Abstract
As a potential antimicrobial, the bacteriophage lysin PlyG has been reported to specifically recognize Bacillus anthracis vegetative cells only and to kill B. anthracis vegetative cells and its germinating spores. However, how PlyG interacts with B. anthracis spores remains unclear. Herein, a 60-amino-acid domain in PlyG (residues 106 to 165), located mainly in the previously identified catalytic domain, was found able to specifically recognize B. anthracis spores but not vegetative cells. The exosporium of the spores was found to be the most probable binding target of this domain. This is the first time that a lysin for spore-forming bacteria has been found to have separate domains to recognize spores and vegetative cells, which might help in understanding the coevolution of phages with spore-forming bacteria. Besides providing new biomarkers for developing better assays for identifying B. anthracis spores, the newly found domain may be helpful in developing PlyG as a preventive antibiotic to reduce the threat of anthrax in suspected exposures to B. anthracis spores.
INTRODUCTION
The Gram-positive Bacillus anthracis bacterium, causing the disease anthrax in humans, is a well-known bioterrorism agent requiring worldwide medical attention (5, 15, 17, 20, 44). This spore-forming bacterium is a member of the Bacillus cereus group, along with B. cereus, Bacillus thuringiensis, and Bacillus mycoides (13). The entire spore of B. anthracis is encased by a heavily glycosylated exosporium (10, 43), which functions as a semipermeable barrier and a matrix for binding of molecules involved in defense, germination control, and other interactions of the spores with the environment (18). Under the exosporium is a much thinner coat layer (11, 12, 31, 38), which is critical for resistance properties as well as pathogenic effects (26). Because they are physicochemically stable and resistant to antiseptics, extreme temperature, high pressure, and UV irradiation (3, 33), B. anthracis spores are the foremost agent in anthrax threats. Therefore, the control of the spores is of great importance for combating bioterrorism and for the effective treatment of anthrax.
Lysins, produced by bacteriophages to digest the bacterial cell wall for the release of progeny virions, have been considered effective anti-infective agents for control of Gram-positive bacteria (4, 8, 27, 29, 32). PlyG, produced by gamma phage, has been reported to be an effective and specific agent for killing B. anthracis vegetative cells (40). The C terminus of PlyG (the site from residue 156 to residue 233) was identified as the cell wall binding domain (CBD), which recognizes vegetative cells, and the N terminus of PlyG (the site from residue 1 to residue 155) was previously considered to be the catalytic domain for destroying the target cells (9). Further research verified that the CBD of PlyG can recognize only the germinating form, but not spores, of B. anthracis (9). In vitro truncation analysis indicated that the residues from 190 to 199 of PlyG were necessary and sufficient for vegetative cell binding (21, 39). The critical region for the catalytic activity of PlyG (the site from residue 1 to residue 90) was also characterized (22). PlyG has also been reported to kill the germinating form of B. anthracis spores (40). However, the mechanism behind this remains unclear, and limited evidence is available on how PlyG interacts with B. anthracis spores.
In this study, as a step to understanding how PlyG interacts with B. anthracis spores, in vitro variant truncated fragments of PlyG were constructed and binding assays were performed to identify whether there is a binding domain in PlyG to recognize B. anthracis spores. Surprisingly, a 60-amino-acid fragment located mainly in the previously identified catalytic domain was found to specifically recognize B. anthracis spores but not vegetative cells, indicating that PlyG uses different binding domains to recognize spores and cells. These results provide new insight into the mechanism of interaction between PlyG and B. anthracis spores and therefore may provide further clues to finding or developing new lysins for controlling B. anthracis spores.
MATERIALS AND METHODS
Bacteria.
All experiments involving live B. anthracis spores and vegetative cells were performed in a biosafety level 3 laboratory. The experimenters were equipped with masks, gloves, and exposure suits. Bacillus anthracis A16 was cultured in standard LB medium and harvested as vegetative cells, as reported previously (2, 7, 50). The washed vegetative cells were heat treated for 5 min under boiling water before staining with the truncated proteins and the synthetic peptides. Spores were prepared on modified Difco sporulation medium (DSM), following the procedures described elsewhere (14, 36, 42, 49). The spores were inactivated immediately after harvest with 1% formaldehyde for more than 24 h to make sure of their dormancy during the experiments.
Escherichia coli BL21(DE3) was used for cloning and expression of the recombinant fusion proteins of PlyG and enhanced green fluorescent protein (EGFP). A total of 20 kinds of bacterial strains from 10 genera, comprising 17 bacillus strains and three other Gram-negative bacterial strains, were used in the binding selectivity assays, and a subset of these was used as labeling controls for the fluorescence analysis.
Construction of recombinant proteins.
The original PlyG gene sequence (GenBank accession no. AF536823) was chemically synthesized by Songon Biotech (Shanghai, China). Fragments from PlyG were genetically fused with the N terminus of EGFP, and eight corresponding expression plasmids, pET-EP9, pET-EP0, pET-EP3, pET-EC3, pET-EC5, pET-EC6, pET-EC8, and pET-EG1, were constructed (see Table S1 in the supplemental material). To obtain active PlyG, the plasmid pBAD-plyG was created (see Table S1). All the resulting clones were confirmed by sequencing.
Synthetic peptides.
Three synthetic peptides (see Table S1) were used in the binding assay, with an N-terminal fluorescein isothiocyanate (FITC) modification. One peptide with an N-terminal cysteine was used to react with 20 nm Au nanoparticles (BBI).
Protein expression and purification.
The recombinant proteins were expressed in the E. coli BL21(DE3) strain in standard LB medium and purified following procedures described previously (40, 54), with minor modifications. For pBAD-containing strains, protein was induced with 0.2% l-arabinose at 16°C for 8 h. Cells were washed, resuspended in 20 mM phosphate buffer at pH 6.6, and lysed with sonication. PlyG, which passed through a HiTrap Q Sepharose FF column (GE Healthcare), bound to a HiTrap SP Sepharose FF column (GE Healthcare) and was eluted in a linear gradient containing 1 M NaCl. For pET-containing strains, protein expression was induced with 1 mM isopropyl β-d-thiogalactoside (IPTG) when an optical density of 0.6 to 0.8 was reached. After induction, the bacteria were incubated overnight at 16°C to allow expression. Purification was achieved through the His6 tag, following the general protocol using a nickel nitrilotriacetic acid column, washing and eluting with imidazole solutions with concentrations of 60 and 265 mM, respectively. Collected fractions were dialyzed against 1× phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4 · H2O, 1.4 mM KH2PO4, pH 7.4) and then stored at −80°C until use after being quantitated by the Bradford assay using bovine serum albumin (BSA) as a standard.
Staining and microscopy of bacterial cells and spores.
To study the binding capabilities of the recombinant proteins, all the bacterial cells and spores tested were stained with the EGFP-tagged truncated proteins and EGFP (as a control), respectively. For bacterial cells, harvested cells were washed twice with 1× PBS by centrifuging (12,000 × g for 1 min) and then inactivated by a heat treatment (5 min under boiling water). Then, the cells were reacted with an excess of the EGFP-tagged proteins at 37°C for 30 min to ensure completed labeling. After washing three times with PBSB (1× PBS and 0.1% BSA) by centrifuging (12,000 × g for 1 min), the labeled cells were analyzed by using a fluorescence microscope (Delta Vision Personal DV; Applied Precision) and/or a laser scanning confocal microscope (TCS-SP2; Leica, Germany).
For the spores, all were preblocked with PBSM (1× PBS, pH 7.4, and 5% defatted milk powder) at 37°C for 2 h and then washed three times with PBST buffer (1× PBS and 0.05% Tween 20) by centrifuging (12,000 × g for 1 min) before staining with PBSB, EGFP, and EGFP-tagged proteins with or without l-alanine and d-alanine, respectively. All B. anthracis spores were completely inactivated by 1% formaldehyde after harvest to ensure biosafety and their dormancy state during the staining process, unless otherwise indicated. After reacting with an excess of EGFP and the EGFP-tagged proteins at 37°C for 30 min, the spores were washed three times with PBST buffer by centrifuging (12,000 × g for 1 min) before analysis by using the Delta fluorescence microscope. For fluorescence quantitative analysis, the total fluorescence intensity of a special defined region containing only one spore was calculated by the instrument's software automatically. The area of each defined region was fixed in all of the assays, and all the images were captured under the same instrument conditions. The specificity of EP0 was statistically evaluated by comparing the average fluorescence intensity of each spore (>300 spores) with the background fluorescence staining with the buffer PBSB and the control fluorescence staining with EGFP.
SPF assay.
A subtraction assay was developed to evaluate the binding affinities of the EGFP-fused truncated proteins for B. anthracis cells and spores individually. To achieve this, pretreated B. anthracis cells and spores were diluted 10-fold serially and stained with the recombinant truncated proteins for different times at 37°C. Then, the mixtures were centrifuged at 12,000 × g for 1 min, and the supernatants (100 μl) were analyzed using a spectrophotofluorometer (SPF) (LS55; Perkin-Elmer) with an excitation wavelength of 488 nm and an emission wavelength of 510 nm. EGFP-fused truncated proteins mixed with PBSB buffer were used as blank controls.
Kinetic analysis of truncated proteins with B. anthracis spores.
Approximately 5 × 105 biotinylated B. anthracis spores were reacted with the streptavidin sensors (ForteBio) at room temperature overnight to ensure complete conjugation prior to analysis. Four truncated proteins, EP0, EP3, EC5, and EC6, and EGFP were tested simultaneously with the spore-modified sensors using an Octet apparatus and its software (ForteBio). It is notable that an on-line blockage strategy with PBSM (1× PBS and 5% defatted milk powder) was used for each sensor (blocked for 10 min, followed by washing with PBS for 3 × 10 min) to reduce nonspecific interactions. Each assay was repeated three times.
Scatchard plot assay.
B. anthracis spores with a constant concentration of 3.5 × 105 were stained by EC6 with a series of concentrations, ranging from 0.11 to 1.82 μM, at 37°C for 120 min. After centrifugation at 12,000 × g for 1 min, the fluorescence of the supernatant of each mixture was read by the SPF assay immediately, using spore-free EC6 solutions as controls. The initial total molar concentration of EC6 was defined as Ct, and the molar concentration of EC6 in the supernatant was defined as Cf, which represents the free proteins that do not bind with the spores. Thus, the molar concentration of the bound protein (Cb) was calculated as (Ct − Cf). For calculation, we assumed that a single spore could be considered a macromolecule, which had a constant mass and volume during the interaction. Therefore, the molar concentration of the spores was defined as Cs, and the binding ratio (ν) was calculated as Cb/Cs. The saturation binding curve was created by fitting the results obtained under a series of dilutions of EC6. Meanwhile, the data were displayed as a Scatchard plot in which the x axis was the binding ratio (ν) and the y axis was the binding ratio (ν) divided by the free ligand concentration (Cf). According to the principle of this method (34, 52), it is possible to estimate Bmax and Kd from the Scatchard plot (Bmax is the x intercept, and Kd is the negative reciprocal of the slope). Bmax represents the maximum ligand number located on a spore for EC6, while Kd represents the binding constant of EC6 interaction with B. anthracis spores.
Labeling peptides with AuNPs.
Peptide N21 was immobilized on Au nanoparticles (AuNPs) using a procedure described previously (16), with some modifications. Briefly, AuNPs (20 nm, with the pH adjusted to 9.2 before use) reacted with N21 (in H2O) under vigorous stirring for 12 h to form Au-S bonds that link N21 to Au. After washing three times with a washing buffer (10 mM phosphate buffer, 150 mM NaCl, pH 7.4) to remove unreacted peptides, the N21-AuNP conjugate was stored at 4°C until use.
Transmission electron microscopy.
PBSM-blocked B. anthracis spores, with or without sonication pretreatment, were reacted with the gold-labeled peptide, N21-AuNPs. The sonication treatment was used to detach part of the exosporium from the spores, and ultracentrifugation was used to purify the sonicated spores from the detached exosporium as described previously (49, 37). After washing three times to remove unbound N21-AuNPs, the sample was analyzed by using a transmission electron microscope (Tecnai G2 20 Twin; Fei).
In vitro lysin activity.
Lysin activity was measured as previously described (32), with some modifications. Briefly, B. anthracis strain A16 was grown to an optical density at 600 nm (OD600) of 0.2 to 0.3, centrifuged, and resuspended in phosphate buffer to a final OD600 of 0.8 to 1.0. Twofold serial dilutions of purified PlyG (100 μl) were added to a 100-μl bacterial suspension in 96-well plates (Perkin-Elmer), and the drop in the OD600 was monitored by using a Synergy H1 spectrophotometer (BioTek) for 30 min at 37°C. A unit of PlyG activity was defined as the highest dilution that decreased the absorbance by 50% within 15 min (32).
Spore killing assay.
The PlyG-mediated spore killing assay was tested in the presence of l-alanine and d-alanine as described previously (40). In brief, aliquots of spores were heat activated at 65°C for 5 min and suspended in 1 ml tryptic soy broth with either 100 mM l-alanine or d-alanine for 5 min at 37°C. Samples were then treated with 10 U PlyG for up to 2 h and finally plated for counting after washing three times with PBS.
RESULTS
Construction and expression of recombinant proteins.
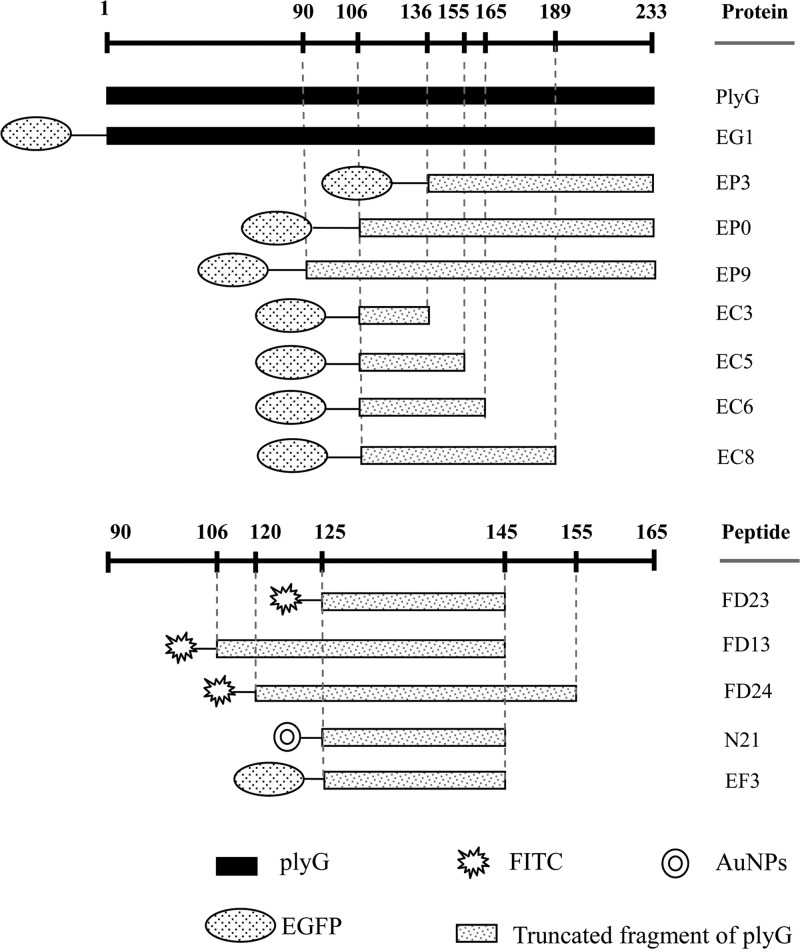
The whole PlyG protein contains 233 amino acids. Using standard genetic engineering methods, a series of recombinant proteins and their fusion proteins with EGFP were constructed and expressed as shown in Fig. 1. It was found that these proteins could be well expressed in E. coli. After purification and dialysis, the proteins were used directly for the binding assays. Meanwhile, four small peptide fragments were synthesized and labeled with or without FITC to confirm their binding activity.
Fig 1.
Schematic positions of the truncated proteins and synthetic peptides used in the study.
Recognition of B. anthracis cells.
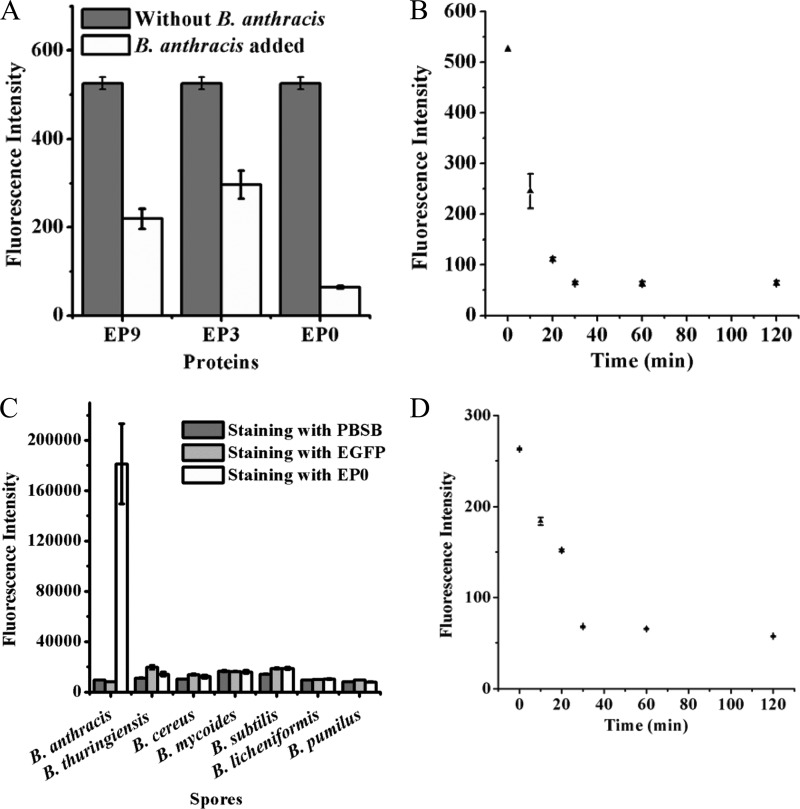
To test if the recombinant truncated fusion proteins retained the ability to recognize B. anthracis cells, equal numbers of B. anthracis cells were stained with the three recombinant proteins EP9, EP0, and EP3 at 37°C for 30 min. The concentrations of the recombinant proteins were chosen within its linear range with the fluorescence intensity. After centrifugation, the supernatants were analyzed using the spectrophotofluorometric (SPF) assay. As shown in Fig. 2A, all three proteins could bind to B. anthracis cells, but EP0 displayed a higher binding affinity than the other two.
Fig 2.
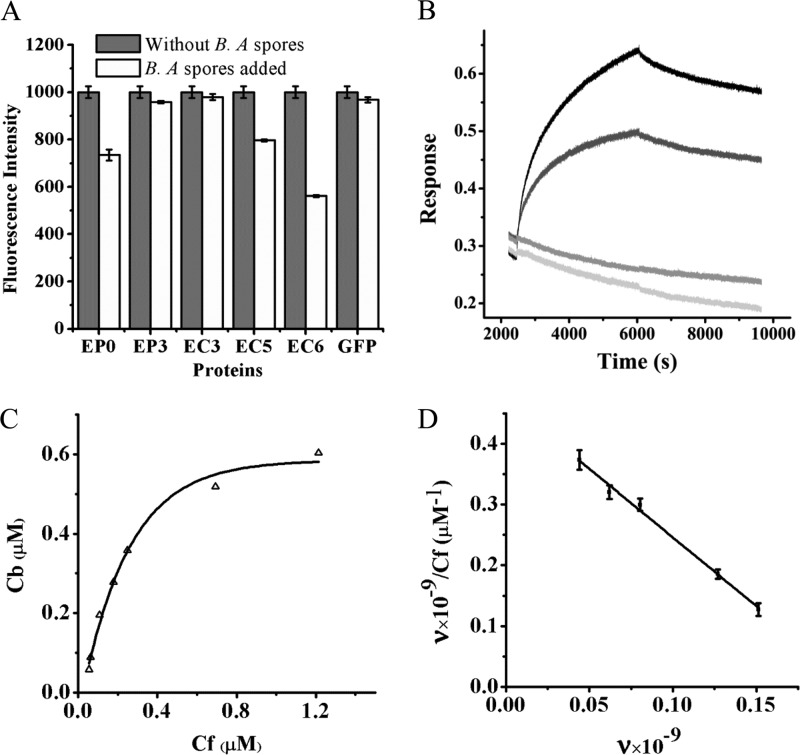
Binding characteristics of the recombinant proteins for B. anthracis vegetative cells and spores. (A) Changes in residual fluorescence intensities of EP9, EP3, and EP0 after reaction with B. anthracis cells for 30 min. (B) Time gradient of fluorescence changes of the supernatant during reaction of EP0 with B. anthracis cells. (C) Specificity of EP0 for B. anthracis spores. After staining with EP0, spores were analyzed by the fluorescence microscope system under the same instrument conditions. (D) Time gradient of fluorescence changes of the supernatant during reaction of EP0 with B. anthracis spores.
To optimize the staining time required for complete labeling of B. anthracis cells, B. anthracis vegetative cells were incubated with EP0 for 0, 10, 20, 30, 60, and 120 min in parallel. Figure 2B shows that 30 min was enough to ensure complete labeling of B. anthracis cells with EP0 under the experimental conditions. For a further confirmation of the staining time of B. anthracis cells, the corresponding cells of each sample, thoroughly washed three times with PBST buffer (1× PBS and 0.05% Tween 20), were imaged by a fluorescence microscope (see Fig. S1 in the supplemental material). The results showed that after more than 30 min of staining, the fluorescence intensity of each cell reached a maximum. This result was consistent with that of the SPF assays, suggesting that 30 min of staining was enough for complete staining of B. anthracis cells by EP0.
Confocal microscopy was also used to determine the binding location of EP0 on B. anthracis cells (see Fig. S2 in the supplemental material). The image clearly showed that EP0 can bind directly to the cell wall of B. anthracis cells, mediated by CBD of PlyG, rather than permeating the cell.
The lysin PlyG can specifically kill B. anthracis cells and has no effect on other Bacillus strains, which was putatively attributed to the high selectivity of its CBD. To confirm if the selectivity of EP0 was the same as that of PlyG, fluorescence microscopy was used to analyze different bacteria, including seven B. thuringiensis strains, five B. cereus strains, one B. subtilis strain, one B. mycoides strain, one B. licheniformis strain, one B. pumilus strain, and three other Gram-negative bacterial strains as controls (see Table S2 in the supplemental material). All the above results showed that the truncated fragments of PlyG fused with EGFP still specifically recognized B. anthracis cells, consistent with results for the full PlyG protein. Therefore, EP0 and EP3 were used in the following work to study if the truncated fragments of PlyG could recognize B. anthracis spores.
Recognition of B. anthracis spores.
To test the ability of the recombinant truncated fragments to recognize B. anthracis spores, equal numbers of B. anthracis spores were mixed with EP0, EP3, and EGFP. As shown in Fig. S3 in the supplemental material, only EP0 displayed an obvious ability to recognize B. anthracis spores. In contrast, EP3 and EGFP could not bind to these spores.
Further experiments found that only B. anthracis spores showed significantly increased fluorescence after staining with EP0, while other closely related spores, including B. thuringiensis spores and B. cereus spores, did not show any appreciable increase in fluorescence (data not shown). The specificity of EP0 for B. anthracis spores was also analyzed statistically by measuring the total fluorescent intensity of each spore (Fig. 2C), demonstrating that only EP0 could significantly enhance the fluorescence, attributable to its direct recognition of and conjugation to the B. anthracis spores.
The binding of EP0 to B. anthracis spores showed a rate similar to that of binding to B. anthracis cells. After 30 min of staining, the fluorescence of each spore reached a maximum (Fig. 2D).
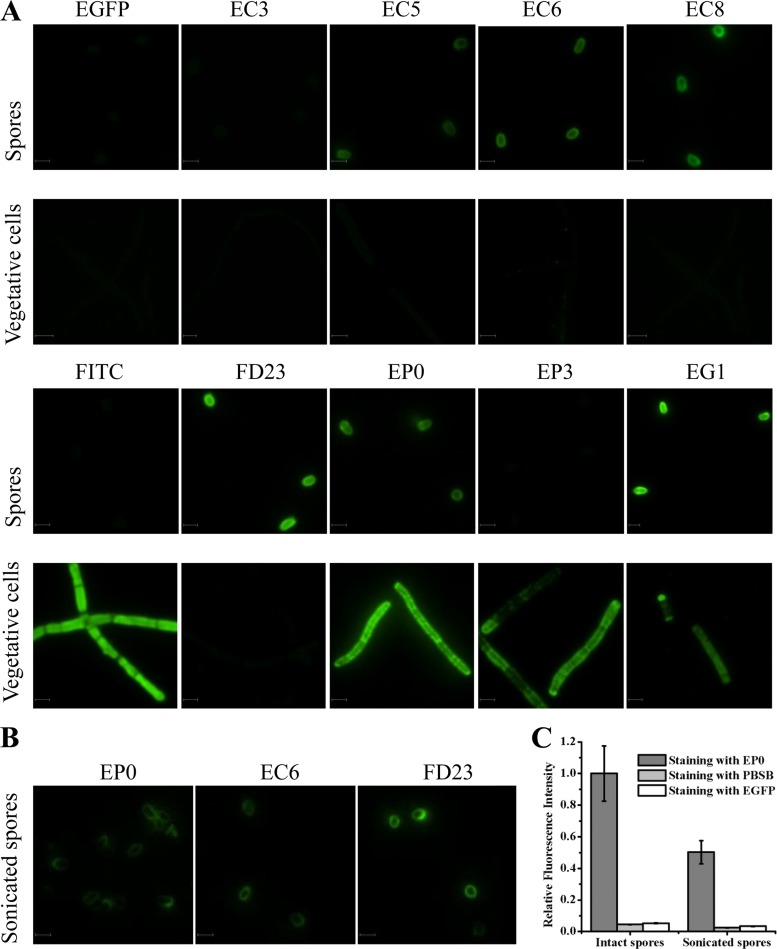
Previous research found that residues 190 to 199 of PlyG were necessary for its binding activity to B. anthracis cells (21). The discovery in our work that only EP0 but not EP3 recognized B. anthracis spores implied that the 30 amino acids (CSR30, located from 106 to 135) in EP0 or a portion of them may play a key role in recognizing the spores. To confirm whether a spore binding domain (SBD) exists in PlyG, a further four truncated fragments fused with EGFP, EC3, EC5, EC6, and EC8, and three synthesized peptides, FD13, FD23, and FD24, labeled with FITC, were constructed to stain the spores and vegetative cells, respectively. As shown in Fig. 3A, the proteins EC5, EC6, and EC8 and the synthetic peptide FD23 could recognize spores but not the vegetative form of B. anthracis. The longer peptides, FD13 and FD24, displayed a staining similar to that of FD23 (see Fig. S3 in the supplemental material). To further ensure that the binding between peptide and B. anthracis spore is specific but not through charge or hydrophobic interactions, an N-terminally EGFP-fused FD23 peptide (EF3) was constructed and used to stain the spores. The results showed that EF3 could bind tightly to B. anthracis spores similarly to the way in which FD23 did (see Fig. S3). What is more, 500 mM Na+ and 200 mM Mg2+, as well as 200 mM Ca2+, had no influence on binding between EC6 and the spores (data not shown). It is notable that for staining of vegetative cells, FITC can penetrate into heat-treated cells because of its smaller size (Fig. 3A), while FD23 and EGFP cannot.
Fig 3.
Binding features of the truncated proteins and the peptide for B. anthracis spores, heat-treated vegetative cells, and sonicated spores. (A) Binding profiles with B. anthracis spores and vegetative cells. (B) Binding profiles with sonicated spores. (C) Relative fluorescence intensities of intact spores and sonicated spores after staining with EP0. All images were taken under the same instrument conditions. Bar = 2 μm.
Additional experiments showed that the full-length PlyG protein fused with EGFP (EG1) recognized the spores and the cells (Fig. 3A), which indicated that the catalytic domain of PlyG did not affect the binding of the truncated PlyG proteins to the spores or vegetative cells.
Further experiments showed that the proteins EP0, EC6, and FD23 also could bind to the sonicated spores, as shown in Fig. 3B. However, the total fluorescence intensity of each sonicated spore stained with EP0 was much lower than that of an intact spore (Fig. 3C). Since sonication can remove part of the exosporium from the spores, the decreased fluorescence may be due to the loss of the exosporium, which indicates that the exosporium of the spores may be the binding target of EP0.
As summarized in Fig. 4, our results showed that recognition of B. anthracis vegetative cells and that of its spores were mediated by different domains in PlyG, and a small fragment (residues 125 to 145) was able to bind to the spores.
Fig 4.
Structure-based analysis of the truncated fragments within the SBD. (A) Structure of the PlyG SBD based on the structure of the catalytic domain of PlyG (PDB 2L47_A; GI 350610326). All of the truncated sites are labeled with different colors. (B) Summarized binding results for the truncated fragments within the SBD.
Analysis of kinetics of binding.
Because less is known about the binding characteristics of the spore binding domain (SBD) for B. anthracis spores, a fluorescence-based Scatchard plot assay was developed to determine the binding constant and the number of binding sites on a spore. As shown in Fig. 5A, under the same concentration and adding the same number of the spores, of all the truncated proteins, EC6 showed the greatest decrease in fluorescence, which was quite close to that of EG1. This indicated that EC6 had the highest affinity for the spores, and it was identified as the SBD. To create the Scatchard plot, EC6 at a series of concentrations ranging from 0.11 to 1.82 μM was reacted with a constant number of B. anthracis spores. The saturation binding data were analyzed using nonlinear regression (Fig. 5C) and displayed in a Scatchard plot, as shown in Fig. 5D, giving a slope of −2.26 μM−1 and a y intercept value of 0.47 μM−1. Therefore, the maximum number of binding sites for EC6 on a spore was found to be 2.1 × 108, and the Kd value of the interaction was 4.4 × 10−7 M.
Fig 5.
Characteristics of EC6 association with B. anthracis (B. A) spores. (A) Differences in binding of the truncated proteins to B. anthracis spores with the same concentrations and conditions. (B) Kinetics of EC6 association with B. anthracis spores at concentrations of 674.6 nM (black) and 337.3 nM (dark gray). EGFP (304.4 nM; gray) and PBS (light gray) were used as the control and the blank, respectively. (C) Saturation binding curve of EC6, shown by the molar concentration of Cf relative to Cb. (D) Simulated Scatchard plot for EC6 association with B. anthracis spores (adjusted r2 = 0.9918).
To analyze the time-resolved dynamic of the interaction between EC6 and B. anthracis spores, an Octet apparatus was used to observe the association and dissociation processes. As shown in Fig. 5B, the interaction process presented a slow-association-and-slow-disassociation profile. In addition, the interactions between the truncated proteins EC5, EP0, and EP3 and B. anthracis spores were also tested in the Octet system (see Fig. S4 in the supplemental material). Consistent with the SPF data, EC6 displayed a higher affinity for the spores.
PlyG-mediated spore killing.
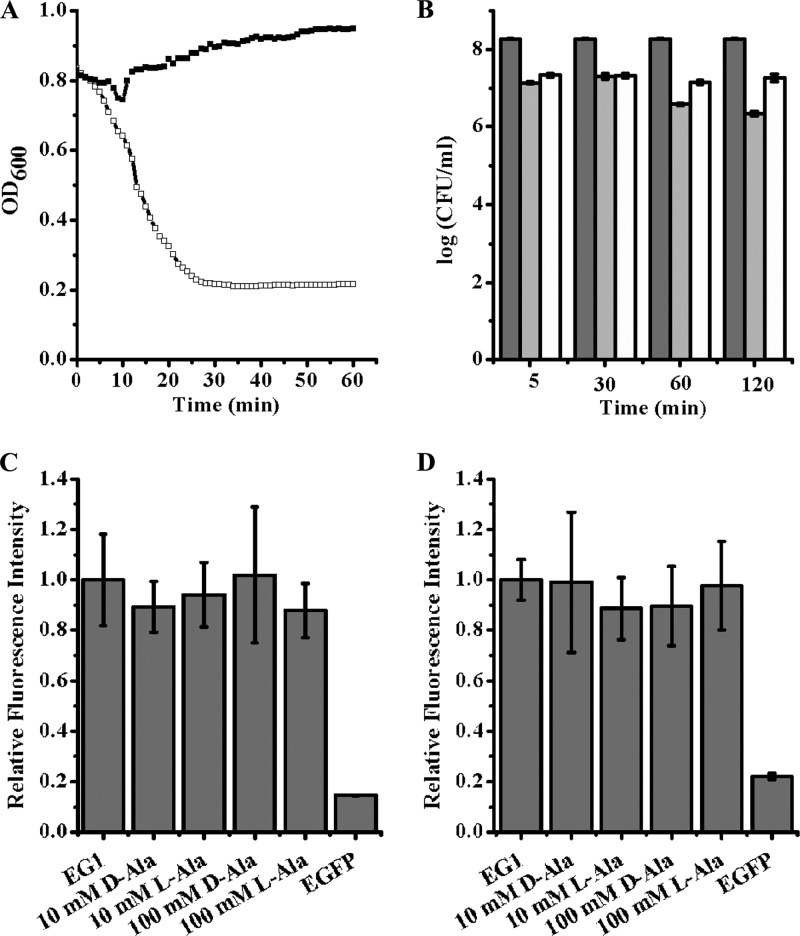
To explore the possible role of SBD on PlyG activity, PlyG with a high lysin activity against B. anthracis vegetative cells was obtained after purification by two-step ion-exchange chromatography (Fig. 6A). In the spore killing assay, the heat-activated spores were mixed with 10 U of PlyG in the presence of 100 mM l-alanine or d-alanine for up to 2 h. When the treatment time was 5 min or 30 min, the decrease in the number of CFU/ml was about 1 log in the presence of either l-alanine or d-alanine. These results indicate that PlyG can bind to the spores, germinating (in the presence of l-alanine) or not (in the presence of d-alanine), hence resulting in almost the same reduced number of CFU (Fig. 6B). Although the reduction in the number of CFU/ml after treatment for 2 h was about 2 logs in the presence of l-alanine, this may be because some of the spores had germinated to vegetative cells and were killed during the exposure. To eliminate the possible influence of l-Ala and d-Ala on the killing efficiency of PlyG through interference with binding of PlyG to B. anthracis spores, spores, inactivated or not, were stained with EG1 in the presence of different concentrations of l-alanine and d-alanine, respectively. The statistical fluorescence intensity of alanine-treated spores had no obvious difference from that of the untreated control, either inactivated (Fig. 6C) or not (Fig. 6D). These results showed that alanine does not interfere with the binding of PlyG to the spores.
Fig 6.
PlyG-mediated spore binding and killing. (A) Lysin activity of PlyG on B. anthracis vegetative cells. One unit of two-step-purified PlyG (open squares) displays high activity against B. anthracis cells, in contrast to the buffer control (solid squares). (B) Effect of PlyG on B. anthracis spores. Heat-activated B. anthracis spores were treated with PlyG (10 U) in the presence of either 100 mM l-alanine (light gray) or d-alanine (white) for different times. The resulted CFU were counted and are contrasted with results for the blank control (gray). EG1 can bind to inactivated (C) or live (D) B. anthracis spores in the presence of either l-alanine or d-alanine, and the relative fluorescence intensity is not obviously different from that of the buffer control.
Transmission electron microscopy.
To further identify the binding sites of SBD on B. anthracis spores, the N21 peptide-labeled AuNPs, N21-AuNPs, were used to stain spores. The maximum absorption of the conjugated N21-AuNPs had a 10-nm red shift relative to that of the bare AuNPs (see Fig. S5 in the supplemental material). Transmission electron microscopy showed that N21-AuNP particles were located mainly on the exosporium of the spores (Fig. 7). Together with the fluorescence results for the sonicated spores in Fig. 3B and C, the exosporium is identified as the most probable binding target of the SBD.
Fig 7.
Transmission electron microscopy pictures of B. anthracis spores, sonicated spores, and the detached exosporium labeled with bare AuNPs and N21-AuNPs, respectively. Bar = 500 nm.
DISCUSSION
Possible impact on mechanism of PlyG activity.
As shown in Fig. 3A, our study shows that PlyG uses different domains to recognize the spores and the vegetative cells. The SBD of PlyG contains 60 amino acids, from residue 106 to residue 165. According to the recently reported structure of the catalytic domain of PlyG (PDB 2L47_A; GI 350610326), the SBD consists of three helixes, 106 to 118 (Hex1), 138 to 145 (Hex2), and 147 to 156 (Hex3), and a sheet from 125 to 129 (Fig. 4A). The fact that EC5, EC6, and FD23 but not EC3 and EP3 retain the ability to recognize B. anthracis spores implies that the sheet and Hex2 are the inner core of SBD, while Hex1 and Hex3 are expanded elements that help to bind the spore tighter.
The transmission electron microscopy assays and fluorescence assays of the sonicated spores indicated that the binding sites of the newly identified SBD are most probably on the exosporium of the spores. This result differs from that in a previous report, which suggested that PlyG could recognize only the germinating form, but not spores, of B. anthracis (9). This contradiction may be because only a shorter C-terminal fragment of PlyG (residues 156 to 233) was studied as the binding domain in the previous research, and the newly identified spore binding domain is located mainly within the catalytic domain previously considered. This finding may offer an important insight for understanding the way PlyG interacts with spores and its use in control of anthrax.
Since PlyG can bind to spores, this also implies that PlyG could be used immediately to reduce the threat of anthrax in suspected exposures to B. anthracis spores without the need to wait until spore germination. Otherwise, if PlyG lacked the ability to bind to spores, early administration of PlyG might not be effective. However, our results also showed that PlyG can bring an obvious but only moderate reduction in the number of CFU of B. anthracis spores within 30 min, even in the presence of l-alanine, implying that some modification may be needed for PlyG to further enhance its spore-killing activity.
Biological significance of the spore recognition property of PlyG.
The finding that PlyG uses different domains to recognize the spores and vegetative cells might also be a great help in understanding the coevolution of gamma phage with B. anthracis. since lysins are used by bacteriophages to digest the bacterial cell wall for the release of progeny virions, it is easy to understand why PlyG can lyse the vegetative cells of B. anthracis through the specific recognition of its CBD. However, it is intriguing that PlyG has a special domain for recognizing the spores. It is well understood that spore formation is a special strategy used by bacteria to escape difficult survival conditions. B. anthracis bacteria are in a dormant state after forming spores, which is not suitable for replication of gamma phage. Given the fact that the SBD of PlyG is located near the catalytic domain and has a good binding affinity for spores, one possible reason may be that binding of PlyG to spores limits the free diffusion and hence the activity of PlyG for other vegetative cells. This may be helpful in keeping an ecological balance between gamma phage and B. anthracis when PlyG is overreleased in the moment of progeny virions' outburst, which could be a new defense mechanism of the exosporium to help the spore survive. Furthermore, it is worth examining whether bacteriophage lysins for other spore-forming bacteria have similar separate domains for recognizing spores and cells.
High specificity of SBD enables rapid identification of B. anthracis spores.
The newly discovered spore binding domain also provides a good biomarker for identifying the structure of B. anthracis spores and for specific detection of the spores. Through studying the targets of the SBD, the cell changes during spore formation in B. anthracis could be identified, and hence the structural differences between B. anthracis spores and closely related spores, such as B. thuringiensis spores and B. cereus spores, could be isolated.
Given the potentially lethal threats, rapid identification of B. anthracis vegetative cells and its spores is of critical importance. Many methods have been developed in order to do this, including conventional culture assays (45–47), peptide-based assays (1, 19, 24, 53), single-chain antibody-based assays (28, 30, 48, 51), and PCR-based assays (6, 23, 25, 35, 41). However, most of these methods are not able to discriminate between spores and vegetative cells. According to our results, the SBD of PlyG provides new opportunities for developing better assays for identifying B. anthracis spores in view of its high specificity and good affinity. Taking it together with the CBD, one could use either one or two of the domains to recognize spores, the vegetative form of B. anthracis, or both in tandem. Furthermore, these highly evolved binding domains target the elements essential for viability, implying a low chance of bacterial mutation to avoid these recognitions.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the basic research program of the Ministry of Science and Technology of China (2012CB721102) to J. Yu and H. Wei, the Infectious Disease Control Research Program of the Ministry of Health of China (2008ZX10004-004 and 2009ZX10003-019), and the National Natural Science Foundation of China (2077508).
We thank Jin He of Huazhong Agricultural University for his kind help in preparation of materials and Simon Rayner at Wuhan Institute of Virology, Chinese Academy of Sciences, for his suggestions and kind help in checking the English of the manuscript.
Footnotes
Published ahead of print 16 July 2012
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Acharya G, et al. 2007. Label-free optical detection of anthrax-causing spores. J. Am. Chem. Soc. 129:732–733 [DOI] [PubMed] [Google Scholar]
- 2. Brumlik MJ, et al. 2001. Use of long-range repetitive element polymorphism-PCR to differentiate Bacillus anthracis strains. Appl. Environ. Microbiol. 67:3021–3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Clery-Barraud C, Gaubert A, Masson P, Vidal D. 2004. Combined effects of high hydrostatic pressure and temperature for inactivation of Bacillus anthracis spores. Appl. Environ. Microbiol. 70:635–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Daniel A, et al. 2010. Synergism between a novel chimeric lysin and oxacillin protects against infection by methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 54:1603–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dixon TC, Meselson M, Guillemin J, Hanna PC. 1999. Anthrax. N. Engl. J. Med. 341:815–826 [DOI] [PubMed] [Google Scholar]
- 6. Drago L, Lombardi A, Vecchi ED, Gismondo MR. 2002. Real-time PCR assay for rapid detection of Bacillus anthracis spores in clinical samples. J. Clin. Microbiol. 40:4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dwyer KG, et al. 2004. Identification of Bacillus anthracis specific chromosomal sequences by suppressive subtractive hybridization. BMC Genomics 5:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fischetti VA, Nelson D, Schuch R. 2006. Reinventing phage therapy: are the parts greater than the sum? Nat. Biotechnol. 24:1508–1511 [DOI] [PubMed] [Google Scholar]
- 9. Fujinami Y, Hirai Y, Sakai I, Yoshino M, Yasuda J. 2007. Sensitive detection of Bacillus anthracis using a binding protein originating from gamma-phage. Microbiol. Immunol. 51:163–169 [DOI] [PubMed] [Google Scholar]
- 10. Garcia-Patrone M, Tandecarz JS. 1995. A glycoprotein multimer from Bacillus thuringiensis sporangia: dissociation into subunits and sugar composition. Mol. Cell. Biochem. 145:29–37 [DOI] [PubMed] [Google Scholar]
- 11. Gerhardt P, Ribi E. 1964. Ultrastructure of the exosporium enveloping spores of Bacillus cereus. J. Bacteriol. 88:1774–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hachisuka Y, Kojima K, Sato T. 1966. Fine filaments on the outside of the exosporium of Bacillus anthracis spores. J. Bacteriol. 91:2382–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Helgason E, et al. 2000. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis-–one species on the basis of genetic evidence. Appl. Environ. Microbiol. 66:2627–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Henriques AO, Beall BW, Roland K, Moran CP., Jr 1995. Characterization of cotJ, a sigma E-controlled operon affecting the polypeptide composition of the coat of Bacillus subtilis spores. J. Bacteriol. 177:3394–3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Higgins JA, et al. 2003. A field investigation of Bacillus anthracis contamination of U.S. Department of Agriculture and other Washington, D.C., buildings during the anthrax attack of October 2001. Appl. Environ. Microbiol. 69:593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hill HD, Mirkin CA. 2006. The bio-barcode assay for the detection of protein and nucleic acid targets using DTT-induced ligand exchange. Nat. Protoc. 1:324–336 [DOI] [PubMed] [Google Scholar]
- 17. Inglesby TV, et al. 1999. Anthrax as a biological weapon: medical and public health management. Working Group on Civilian Biodefense. JAMA 281:1735–1745 [DOI] [PubMed] [Google Scholar]
- 18. Kailas L, et al. 2011. Surface architecture of endospores of the Bacillus cereus/anthracis/thuringiensis family at the subnanometer scale. Proc. Natl. Acad. Sci. U. S. A. 108:16014–16019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kaman WE, et al. 2011. Peptide-based fluorescence resonance energy transfer protease substrates for the detection and diagnosis of Bacillus species. Anal. Chem. 83:2511–2517 [DOI] [PubMed] [Google Scholar]
- 20. Keim P, et al. 2001. Molecular investigation of the Aum Shinrikyo anthrax release in Kameido, Japan. J. Clin. Microbiol. 39:4566–4567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kikkawa H, Fujinami Y, Suzuki S, Yasuda J. 2007. Identification of the amino acid residues critical for specific binding of the bacteriolytic enzyme of gamma-phage, PlyG, to Bacillus anthracis. Biochem. Biophys. Res. Commun. 363:531–535 [DOI] [PubMed] [Google Scholar]
- 22. Kikkawa HS, Ueda T, Suzuki S, Yasuda J. 2008. Characterization of the catalytic activity of the gamma-phage lysin, PlyG, specific for Bacillus anthracis. FEMS Microbiol. Lett. 286:236–240 [DOI] [PubMed] [Google Scholar]
- 23. Kim K, et al. 2005. Rapid genotypic detection of Bacillus anthracis and the Bacillus cereus group by multiplex real-time PCR melting curve analysis. FEMS Immunol. Med. Microbiol. 43:301–310 [DOI] [PubMed] [Google Scholar]
- 24. Knurr J, et al. 2003. Peptide ligands that bind selectively to spores of Bacillus subtilis and closely related species. Appl. Environ. Microbiol. 69:6841–6847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kumar S, Tuteja U. 2009. Detection of virulence-associated genes in clinical isolates of Bacillus anthracis by multiplex PCR and DNA probes. J. Microbiol. Biotechnol. 19:1475–1481 [DOI] [PubMed] [Google Scholar]
- 26. Lai EM, et al. 2003. Proteomic analysis of the spore coats of Bacillus subtilis and Bacillus anthracis. J. Bacteriol. 185:1443–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Loeffler JM, Nelson D, Fischetti VA. 2001. Rapid killing of Streptococcus pneumoniae with a bacteriophage cell wall hydrolase. Science 294:2170–2172 [DOI] [PubMed] [Google Scholar]
- 28. Long GW, O'Brien T. 1999. Antibody-based systems for the detection of bacillus anthracis in environmental samples. J. Appl. Microbiol. 87:214. [DOI] [PubMed] [Google Scholar]
- 29. Low LY, Yang C, Perego M, Osterman A, Liddington RC. 2005. Structure and lytic activity of a Bacillus anthracis prophage endolysin. J. Biol. Chem. 280:35433–35439 [DOI] [PubMed] [Google Scholar]
- 30. Mabry R, et al. 2006. Detection of anthrax toxin in the serum of animals infected with Bacillus anthracis by using engineered immunoassays. Clin. Vaccine Immunol. 13:671–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Moberly BJ, Shafa F, Gerhardt P. 1966. Structural details of anthrax spores during stages of transformation into vegetative cells. J. Bacteriol. 92:220–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nelson D, Loomis L, Fischetti VA. 2001. Prevention and elimination of upper respiratory colonization of mice by group A streptococci by using a bacteriophage lytic enzyme. Proc. Natl. Acad. Sci. U. S. A. 98:4107–4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nicholson WL, Galeano B. 2003. UV resistance of Bacillus anthracis spores revisited: validation of Bacillus subtilis spores as UV surrogates for spores of B. anthracis Sterne. Appl. Environ. Microbiol. 69:1327–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Norby JG, Ottolenghi P, Jensen J. 1980. Scatchard plot: common misinterpretation of binding experiments. Anal. Biochem. 102:318–320 [DOI] [PubMed] [Google Scholar]
- 35. Qi Y, et al. 2001. Utilization of the rpoB gene as a specific chromosomal marker for real-time PCR detection of Bacillus anthracis. Appl. Environ. Microbiol. 67:3720–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Qiao YM, et al. 2007. Loop-mediated isothermal amplification for rapid detection of Bacillus anthracis spores. Biotechnol. Lett. 29:1939–1946 [DOI] [PubMed] [Google Scholar]
- 37. Redmond C, Baillie LW, Hibbs S, Moir AJ, Moir A. 2004. Identification of proteins in the exosporium of Bacillus anthracis. Microbiology 150:355–363 [DOI] [PubMed] [Google Scholar]
- 38. Roth IL, Williams RP. 1963. Comparison of the fine structure of virulent and avirulent spores of Bacillus Anthracis. Tex. Rep. Biol. Med. 21:394–399 [PubMed] [Google Scholar]
- 39. Sainathrao S, Mohan KV, Atreya C. 2009. Gamma-phage lysin PlyG sequence-based synthetic peptides coupled with Qdot-nanocrystals are useful for developing detection methods for Bacillus anthracis by using its surrogates, B. anthracis-Sterne and B. cereus-4342. BMC Biotechnol. 9:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schuch R, Nelson D, Fischetti VA. 2002. A bacteriolytic agent that detects and kills Bacillus anthracis. Nature 418:884–889 [DOI] [PubMed] [Google Scholar]
- 41. Sohni Y, Kanjilal S, Kapur V. 2008. Performance evaluation of five commercial real-time PCR reagent systems using TaqMan assays for B. anthracis detection. Clin. Biochem. 41:640–644 [DOI] [PubMed] [Google Scholar]
- 42. Sonenshein AL, Cami B, Brevet J, Cote R. 1974. Isolation and characterization of rifampin-resistant and streptolydigin-resistant mutants of Bacillus subtilis with altered sporulation properties. J. Bacteriol. 120:253–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sylvestre P, Couture-Tosi E, Mock M. 2002. A collagen-like surface glycoprotein is a structural component of the Bacillus anthracis exosporium. Mol. Microbiol. 45:169–178 [DOI] [PubMed] [Google Scholar]
- 44. Takahashi H, et al. 2004. Bacillus anthracis incident, Kameido, Tokyo, 1993. Emerg. Infect. Dis. 10:117–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Titball RW, Turnbull PC, Hutson RA. 1991. The monitoring and detection of Bacillus anthracis in the environment. Soc. Appl. Bacteriol. Symp. Ser. 20:9S–18S [PubMed] [Google Scholar]
- 46. Turnbull PC. 1991. Anthrax vaccines: past, present and future. Vaccine 9:533–539 [DOI] [PubMed] [Google Scholar]
- 47. Turnbull PC. 1999. Definitive identification of Bacillus anthracis—a review. J. Appl. Microbiol. 87:237–240 [DOI] [PubMed] [Google Scholar]
- 48. Wang DB, et al. 2009. Label-free detection of B. anthracis spores using a surface plasmon resonance biosensor. Analyst 134:738–742 [DOI] [PubMed] [Google Scholar]
- 49. Wang DB, et al. 2009. Detection of B. anthracis spores and vegetative cells with the same monoclonal antibodies. PLoS One 4:e7810 doi:10.1371/journal.pone.0007810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang J, et al. 2005. 2-D reference map of Bacillus anthracis vaccine strain A16R proteins. Proteomics 5:4488–4495 [DOI] [PubMed] [Google Scholar]
- 51. Wang SH, et al. 2006. Construction of single chain variable fragment (ScFv) and BiscFv-alkaline phosphatase fusion protein for detection of Bacillus anthracis. Anal. Chem. 78:997–1004 [DOI] [PubMed] [Google Scholar]
- 52. Weder HG, Schildknecht J, Lutz RA, Kesselring P. 1974. Determination of binding parameters from Scatchard plots. Theoretical and practical considerations. Eur. J. Biochem. 42:475–481 [DOI] [PubMed] [Google Scholar]
- 53. Williams DD, Benedek O, Turnbough CL., Jr 2003. Species-specific peptide ligands for the detection of Bacillus anthracis spores. Appl. Environ. Microbiol. 69:6288–6293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yang H, He J, Hu F, Zheng C, Yu Z. 2010. Detection of Escherichia coli enoyl-ACP reductase using biarsenical-tetracysteine motif. Bioconjug. Chem. 21:1341–1348 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.