Abstract
Salmonella genomic island 1 (SGI1) is a 43-kb integrative mobilizable element that harbors a great diversity of multidrug resistance gene clusters described in numerous Salmonella enterica serovars and also in Proteus mirabilis. The majority of SGI1 variants contain an In104-derivative complex class 1 integron inserted between resolvase gene res and open reading frame (ORF) S044 in SGI1. Recently, the international spread of ciprofloxacin-resistant S. enterica serovar Kentucky sequence type 198 (ST198) containing SGI1-K variants has been reported. A retrospective study was undertaken to characterize ST198 S. Kentucky strains isolated before the spread of the epidemic ST198-SGI1-K population in Africa and the Middle East. Here, we characterized 12 ST198 S. Kentucky strains isolated between 1969 and 1999, mainly from humans returning from Southeast Asia (n = 10 strains) or Israel (n = 1 strain) or from meat in Egypt (n = 1 strain). All these ST198 S. Kentucky strains did not belong to the XbaI pulsotype X1 associated with the African epidemic clone but to pulsotype X2. SGI1-J subgroup variants containing different complex integrons with a partial transposition module and inserted within ORF S023 of SGI1 were detected in six strains. The SGI1-J4 variant containing a partially deleted class 1 integron and thus showing a narrow resistance phenotype to sulfonamides was identified in two epidemiologically unrelated strains from Indonesia. The four remaining strains harbored a novel SGI1-J variant, named SGI1-J6, which contained aadA2, floR2, tetR(G)-tetA(G), and sul1 resistance genes within its complex integron. Moreover, in all these S. Kentucky isolates, a novel insertion sequence related to the IS630 family and named ISSen5 was found inserted upstream of the SGI1 complex integron in ORF S023. Thus, two subpopulations of S. Kentucky ST198 independently and exclusively acquired the SGI1 during the 1980s and 1990s. Unlike the ST198-X1 African epidemic subpopulation, the ST198-X2 subpopulation mainly from Asia harbors variants of the SGI1-J subgroup that are encountered mainly in the Far East, as previously described for S. enterica serovars Emek and Virchow.
INTRODUCTION
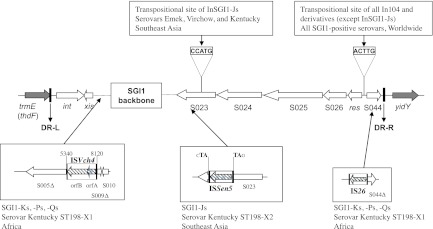
The Salmonella genomic island 1 (SGI1) is a 43-kb integrative mobilizable element initially described in worldwide epidemic multidrug-resistant (MDR; resistance to two or more antibiotic families) Salmonella enterica serovar Typhimurium phage type DT104 (here referred to as S. Typhimurium DT104) (1, 5, 11, 19). SGI1 is the first MDR genomic island identified in S. enterica, which contains a complex class 1 integron, named In104 (11, 19). Since the identification of SGI1 in S. Typhimurium DT104, variant SGI1 MDR complex integrons have been described in a wide variety of S. enterica serovars (>25) and more recently also in Proteus mirabilis. According to the SGI1 nomenclature, variants are classified from SGI1-A to SGI1-V (11, 21, 24). The complex In104 integron variants classically possess one or two cassette attachment sites (attI1) carrying various resistance gene cassette arrays, contain an IS6100 element, may contain additional resistance genes, and are bound by 25-bp inverted repeats IRi and IRt (1). In the great majority of these variants, except the SGI1-J subgroup, the complex In104 integron or variants of it are found always at the same position in the SGI1 scaffold, i.e., between the resolvase gene res (also named tnpR) and the open reading frame (ORF) S044 at the 3′ end of SGI1 (Fig. 1) (3, 4, 11, 19).
Fig 1.
Schematic view of the SGI1 backbone and its specific features encountered in various Salmonella enterica serovars. Gray arrows correspond to chromosomal genes flanking SGI1. DR-L and DR-R are the 18-bp left and right direct repeats, respectively, bracketing SGI1. The 5-bp transpositional target sites of complex integrons InSGI1-J and all others are indicated in ORF S023 and between the res gene and ORF S044, respectively. The three different IS elements are indicated by boxes containing black hatched arrows representing the transposase genes. Extended views of the insertion site in the SGI1 backbone are schematized below the map within boxes. The SGI1 variants, the Salmonella enterica serovars, and geographical origins are indicated for each genetic feature.
The SGI1-J subgroup is represented by the unique and peculiar SGI1 variants that have been characterized in two S. enterica serovars, i.e., Emek and Virchow, isolated from Southeast Asia and Oceania from 1993 to 2006 (3, 4, 14, 15, 24). The main characteristic of SGI1-J relies in that different complex integrons are inserted by transposition within the ORF S023 and not in the classical SGI1 integron insertion site, i.e., between the res gene and ORF S044 that is left empty (Fig. 1) (3, 4, 14, 15, 24). Levings et al. have proposed to classify this subgroup as a derivative named SGI2; nevertheless, according to high nucleotide identity of the SGI1 backbone, the classification as in SGI1-J1 to -J5 is more advisable and is related to different complex integrons, genetic rearrangements, and the presence of resistance genes floR2 (also called cmlA9) for chloramphenicol resistance, tetR(G) and tetA(G) for tetracycline resistance, and sul1 and dfrA1 for sulfamethoxazole/trimethoprim resistance (3, 4, 14, 15).
Since 2002, there is a worrying emergence of MDR and/or ciprofloxacin-resistant (CIPr) S. enterica serovar Kentucky isolates from European travelers returning from Africa (7, 12, 23). Recently, a multinational molecular epidemiologic study on human and nonhuman isolates demonstrated that this emergence concerned mainly a single S. Kentucky clone defined as the multilocus sequence type (MLST) ST198 and by pulsed-field gel electrophoresis (PFGE) pulsotype cluster X1. To date, this clone has infected hundreds of European travelers returning from various African and the Middle East countries (12). This ST198-X1 CIPr S. Kentucky clone was also isolated from chickens and turkeys in different African countries, suggesting that poultry is an important vehicle for human infection by this strain population (12). Interestingly, the majority of the isolates studied possessed SGI1 variants that presented two specific genetic rearrangements in the SGI1 backbone (Fig. 1). The first one corresponded to a deletion of an SGI1 5′ segment and its replacement by an ISVch4 element, and the second genetic characteristic was the presence of an IS26 element found inserted at the same precise position within ORF S044 of the SGI1 backbone (Fig. 1) (7). The presence of this IS26 element in ORF S044 is probably the key feature in the generation of variant SGI1-K, -P, and -Q MDR regions. These insertions of ISVch4 and IS26 indicate that all SGI1 islands of ST198-X1 S. Kentucky strains probably evolved from a common SGI1 ancestor. The earliest ST198-X1 strains possessing SGI1-K were isolated from French travelers returning from Egypt in 1996 (Table 1) (12).
Table 1.
Characteristics of S. enterica serovar Kentucky isolates from the French National Reference Center collection, 1959 to 1999
| Strain | Yr of isolation | Source | Country of origin | Antimicrobial resistance profilea | PFGE typeb | MLST typec | SGI1 |
|---|---|---|---|---|---|---|---|
| 98K | 1937 | Chicken | USA | Susceptible | X4 | ST198 | Absence |
| 2-59 | 1959 | Human | Tunisia | Susceptible | NT | ST728 | Absence |
| 3-59 | 1959 | Human | Angola | Susceptible | NT | ST152 | Absence |
| 1-61 | 1961 | Human | Senegal | Susceptible | X1d | ST198 | Absence |
| 1-63 | 1963 | Human | Vietnam | Susceptible | X19 | ST198 | Absence |
| 2-64 | 1964 | Fish meal | Portugal | Susceptible | X6 | ST152 | Absence |
| 1-66 | 1966 | Human | Vietnam | AMX | X18 | ST198 | Absence |
| 1-67 | 1967 | Fish meal | Portugal | Susceptible | NT | ST152 | Absence |
| 1-68 | 1968 | Human | Senegal | STR SMX | X1b | ST727 | Absence |
| 61-69 | 1969 | Turkey | France | TET | X2a | ST198 | Absence |
| 6-71 | 1971 | Human | Tahiti (French Polynesia) | Susceptible | NT | ST152 | Absence |
| 14-72 | 1972 | Human | Martinique (French West Indies) | Susceptible | X16 | ST198 | Absence |
| 2-75 | 1975 | Human | Senegal | Susceptible | X1a | ST198 | Absence |
| 3-75 | 1975 | Human | Djibouti | Susceptible | X11 | ST723 | Absence |
| 5-76 | 1976 | Soya bean | France | Susceptible | X4 | ST198 | Absence |
| 8-76 | 1976 | Superficial water | Indonesia | STR | NT | ST198 | Absence |
| 9-76 | 1976 | Meat flour | France | Susceptible | X9 | ST198 | Absence |
| 4-77 | 1977 | Poultry meal | France | STR | NT | ST198 | Absence |
| 12-78 | 1978 | Fish meal | Turkey | STR | X15 | ST198 | Absence |
| 18-78 | 1978 | Human | Iraq | STR | X17 | ST198 | Absence |
| 2-79 | 1979 | Human | Guinea | Susceptible | X9 | ST198 | Absence |
| 6-79 | 1979 | Human | Burkina Faso | Susceptible | X9 | ST724 | Absence |
| 4-82 | 1982 | Human | Morocco | Susceptible | X20 | ST393 | Absence |
| 1-83 | 1983 | Human | Gabon | Susceptible | X12 | ST198 | Absence |
| 8-84 | 1984 | Human | Ivory Coast | Susceptible | X9 | ST724 | Absence |
| 17-85 | 1985 | Human | Egypt | STR SMX | X14 | ST198 | Absence |
| 19-85 | 1985 | Meat | Egypt | STR SPE CHL SMX TET | X2b | ST198 | SGI1-J6 |
| 1-86 | 1986 | Human | Vietnam | STR SPE CHL SMX TET | X2b | ST198 | SGI1-J6 |
| 7-89 | 1989 | Human | Ivory Coast | Susceptible | NT | ST696 | Absence |
| 12-89 | 1989 | Human | Thailand | STR SPE CHL SMX TET | X2b | ST198 | SGI1-J6 |
| 4-90 | 1990 | Human | Israel | STR SPE CHL SMX TET | X2a | ST198 | Absence |
| 10-90 | 1990 | Human | Indonesia | SMX | X2d | ST198 | SGI1-J4 |
| 92-6276 | 1992 | Human | Indonesia | STR SPE CHL SMX TET | X2e | ST198 | SGI1-J6 |
| 92-6923 | 1992 | Human | India-Nepal | Susceptible | X2g | ST198 | Absence |
| 93-6429 | 1993 | Human | Indonesia | SMX | X2c | ST198 | SGI1-J4 |
| 93-10205 | 1993 | Human | Réunion Islande | Susceptible | X12 | ND | Absence |
| 94-1725 | 1994 | Human | Senegal | AMP SUL | X1a | ND | Absence |
| 94-3543 | 1994 | Human | East Africa | Susceptible | X1c | ND | Absence |
| 94-7021 | 1994 | Human | India | Susceptible | X2h | ST198 | Absence |
| 94-7842 | 1994 | Human | India | Susceptible | X2a | ST198 | Absence |
| 94-9173 | 1994 | Human | India | Susceptible | X2j | ST198 | Absence |
| 95-5335 | 1995 | Human | Mauritania | Susceptible | X1c | ND | Absence |
| 96-8196 | 1996 | Human | Africa | Susceptible | X1a | ND | Absence |
| 96-11313 | 1996 | Human | Egypt | STR SPE GEN SMX TET | X1a | ST198 | SGI1-Ks |
| 97-0568 | 1997 | Human | Africa | STR SMX TMP | X1a | ND | Absence |
| 97-2801 | 1997 | Human | Tunisia | Susceptible | X8 | ND | Absence |
| 97-2858 | 1997 | Human | Tunisia | Susceptible | X20 | ST393 | Absence |
| 97-6819 | 1997 | Human | Egypt | AMX STR SPE SMX TET | X1a | ST198 | SGI1-Ks |
| 97-11473 | 1997 | Human | Egypt | AMX STR SPE GEN SMX TET NAL | X1k | ST198 | SGI1-Ks |
| 99-2998 | 1999 | Human | Egypt | AMX CRO CAZ FOX STR CHL SMX TMP TET NALd | X1d | ND | Absence |
AMX, amoxicillin; CRO, ceftriaxone; CAZ, ceftazidime; FOX, cefoxitin; CHL, chloramphenicol; GEN, gentamicin; KAN, kanamycin; SPE, spectinomycin; NAL, nalidixic acid; STR, streptomycin; SMX, sulfamethoxazole; TET, tetracycline; and TMP, trimethoprim. Bold antibiotic resistances are conferred by SGI1.
NT, not typeable due to lysis.
ND, not done.
This strain contains a blaCMY-2 gene carried by an IncA/C 150-kb plasmid.
French territory in the Indian Ocean.
In the present work, we conducted a retrospective study of S. Kentucky strains collected by the French National Reference Centre for Salmonella (Institut Pasteur, Paris, France) acquired worldwide before the emergence of the African ST198-X1 CIPr S. Kentucky population. These isolates were typed by MLST and XbaI-PFGE, and SGI1 variants were characterized for those that were antibiotic resistant.
MATERIALS AND METHODS
Bacterial strains and antibiotic susceptibility testing.
Fifty S. Kentucky strains were used for this study. Forty-nine were from the collection of the French National Reference Centre for Salmonella at the Institut Pasteur and were isolated from 1959 to 1999, mainly from humans (stool) who acquired salmonellosis during or immediately after foreign travel (Table 1). The S. Kentucky reference strain 98K from the collection of the WHO Collaborative Centre for Reference and Research on Salmonella at the Institut Pasteur was isolated from a chick in the United States in 1937 (8). These strains were partially characterized in a previous study reporting the emergence of the epidemic ST198-X1 CIPr S. Kentucky population (12). The three SGI1-positive ST198-X1 MDR S. Kentucky isolates (96-11313, 97-11473, 97-6819) represented the first strains of the ST198-X1 MDR clonal population harboring SGI1-Ks variants and isolated from humans in France who acquired salmonellosis during or immediately after travel to Egypt (12). The S. Kentucky strains and control strains S. Typhimurium DT104 BN9181 (SGI1) and S. Virchow SV20 (SGI1-J4) and B98 (SGI1-J3) were grown at 37°C in brain heart infusion or Luria-Bertani broth or on agar plates. The strains were tested for their antibiotic susceptibilities by the disk diffusion assay on Mueller-Hinton plates according to the guidelines of the Antibiogram Committee of the French Society for Microbiology (18). Susceptibility was determined using disks containing 32 antibiotics as previously described (7). All antibiotic disks except for florfenicol were purchased from Bio-Rad (Marnes-la-Coquette, France). Florfenicol disks were obtained from Schering-Plough Animal Health (Kenilworth, NJ).
Molecular typing.
PulseNet standard PFGE of XbaI-digested chromosomal DNA was performed on all S. enterica serovar Kentucky strains (20). Additional representative strains of different pulsotypes were included for genetic relatedness comparison and to assess the genetic diversity of the S. Kentucky strains. PFGE profiles were compared by using Bionumerics software, version 6.6 (Applied Maths, Sint-Martens-Latem, Belgium). MLST was performed on 42 isolates as described previously (12). Data were submitted to the MLST database website (http://mlst.ucc.ie/mlst/dbs/Senterica).
SGI1 characterization, PCR mapping, and Southern blot hybridization.
SGI1 and its chromosomal location were detected using primers corresponding to the left and right junctions (DR-L and DR-R) in the chromosome as described previously (4–7). PCR mapping of the SGI1 scaffold (outside complex integron) was performed using PCRs and primers previously described (3, 4, 7). The atypical insertion site of the complex integrons InSGI1-J (in ORF S023) was further assessed by Southern blot hybridization of whole genomic DNA cut by XbaI (Promega, Charbonnières, France) by using probe p1-9 as previously described (1, 4, 5, 7). This probe corresponds to a 2-kb EcoRI fragment comprising parts of the S023 and S024 ORFs. To assess variant SGI1 MDR regions and their insertion sites identified in this study, a complete integron PCR mapping and integron junction PCR were also performed using primers previously described (3, 4, 7). Integron cassette arrays were amplified by PCR using primers CS1 and CS2 (13).
Nucleotide sequence accession number.
A partial nucleotide sequence from the SGI1-J6 variant reported in this paper has been deposited in the GenBank database under accession number JQ796373. The novel insertion sequence ISSen5 of the IS630 family has been deposited in the IS Finder database (http://www-is.biotoul.fr).
RESULTS AND DISCUSSION
S. enterica serovar Kentucky, 1959 to 1999: population structure and antimicrobial susceptibility.
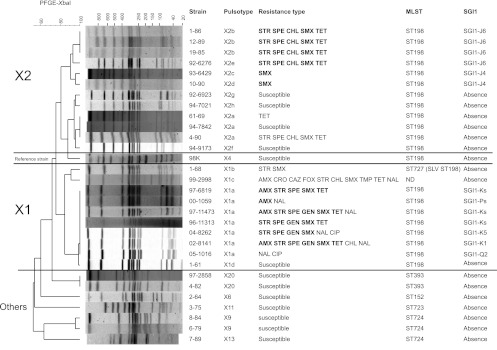
The genetic relationship between strains was studied by MLST and PFGE (Table 1 and Fig. 2). MLST separated the strains into 8 STs grouped into 6 ST complexes (containing STs that share all but one or two alleles) that often correspond to monophyletic lineages. The ST198 complex was the most prevalent (31/42 strains tested, 74%). By XbaI-PFGE, most of the ST198 strains clustered into two groups, X1 and X2 (with a Dice similarity coefficient of >80%) (Fig. 2). The PFGE grouping was correlated with the geographic origin of the strains, i.e., different African countries for X1 and mainly different Asian countries (India, Thailand, Indonesia, Vietnam) for X2 (Table 1 and Fig. 2). Before the 2000s, the majority of ST198 S. Kentucky strains were susceptible to all antibiotics tested (Table 1). The first isolations of ST198 MDR S. Kentucky occurred during the 1980s and consisted of strains from human in Vietnam, Thailand, Israel, and India and from food in Egypt. These strains belonged to the highly related X2a, X2b, and X2c pulsotypes (Table 1). As recently described, the first ST198-X1 MDR S. Kentucky isolates appeared in Egypt during the second part of the 1990s. These ST198-X1 MDR strains had acquired the genomic island SGI1-K. Then sequential acquisition of mutations in gyrA and parC topoisomerase subunits resulted in an epidemic CIPr-MDR S. Kentucky clonal population spreading in Africa and the Middle East (12).
Fig 2.
Analysis of XbaI-PFGE profiles obtained among 30 representative Salmonella enterica serovar Kentucky strains from humans and nonhumans during the period 1937 to 2004. Strain code, pulsotype, resistance type, MLST type, and the presence of the SGI1 are indicated to the right of the PFGE profiles. Antibiotics: AMX, amoxicillin; CRO, ceftriaxone; CAZ, ceftazidime; FOX, cefoxitin; CHL, chloramphenicol; CIP, ciprofloxacin; GEN, gentamicin; NAL, nalidixic acid; SMX, sulfamethoxazole; SPE, spectinomycin; STR, streptomycin; TET, tetracycline; TMP, trimethoprim. Bold antibiotic resistances are conferred by SGI1.
Detection of SGI1 in the ST198-X2 MDR S. Kentucky population, Southeast Asia.
Fifty S. Kentucky strains were analyzed for the presence of SGI1 by junction PCRs with the Salmonella chromosome (Table 1). SGI1 was thus detected in 9 ST198 S. Kentucky strains isolated between 1985 and 1997. It was found integrated in the 3′ end of the chromosomal gene trmE (also called thdF). Three SGI1-positive ST198-X1 MDR S. Kentucky strains were previously studied (12). The six remaining SGI1-positive S. Kentucky strains clustered into the second main PFGE group X2 (Table 1 and Fig. 1). Interestingly, these 6 ST198-X2 SGI1-positive S. Kentucky strains showed different antimicrobial resistance phenotypes never seen as encoded by SGI1 (Table 1). In all strains studied here, the second attachment site of SGI1, i.e., between the chromosomal genes sodB and purR, was found empty (data not shown) (6).
Genetic characterization of SGI1 in ST198-X2 MDR S. Kentucky.
A genetic characterization of SGI1 in ST198-X2 S. Kentucky was performed to compare the MDR genomic islands found in both major pulsotypes of ST198 S. Kentucky before the emergence of the epidemic ST198-X1 CIPr-MDR population in Africa. All ST198-X2 S. Kentucky strains yielded fragments from the integrase gene int to the conjugal transfer gene traG (overlaying the first 10 kb of SGI1) of the sizes expected from S. Typhimurium DT104 control strain BN9181 harboring SGI1. These results indicated that the SGI1-positive ST198-X2 strain did not show the insertion/deletion of ISVch4 in the region spanning from ORFs S005 to S009 as previously described for SGI1-Ks, -Ps, and -Qs from ST198-X1 S. Kentucky from Africa (GenBank accession no. EU664602) (7). The other typical feature of the SGI1-Ks, -Ps, and -Qs is the insertion of an IS26 element within the 5′ left end of ORF S044 that represented the conserved 3′ right end of these variants. All ST198-X2 S. Kentucky strains were negative for the presence of an IS26 element within ORF S044 by PCR using previously described primers (7). Thus, in addition to their specific antimicrobial resistance phenotypes, the ST198-X2 S. Kentucky strains seemed to contain specific SGI1 variants that differ from those found in the epidemic ST198-X1 population from Africa.
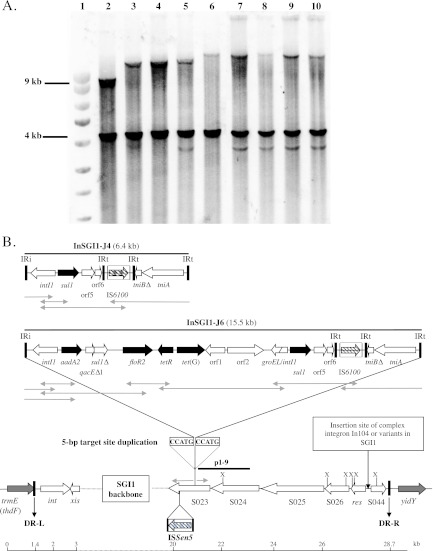
A large central region of SGI1 from positions 11949 to 24937 (GenBank accession no. AF261825) was assessed by Southern blot hybridization with probe p1-9 as previously described (1, 4–7). This probe revealed two different XbaI hybridization profiles for the ST198-X2 S. Kentucky isolates (Fig. 3A). In all ST198-X2 isolates, the p1-9 probe revealed the expected 4-kb size as in the SGI1-carrying control strains, indicating that the ORFs S024 to S026 were conserved (Fig. 3). However, these isolates showed second fragments larger than the expected 9-kb size (Fig. 3A). Similar p1-9 hybridization patterns have been previously described in the SGI1-J variant subgroup and were due to the insertion of various complex integrons in S023 (Fig. 3A) (3, 4). Thus, to localize the complex integron in SGI1 from ST198-X2 S. Kentucky strains, a first PCR was carried out to assess the classical insertion site of complex integron In104 or variants, i.e., between the resolvase gene res and ORF S044 using previously described primers (3, 4). The six SGI1-positive ST198-X2 S. Kentucky strains yielded an approximately 750-bp PCR product that indicated the absence of insertion between the res gene and ORF S044 (Fig. 3B).
Fig 3.
(A) Southern blot hybridization with the p1-9 probe of XbaI-digested genomic DNAs of S. enterica serovar Typhimurium DT104 control strain BN9181 carrying SGI1 (lane 2), serovar Virchow control strain SV20 carrying SGI1-J4 (lane 3), serovar Kentucky strain 10-90 (lane 4), serovar Kentucky strain 93-6429 (lane 5), serovar Virchow control strain B98 carrying SGI1-J3 (lane 6), serovar Kentucky strain 19-85 (lane 7), serovar Kentucky strain 1-86 (lane 8), serovar Kentucky strain 12-89 (lane 9), and serovar Kentucky strain 92-6276 (lane 10). Kbp DNA marker (Smartladder, Eurogentec, Seraing, Belgium) (lane 1). (B) Genetic organization of the complex class 1 integron in SGI1-J4 and SGI1-J6 from S. Kentucky strains. Black and gray arrows correspond to SGI1 antibiotic resistance genes and chromosomal genes flanking SGI1, respectively. IS elements are indicated by boxes containing black hatched arrows representing the transposase genes. DR-L and DR-R are the 18-bp left and right direct repeats, respectively, bracketing SGI1. IRi and IRt are 25-bp imperfect inverted repeats defining the left and right ends of complex class 1 integrons. The usual insertion point of the complex integron In104 or variants of it in SGI1 and the 5-bp target site duplication flanking the complex integron (InSGI1-J4 and -J6) in ORF S023 are indicated. The p1-9 probe and XbaI restriction sites (X) are indicated. PCRs carried out to map the InSGI1-J complex integrons are indicated by light-gray arrows under each map.
A set of junction PCRs specific for the SGI1-J complex integron insertion site was performed using primers previously described (3). The right junction PCR yielded a positive result for all SGI1-positive ST198-X2 S. Kentucky strains of the expected size as in SGI1-J S. Virchow control strains (data not shown). This result indicated that the complex integron was inserted within ORF S023 (Fig. 3B). The left PCR junction between the SGI1 backbone (S023) and the complex integron revealed a specific feature of SGI1 found in all ST198-X2 S. Kentucky strains. This junction was positive, but the amplified fragment was ca. 1.2 kb larger than that of SGI1-J S. Virchow control strains (data not shown) (3, 4). This 3.5-kb PCR product was sequenced (GenBank accession no. JQ796373). Sequence analysis using BLASTN confirmed the precise insertion site of the complex integron InSGI1-J at position 19747 of the SGI1 backbone (GenBank accession no. AF261825). Moreover, sequence analysis identified an insertion sequence element of 1,153 bp inserted in ORF S023 at position 19054 ca. 690 bp upstream of InSGI1-J (GenBank accession no. AF261825 and JQ796373) (Fig. 3B).
Novel ISSen5 element.
This novel IS element showed 84% nucleotide identity to ISSod16 (in the IS Finder database at http://www-is.biotoul.fr) described for Shewanella oneidensis strain MR-1 (GenBank accession no. AE014299). This IS belongs to the IS630 family and shares all its characteristics: (i) its length between 1,100 to 1,200 bp, (ii) a single long reading frame coding for the transposase (87% and 65% amino acid identities to the transposase of ISSod16 and IS630, respectively), (iii) its preferred target sequence (5′-CTAG-3′), (iv) and a target TA dinucleotide duplication created upon insertion in SGI1 (Fig. 2B) (2, 17). This novel IS of the IS630 family has been deposited in the IS Finder database and named ISSen5 (GenBank accession no. JQ796373). Interestingly, the insertion of ISSen5 in SGI1 seems to be specific to the SGI1-J variants encountered in ST198-X2 S. Kentucky, as this IS element is absent in SGI1-J from S. Virchow and S. Emek from Taiwan and Thailand, respectively (3, 4, 14, 15, 24).
New SGI1-J complex integron variant.
The gene cassette arrays of ST198-X2 S. Kentucky strains were determined by PCR using previously described primers CS1 and CS2 (13). Four MDR SGI1-positive isolates harbored a single 1-kb cassette array, and no amplification products were observed in the two sulfamethoxazole-resistant isolates (data not shown). This result suggested the occurrence of the streptomycin/spectinomycin resistance gene cassette aadA2 often found at the first cassette attachment site attI1 of In104. The presence of the gene cassette aadA2 was confirmed by two PCRs using a forward primer in S023 or in the intI1 gene (Fig. 3B). In these four MDR ST198-X2 S. Kentucky strains, the InSGI1-J integron PCR mapping confirmed the presence of the resistance genes floR2 and tetR(G)-tetA(G), a specific deletion that removes part of the second attI1 site and part of the 3′-CS on the right, the IRt-flanked IS6100 element, and the right outer end of the mercury resistance transposon Tn5058 (tniBΔ, tniA, and a third IRt copy, Fig. 3B) (3, 4). Together, all these results allowed the identification of a novel SGI1-J variant, which was named SGI1-J6 according to the SGI1 nomenclature (3, 4).
The two remaining SGI1-positive ST198-X2 S. Kentucky strains were resistant only to sulfamethoxazole and were negative for floR2 and tetR(G)-tetA(G). Two additional PCRs were carried out using a forward primer in the intI1 gene and reverse primers in the sul1 gene or IS6100 element (Fig. 3B). The resulting PCR products were of the expected sizes, as in InSG1-J4 (data not shown). The variant InSGI1-J4 that harbored a deletion that removed part of the attI1 site, the gene cassette, and part of the 3′-conserved segment may be due to a recombinational event between the 2 copies of the intI1 sequences (Fig. 2B) (3).
Concluding remarks.
The present study revealed specific characteristics encountered among S. Kentucky ST198 strains from Asian countries. These strains clustered into a specific XbaI-PFGE group, X2, and since the mid 1980s have been show to harbor SGI1 variants (SGI1-J) different from those (SGI1-K, -P, and -Q) identified in the African ST198-X1 S. Kentucky clone. In Asian MDR ST198-X2 strains, SGI1-J variants presented, as the first and specific characteristic, the insertion of the novel ISSen5 element described in the present study within ORF S023 (Fig. 1 and 3B). The second feature is the transposition of the complex integron InSGI1-J within ORF S023 (Fig. 1 and 3B). This feature was also identified with an identical insertion site into other SGI1-J variants carrying S. Emek and S. Virchow with Asian travel history (Fig. 1) (4, 14, 15). The ISVch4 and ISSen5 elements in SGI1 variants of the ST198-X1 and ST198-X2 populations, respectively, suggested that the aquatic ecosystem probably plays a role in genetic exchanges between environmental bacteria and human enteric pathogens. Interestingly, one SGI1-J6 strain carrying S. Kentucky was isolated from meat from Egypt (Table 1), indicating that in this same period, the SGI1-J-positive ST198-X2 S. Kentucky population was not restrictively limited to Southeast Asia. A similar description of the SGI1-K1 variant in one S. Kentucky strain, SRC73 isolated in 2001 from spice imported into Australia from India, suggested also the spread of the ST198-X1 clone outside Africa (16). The recent expansion of the ST198-X1-SGI1-K population in Asia is also observed in travelers at the French National Reference Centre (S. Le Hello and F. X. Weill, unpublished data).
It is noteworthy to mention that the ST198-X2 S. Kentucky strains isolated in 1985 and 1986 represent some of the oldest S. enterica isolates harboring SGI1 concomitantly to the first isolations of the MDR S. enterica serovar Typhimurium DT104 from gulls and exotic birds in the United Kingdom (22).
In conclusion, ST198 S. Kentucky represents a propitious serotype for the acquisition of the SGI1, as during the 1980s and the 1990s, two subpopulations of ST198 S. Kentucky acquired independently the SGI1 of 2 different types. Moreover, ST198 S. Kentucky carrying SGI1 confirmed the spread of this MDR genomic island in other S. enterica serovars before the worldwide epidemic spread of the MDR S. Typhimurium DT104 clone. Different sources, i.e., geographic, environmental ecosystem, bacterial species, animal species, etc., may have been implicated in the generation of different lineages of SGI1. Thorough epidemiological studies at the level of the bacteria and of the MDR genetic element allowed fine identifications of clonal populations geographically related and/or of related MDR genetic elements.
Moreover, the most recently described variant of P. mirabilis, SGI1-V, contained for the first time an extended-spectrum β-lactamase gene, blaVEB-6, and a quinolone resistance determinant, qnrA1, and thus represents an alarming feature in regard to the remaining treatment choices (21). In the same public health concern, the SGI1-S variant from S. enterica serovar Virchow SL491 isolated in the United States from children returning from India harbored a 16S rRNA methyltransferase-encoding gene, rmtC, conferring wide-range aminoglycoside resistance (9, 10, 24). The great plasticity of the MDR region of SGI1 associated with its ability of horizontal transfer could contribute to its spread and persistence among other clinical pathogenic bacteria and could have significant implications for physicians.
ACKNOWLEDGMENTS
This work was supported by the Institut Pasteur and by public funds from the French National Institute of Agronomic Research.
Footnotes
Published ahead of print 16 July 2012
REFERENCES
- 1. Boyd DA, et al. 2001. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 183:5725–5732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chandler M, Mahillon J. 2002. Insertion sequences revisited, p 305–366 In Craig NL, Craigie R, Gellert M, Lambowitz AM. (ed), Mobile DNA II. ASM Press, Washington, DC [Google Scholar]
- 3. Chu C, et al. 2012. Salmonella genomic island 1-J variants associated with change in the antibiotic resistance gene cluster in multidrug-resistant Salmonella enterica serovar Virchow isolated from humans, Taiwan, 2004-2006. Clin. Microb. Infect. 18:47–53 [DOI] [PubMed] [Google Scholar]
- 4. Doublet B, Chu C, Chiu C-H, Fan Y-C, Cloeckaert A. 2009. Truncated tni module adjacent to the complex integron of Salmonella genomic island 1 in Salmonella enterica serovar Virchow. Antimicrob. Agents Chemother. 53:824–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Doublet B, Boyd DA, Mulvey MR, Cloeckaert A. 2005. The Salmonella genomic island 1 is an integrative mobilizable element. Mol. Microbiol. 55:1911–1924 [DOI] [PubMed] [Google Scholar]
- 6. Doublet B, Golding GR, Mulvey MR, Cloeckaert A. 2008. Secondary chromosomal attachment site and tandem integration of the mobilizable Salmonella genomic island 1. PLoS One 3:e2060 doi:10.1371/journal.pone.0002060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Doublet B, et al. 2009. Novel insertion sequence and transposon-mediated genetic rearrangements in genomic island SGI1 of Salmonella enterica serovar Kentucky. Antimicrob. Agents Chemother. 53:3745–3754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Edwards PR. 1938. A new Salmonella type: Salmonella Kentucky. J. Hyg. (Lond.) 38:306–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Folster JP, Rickert R, Barzilay EJ, Whichard JM. 2009. Identification of the aminoglycoside resistance determinants armA and rmtC among non-Typhi Salmonella isolates from humans in the United States. Antimicrob. Agents Chemother. 53:4563–4564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fricke WF, et al. 2011. Comparative genomics of 28 Salmonella enterica isolates: evidence for CRISPR-mediated adaptive sublineage evolution. J. Bacteriol. 193:3556–3568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hall RM. 2010. Salmonella genomics islands and antibiotic resistance in Salmonella enterica. Future Microbiol. 5:1525–1538 [DOI] [PubMed] [Google Scholar]
- 12. Le Hello S, et al. 2011. International spread of an epidemic population of Salmonella enterica serotype Kentucky ST198 resistant to ciprofloxacin. J. Infect. Dis. 204:675–684 [DOI] [PubMed] [Google Scholar]
- 13. Lévesque C, Piché L, Larose C, Roy PH. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Levings RS, Lightfoot D, Partridge SR, Hall RM, Djordjevic SP. 2005. The genomic island SGI1, containing the multiple antibiotic resistance region of Salmonella enterica serovar Typhimurium DT104 or variants of it, is widely distributed in other S. enterica serovars. J. Bacteriol. 187:4401–4409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Levings RS, Djordjevic SP, Hall RM. 2008. SGI2, a relative of Salmonella genomic island SGI1 with an independent origin. Antimicrob. Agents Chemother. 52:2529–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Levings RS, Partridge SR, Djordjevic SP, Hall RM. 2007. SGI1-K, a variant of the SGI1 genomic island carrying a mercury resistance region, in Salmonella enterica serovar Kentucky. Antimicrob. Agents Chemother. 51:317–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mahillon J, Chandler M. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Members of the SFM Antibiogram Committee 2003. Comité de l'antibiogramme de la société française de microbiologie report 2003. Int. J. Antimicrob. Agents 21:364–391 [DOI] [PubMed] [Google Scholar]
- 19. Mulvey MR, Boyd DA, Olson AB, Doublet B, Cloeckaert A. 2006. The genetics of Salmonella genomic island 1. Microbes Infect. 8:1915–1922 [DOI] [PubMed] [Google Scholar]
- 20. Ribot EM, et al. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59–67 [DOI] [PubMed] [Google Scholar]
- 21. Siebor E, Neuwirth C. 2011. The new variant of Salmonella genomic island 1 (SGI1-V) from a Proteus mirabilis French clinical isolate harbours blaVEB-6 and qnrA1 in the multiple antibiotic resistance region. J. Antimicrob. Chemother. 66:2513–2520 [DOI] [PubMed] [Google Scholar]
- 22. Threlfall EJ. 2000. Epidemic Salmonella Typhimurium DT 104—a truly international multiresistant clone. J. Antimicrob. Chemother. 46:7–10 [DOI] [PubMed] [Google Scholar]
- 23. Weill F-X, et al. 2006. Ciprofloxacin-resistant Salmonella Kentucky in travelers. Emerg. Infect. Dis. 12:1611–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wilson NL, Hall RM. 2010. Unusual class 1 integron configuration found in Salmonella genomic island 2 from Salmonella enterica serovar Emek. Antimicrob. Agents Chemother. 54:513–516 [DOI] [PMC free article] [PubMed] [Google Scholar]