinical trials examining dual, triple, and quadruple combination approaches for the treatment of patients chronically infected with HCV genotypes 1 and 4 (5, 8, 28). The efficacy and safety of ASV were recently explored in a dual-DAA regimen of ASV combined with daclatasvir in difficult-to-treat genotype 1 null responders in phase 2a studies (8, 28). In one study, 36% of patients achieved SVR following 24 weeks of treatment, providing proof-of-concept that an interferon-sparing regimen can cure HCV infection. This SVR rate is similar to the ∼33% rate reported for a 48-week triple regimen of telaprevir combined with pegylated alfa interferon and ribavirin in genotype 1 null responders (50). Analyses of virologic failures in the dual-DAA regimen demonstrated that all the patients were infected with HCV genotype 1a and that the emergent NS3 protease-resistant variants detected were similar to those obtained in vitro (30). However, in a study in which all patients were infected with genotype 1b, all patients receiving ASV and daclatasvir for 24 weeks achieved SVR, suggesting that genotype 1 subtype can strongly influence virologic outcome (8).
In conclusion, the preclinical profile and subsequent data from early clinical trials support further research to assess the benefits of ASV-containing combination regimens in larger clinical studies.
Supplementary Material
ACKNOWLEDGMENTS
We thank David Stock for his development of the in-house combination study software, Burt Rose for constructing plasmids p50A-1 and p51-1, and Mark Cockett for his continued support of the program. Editorial assistance with the preparation of the manuscript was provided by Andrew Street of Articulate Science and was funded by Bristol-Myers Squibb.
Footnotes
Published ahead of print 6 August 2012
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Bartenschlager R. 1999. The NS3/4A proteinase of the hepatitis C virus: unravelling structure and function of an unusual enzyme and a prime target for antiviral therapy. J. Viral Hepat. 6:165–181 [DOI] [PubMed] [Google Scholar]
- 2. Bartenschlager R, Ahlborn-Laake L, Mous J, Jacobsen H. 1994. Kinetic and structural analyses of hepatitis C virus polyprotein processing. J. Virol. 68:5045–5055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bartenschlager R, Lohmann V, Wilkinson T, Koch JO. 1995. Complex formation between the NS3 serine-type proteinase of the hepatitis C virus and NS4A and its importance for polyprotein maturation. J. Virol. 69:7519–7528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reference deleted.
- 5. Bronowicki J-P, et al. 2012. Asunaprevir (ASV; BMS-650032), an NS3 protease inhibitor, in combination with peginterferon and ribavirin in treatment-naive patients with genotype 1 chronic hepatitis C infection. J. Hepatol. 56(Suppl 2):S431–S432 [Google Scholar]
- 6. Burns C, et al. 2004. Benzofuran compounds, compositions and methods for treatment and prophylaxis of hepatitis C viral infections and associated diseases. PCT international patent application WO2004/041201
- 7. Campbell JA, Good AC. 2002. Preparation of tripeptides as hepatitis C virus inhibitors. PCT international patent application WO2002/060926
- 8. Chayama K, et al. 2012. Dual therapy with the NS5A inhibitor BMS-790052 and the NS3 protease inhibitor BMS-650032 in HCV genotype 1b-infected null responders. Hepatology 55:742–748 [DOI] [PubMed] [Google Scholar]
- 9. Chen S-H, et al. 2005. P1 and P1′ optimization of [3,4]-bicycloproline P2 incorporated terapeptidyl α-ketoamide based HCV protease inhibitors. Lett. Drug Design Discov. 2:118–123 [Google Scholar]
- 10. Chou TC. 2006. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol. Rev. 58:621–681 [DOI] [PubMed] [Google Scholar]
- 11. Clackson T, Winter G. 1989. ‘Sticky feet’-directed mutagenesis and its application to swapping antibody domains. Nucleic Acids Res. 17:10163–10170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Coelmont L, et al. 2006. Ribavirin antagonizes the in vitro anti-hepatitis C virus activity of 2′-C-methylcytidine, the active component of valopicitabine. Antimicrob. Agents Chemother. 50:3444–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Don RH, Cox PT, Wainwright BJ, Baker K, Mattick JS. 1991. ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 19:4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Drexler D, et al. 2006. Development of an on-line automated sample clean-up method and liquid chromatography-tandem mass spectrometry analysis: application in an in vitro proteolytic assay. Anal. Bioanal Chem. 384:1145–1154 [DOI] [PubMed] [Google Scholar]
- 15. Fridell RA, et al. 2011. Distinct functions of NS5A in hepatitis C virus RNA replication uncovered by studies with the NS5A inhibitor BMS-790052. J. Virol. 85:7312–7320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fridell RA, Qiu D, Wang C, Valera L, Gao M. 2010. Resistance analysis of the hepatitis C virus NS5A inhibitor BMS-790052 in an in vitro replicon system. Antimicrob. Agents Chemother. 54:3641–3650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fried MW, et al. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975–982 [DOI] [PubMed] [Google Scholar]
- 18. Gane EJ, et al. 2011. Antiviral activity, safety, and pharmacokinetics of danoprevir/ritonavir plus PEG-IFN alpha-2a/RBV in hepatitis C patients. J. Hepatol. 55:972–979 [DOI] [PubMed] [Google Scholar]
- 19. Gao M, et al. 2010. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature 465:96–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ghany MG, Strader DB, Thomas DL, Seeff LB, and American Association for the Study of Liver Diseases 2009. Diagnosis, management, and treatment of hepatitis C: an update. Hepatology 49:1335–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Grobler JA, et al. 2003. Identification of a key determinant of hepatitis C virus cell culture adaptation in domain II of NS3 helicase. J. Biol. Chem. 278:16741–16746 [DOI] [PubMed] [Google Scholar]
- 22. Jacobson IM, et al. 2011. Telaprevir for previously untreated chronic hepatitis C virus infection. N. Engl. J. Med. 364:2405–2416 [DOI] [PubMed] [Google Scholar]
- 23. Kadow JF, et al. 2012. Discovery of BMS-791325, an allosteric NS5B replicase inhibitor for the treatment of hepatitis C, abstr. MEDI-23. 243rd American Chemical Society National Meeting and Exposition [Google Scholar]
- 24. Kim AI, Saab S. 2005. Treatment of hepatitis C. Am. J. Med. 118:808–815 [DOI] [PubMed] [Google Scholar]
- 25. Kuiken C, Simmonds P. 2009. Nomenclature and numbering of the hepatitis C virus. Methods Mol. Biol. 510:33–53 [DOI] [PubMed] [Google Scholar]
- 26. Lamarre D, et al. 2003. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature 426:186–189 [DOI] [PubMed] [Google Scholar]
- 27. Llinas-Brunet M, et al. 2004. A systematic approach to the optimization of substrate-based inhibitors of the hepatitis C virus NS3 protease: discovery of potent and specific tripeptide inhibitors. J. Med. Chem. 47:6584–6594 [DOI] [PubMed] [Google Scholar]
- 28. Lok AS, et al. 2012. Preliminary study of two antiviral agents for hepatitis C genotype 1. N. Engl. J. Med. 366:216–224 [DOI] [PubMed] [Google Scholar]
- 29. McHutchison JG, et al. 2009. Peginterferon alfa-2b or alfa-2a with ribavirin for treatment of hepatitis C infection. N. Engl. J. Med. 361:580–593 [DOI] [PubMed] [Google Scholar]
- 30. McPhee F, et al. 2012. Resistance analysis of the hepatitis C virus NS3 protease inhibitor asunaprevir. Antimicrob. Agents Chemother. 56:3670–3681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McPhee F, et al. 2009. The discovery and early development of the HCV NS3 protease inhibitor BMS-605339. Global Antiviral J. 5(Suppl 1):51 [Google Scholar]
- 32. Merck & Co 2011. VictrelisTM (boceprevir), package insert. Merck & Co., Whitehouse Station, NJ [Google Scholar]
- 33. Miller RH, Purcell RH. 1990. Hepatitis C virus shares amino acid sequence similarity with pestiviruses and flaviviruses as well as members of two plant virus supergroups. Proc. Natl. Acad. Sci. U. S. A. 87:2057–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Murakami E, et al. 2010. Mechanism of activation of PSI-7851 and its diastereoisomer PSI-7977. J. Biol. Chem. 285:34337–34347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Neurath H. 1989. The diversity of proteolytic enzymes. In Beynon RJ, Bond JS. (ed) Proteolytic enzymes: a practical approach. Oxford University Press, Oxford, United Kingdom [Google Scholar]
- 36. O'Boyle DR, 2nd, et al. 2005. Development of a cell-based high-throughput specificity screen using a hepatitis C virus-bovine viral diarrhea virus dual replicon assay. Antimicrob. Agents Chemother. 49:1346–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pasquinelli C, et al. 2012. Single and multiple ascending dose studies of the NS3 protease inhibitor, asunaprevir, in subjects with or without chronic hepatitis C. Antivir. Agents Chemother. 56:1838–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Perni RB, et al. 2006. Preclinical profile of VX-950, a potent, selective, and orally bioavailable inhibitor of hepatitis C virus NS3-4A serine protease. Antimicrob. Agents Chemother. 50:899–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Poordad F, et al. 2011. Boceprevir for untreated chronic HCV genotype 1 infection. N. Engl. J. Med. 364:1195–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rajagopalan R, et al. 2009. Inhibition and binding kinetics of the hepatitis C virus NS3 protease inhibitor ITMN-191 reveals tight binding and slow dissociative behavior. Biochemistry 48:2559–2568 [DOI] [PubMed] [Google Scholar]
- 41. Raney KD, Sharma SD, Moustafa IM, Cameron CE. 2010. Hepatitis C virus non-structural protein 3 (HCV NS3): a multifunctional antiviral target. J. Biol. Chem. 285:22725–22731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Reagan-Shaw S, Nihal M, Ahmad N. 2008. Dose translation from animal to human studies revisited. FASEB J. 22:659–661 [DOI] [PubMed] [Google Scholar]
- 43. Robertson B, et al. 1998. Classification, nomenclature, and database development for hepatitis C virus (HCV) and related viruses: proposals for standardization. International Committee on Virus Taxonomy. Arch. Virol. 143:2493–2503 [DOI] [PubMed] [Google Scholar]
- 44. Sarrazin C, et al. 2007. Dynamic hepatitis C virus genotypic and phenotypic changes in patients treated with the protease inhibitor telaprevir. Gastroenterology 132:1767–1777 [DOI] [PubMed] [Google Scholar]
- 45. Scola P, et al. 2010. Discovery of BMS-650032, an NS3 protease inhibitor for the treatment of hepatitis C, abstr. MEDI-38. Scientific Abstracts for the 239th American Chemical Society National Meeting and Exposition [Google Scholar]
- 46. Sheaffer AK, et al. 2011. Development of a chimeric replicon system for phenotypic analysis of NS3 protease sequences from HCV clinical isolates. Antivir. Ther. 16:705–718 [DOI] [PubMed] [Google Scholar]
- 47. Taliani M, et al. 1996. A continuous assay of hepatitis C virus protease based on resonance energy transfer depsipeptide substrates. Anal. Biochem. 240:60–67 [DOI] [PubMed] [Google Scholar]
- 48. Tautz N, Kaiser A, Thiel HJ. 2000. NS3 serine protease of bovine viral diarrhea virus: characterization of active site residues, NS4A cofactor domain, and protease-cofactor interactions. Virology 273:351–363 [DOI] [PubMed] [Google Scholar]
- 49. Thibeault D, et al. 2004. Sensitivity of NS3 serine proteases from hepatitis C virus genotypes 2 and 3 to the inhibitor BILN 2061. J. Virol. 78:7352–7359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vertex Pharmaceuticals Inc 2011. IncivekTM (telaprevir), package insert. Vertex Pharmaceuticals Inc., Cambridge, MA [Google Scholar]
- 51. Wakita T, et al. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. World Health Organization 2011. Hepatitis C key facts. http://www.who.int/mediacentre/factsheets/fs164/en/ Accessed July 2012
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.