Abstract
Identifying and understanding the collection of all antibiotic resistance determinants presented in the global microbiota, the antibiotic resistome, provides insight into the evolution of antibiotic resistance and critical information for the development of future antimicrobials. The rifamycins are broad-spectrum antibiotics that target bacterial transcription by inhibition of RNA polymerase. Although mutational alteration of the drug target is the predominant mechanism of resistance to this family of antibiotics in the clinic, a number of diverse inactivation mechanisms have also been reported. In this report, we investigate a subset of environmental rifampin-resistant actinomycete isolates and identify a diverse collection of rifampin inactivation mechanisms. We describe a single isolate, WAC1438, capable of inactivating rifampin by glycosylation. A draft genome sequence of WAC1438 (most closely related to Streptomyces speibonae, according to a 16S rRNA gene comparison) was assembled, and the associated rifampin glycosyltransferase open reading frame, rgt1438, was identified. The role of rgt1438 in rifampin resistance was confirmed by its disruption in the bacterial chromosome, resulting in a loss of antibiotic inactivation and a 4-fold decrease in MIC. Interestingly, examination of the RNA polymerase β-subunit sequence of WAC1438 suggests that it harbors a resistant target and thus possesses dual mechanisms of rifamycin resistance. Using an in vitro assay with purified enzyme, Rgt1438 could inactivate a variety of rifamycin antibiotics with comparable steady-state kinetics constants. Our results identify rgt1438 as a rifampin resistance determinant from WAC1438 capable of inactivating an assortment of rifamycins, adding a new element to the rifampin resistome.
INTRODUCTION
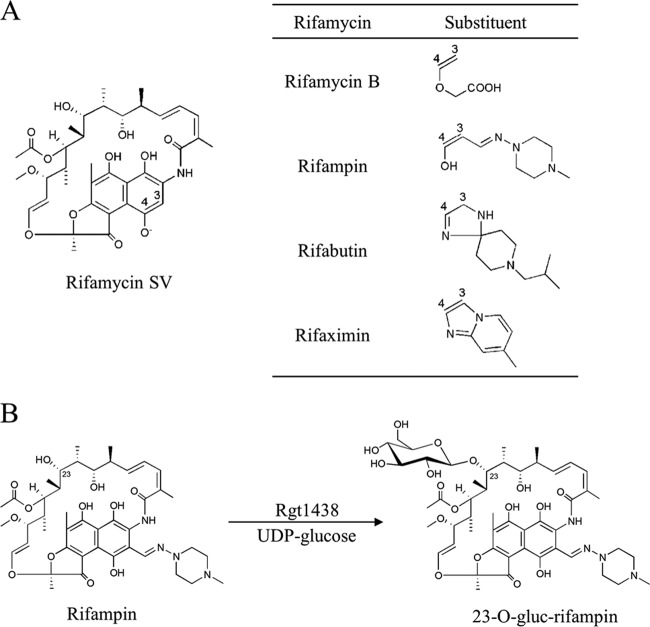
The introduction of antibiotics to the clinic has been one of the most successful forms of chemotherapy in modern medicine. Today, however, the utility of these molecules against many pathogens is threatened because of antibiotic resistance (15). Antibiotic resistance currently impacts all classes of natural, semisynthetic, and completely synthetic antibiotics (50). The rifamycins are broad-spectrum antibiotics produced by Amycolatopsis mediterranei and many other actinomycetes (27, 39). These compounds target bacterial transcription by binding to a highly conserved region of the β-subunit of RNA polymerase, encoded by rpoB (16). Rifampin (RIF) is a semisynthetic rifamycin introduced to the clinic over 50 years ago that remains vital to the treatment of tuberculosis and other mycobacterial infections in addition to its growing use in combination therapy for infections caused by multidrug-resistant Gram-positive pathogens such as methicillin-resistant Staphylococcus aureus (MRSA) (17, 46). Due to the clinical success of RIF, the rifamycin scaffold continues to be explored by semisynthesis for the generation of superior rifamycins that retain activity against rifampin-resistant strains and reduce host toxicity (3, 29, 33). These efforts have focused on one region of the molecule (positions C3 and C4) that tolerates modifications while retaining biological activity (48) (Fig. 1). The broad antimicrobial spectrum of the rifamycins has made this pharmacophore an attractive scaffold for the generation of new analogs effective against a wider array of diseases. These include rifalazil, which demonstrates promise for the treatment of chlamydial infections (37), and rifaximin, administered prophylactically for travelers' diarrhea (22) and as a therapeutic for inflammatory bowel disease (10). The introduction of new rifamycins used to combat a more heterogeneous collection of pathogens is expected to apply strong pressure for the selection of new and diverse resistance mechanisms (45).
Fig 1.
(A) Structures of various rifamycins. (B) The glycosylation of rifampin catalyzed by Rgt1438, yielding 23-O-gluc-rifampin.
A diverse collection of RIF resistance determinants have been described (45). In the clinic, the predominant mechanism of resistance to RIF arises by mutation of a target (rpoB) where single amino acid substitutions in the resultant protein may confer high-level resistance (16, 24, 35). Additionally, a number of pathogenic and environmental bacteria have demonstrated the ability to inactivate rifampin via enzymatic modification, altering the antibiotic to an extent where it can no longer bind its target with high affinity. A total of three group transfer mechanisms of rifampin resistance have been identified: ADP ribosylation, phosphorylation, and glycosylation (31, 51, 52). The targets of these inactivating mechanisms are critical hydroxyls of the ansa chain of RIF (Fig. 1); these groups facilitate hydrogen bond interactions with a number of protein residues of RpoB and are crucial for biological activity (7, 48). Additionally, the decomposition of RIF has been reported as a mechanism of inactivation (2). Recently, the rox gene product of Nocardia farcinica was shown to promote the decomposition of RIF initiated by monooxygenation (21). With the exception of ADP-ribosylation mediated by the Arr enzymes, the remaining group transfer mechanisms of RIF resistance have not been extensively investigated (5).
Recently, we surveyed a collection of approximately 500 soil actinomycetes for their ability to resist an assorted collection of 21 antibiotics (12). Each isolate was on average resistant to 7 to 8 antibiotics, identifying environmental bacteria as a likely reservoir of diverse antibiotic-resistant determinants. A subset of these isolates possessed the ability to inactivate RIF. Here we describe the characterization of these RIF-inactivating isolates and identify their various modes of inactivation. We also discuss the investigation of isolate WAC1438 (Streptomyces speibonae), which inactivates RIF by glycosylation. Using whole-genome sequencing, we identified the genetic sequence encoding a RIF glycosyltransferase (open reading frame [ORF]rgt1438) and investigated its role as a resistance determinant.
MATERIALS AND METHODS
Bacterial strains.
Escherichia coli ET12567/pUZ8002 was used as the donor strain for conjugal transfer for both WAC1438 and Streptomyces albus J1074. Rhodococcus erythropolis L-88 and pTIP-QC1 were used for overexpression of rgt1438 and were gifts from Tomohiro Tamura of the National Institute of Advanced Industrial Science and Technology (AIST), Japan (30, 34). Streptomyces albus J1074 was generously provided by Michael Fischbach.
RIF inactivation assay.
Fresh spores were used to inoculate 5 ml of Streptomyces isolation media (SIM) in culture tubes with one glass bead incubated at 30°C with shaking at 250 rpm as previously described (12). After 3 days, RIF was added at a concentration of 20 μg/ml and a sample was taken for the zero time point. At specified intervals, a sample of each culture was removed and analyzed for residual RIF antimicrobial activity using the Kirby-Bauer disk diffusion assay with Bacillus subtilis 168 as the RIF-sensitive indicator microorganism; plates were incubated for 16 h at 37°C. Samples were also analyzed using liquid chromatography-electrospray ionization-mass spectrometry (LC-ESI-MS), an Agilent 1100 Series LC system (Agilent Technologies Canada, Inc.), and a QTrap LC/MS/MS system (Applied Biosystems/MDS Sciex). Equal volumes of methanol were added to all samples followed by clarification by centrifugation at 15,000 rpm for 10 min, and 10 μl of sample was analyzed by high-performance LC (HPLC) or LC-MS using the method described below.
16S rRNA gene analysis.
Genomic DNA for each isolate was prepared by standard methods (25). PCR was performed (12) using primers F27 [5′-AGAGTTTGATC(A/C)TGGCTCAG-3′] and R1492 [5′-TACGG(C/T)TACCTTGTTACGACTT-3′] (20). Products were sequenced at the Mobix Central Facility (McMaster University, Hamilton, Canada).
WAC1438 genome sequencing and annotation.
WAC1438 genomic DNA was sequenced using a Roche 454 platform. DNA was sheared into approximately 800-bp fragments using a Covaris S220 ultrasonicator (Covaris Inc., Woburn, MA), and libraries were prepared following the XL Rapid Library Preparation Method manual (454 Life Sciences/Roche, Branford, CT). Emulsion PCR and sequencing were carried out according to the manufacturer's instructions for the FLX instrument. Reads were assembled using MIRA version 3.4.0, the 454′ quick-switch parameters (“de novo,” “genome,” “accurate”), and six passes (8). The resulting assembly consisted of 270 contigs totaling 8,201,776 bp with an N50 statistic of 72,334 bp.
Inactivation of RIF glycosyltransferase open reading frame rgt1438 from WAC1438.
The rgt1438 open reading frame was disrupted in WAC1438 by insertional inactivation. A partial internal fragment of rgt1438 (∼550 bp in length) was amplified by PCR using primers rgtPartF (5′-ATGCATGAATTCCGAGGCGGCCGAGGGTTGT-3′) and rgtPartR (5′-ATGCATAAGCTTCCGCGAGCGAGGACCACACG-3′) (see Table S1 in the supplemental material). The PCR product was cloned in the EcoRI and HindIII sites of pSET152. The resulting construct, pSET152-rgt1438part, lacks attP and int (ΦC31) and thus functions as a suicide vector when introduced into Streptomyces spp. pSET152-rgt1438part was transformed into E. coli ET12567/pUZ8002 for conjugal transfer to WAC1438 (25). Transconjugants were selected by growth on apramycin at 25 μg/ml and confirmed by PCR and amplicon sequencing. The rgt1438::aac(3)-IV mutant was named PTS1.
Construction of pIJ8600K-rgt1438 for complementation and heterologous expression.
Using PCR, rgt1438 was amplified using primers rgtF (5′-ATGCATGAATTCCGAGGCGGCCGAGGGTTGT-3′) and rgtR-pIJK (5′-ATGCATGGATCCCCGACGGGGGCTGGACTC-3) and was directionally cloned using NdeI and BamHI into pIJ8600K (pIJ8600 derivative [25] with a neomycin-kanamycin resistance cassette). This construct was transformed into E. coli ET12567/pUZ8002 and transferred to PTS1 by conjugation. Transconjugants were selected on kanamycin at 25 μg/ml, generating strain PTS2. pIJ8600K-rgt1438 was similarly introduced into Streptomyces albus J1074.
Determination of RIF MICs.
RIF MIC determinations were performed as previously described (26). The average titer of bacteria was 107 CFU/ml. Plates were incubated at 30°C for 3 days, and the RIF MIC was determined as the lowest concentration of drug that failed to yield any bacterial colonies.
Overexpression and purification of recombinant Rgt1438.
A 1.3-kb DNA fragment containing rgt1438 was amplified by PCR using genomic DNA of WAC1438 and primers rgtF (5′-ATCGATCATATGCGCATGCTGCTGACCAC-3′) and rgtR (5′-ATGCATAAGCTTCCGACGGGGGCTGGACTC-3′). Using restriction sites incorporated into these primers, rgt1438 was directionally cloned into the pTIP-QC1 vector at NdeI and HindIII sites, introducing an N-terminal His tag (34). The pTIP-QC1-rgt1438 construct was transformed into R. erythropolis L-88 cells by electroporation as previously described (13, 30, 41). A single colony was used to inoculate a starter culture of 25 ml of LB with 30 μg/ml chloramphenicol and incubated for 48 h at 30°C with shaking at 250 rpm. The 25-ml starter culture was used to inoculate 1 liter of LB and grown to an optical density at 600 nm of 0.5 at 30°C with shaking at 250 rpm. Expression of rgt1438 was induced with the addition of thiostrepton at 1 μg/ml, and the culture was grown for an additional 16 h. Cells were harvested by centrifugation at 5,000 × g for 15 min, washed with 0.85% NaCl, and frozen at −20°C. In preparation for lysis, the cell pellet was thawed and resuspended in 20 ml of buffer A {50 mM N-tris(hydroxymethyl)methyl-3-aminopropanesulfonic acid (TAPS) [pH 8.4], 250 mM NaCl, 5 mM imidazole}, lysozyme was added to achieve a final concentration of 1 mg/ml, and the reaction mixture was incubated on ice for 1 h. Phenylmethylsulfonyl fluoride (1 mM final concentration) and 1 μg/ml pancreatic bovine DNase were added to the cell suspension and processed using a Constant Systems continuous-flow cell disruptor (Constant Systems Ltd., Daventry, United Kingdom). The sample was clarified by centrifugation at 50,000 × g for 30 min and the supernatant applied onto a 5-ml nickel-nitrilotriacetic acid column (Qiagen, Valencia, CA) preequilibrated with buffer A at a flow rate of 1 ml/min. The column was washed with 5 column volumes of buffer A, and bound protein was eluted with a stepwise gradient of buffer B (50 mM TAPS [pH 8.4], 250 mM NaCl, 500 mM imidazole) over 25 column volumes. The protein content of the fractions was analyzed using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and fractions containing protein corresponding to the correct molecular weight of Rgt1438 were pooled and dialyzed against 3 liters of buffer C (50 mM TAPS [pH 8.4]) overnight at 4°C. This protein fraction was then applied to a 1-ml Mono Q 5/50 GL column (GE Healthcare) preequilibrated with buffer C at a flow rate of 0.5 ml/min. Proteins were eluted from this column using a linear gradient of 0% to 100% buffer D (50 mM TAPS [pH 8.4], 500 mM NaCl) over 50 column volumes. Protein fractions were analyzed using SDS-PAGE, and fractions containing Rgt1438 were pooled, concentrated, and stored at −20°C, with a final glycerol content of 10% (wt/vol). The protein concentration was determined using the Bradford protein assay (Bio-Rad).
In vitro RIF glycosylation assay and steady-state kinetics.
Assays for Rgt1438 enzyme activity consisted of rifamycin and UDP-glucose substrates incubated with 50 mM TAPS (pH 8.4), 10 mM MgCl2, 1 mM dithiothreitol (DTT), 1 mg/ml bovine serum albumin, and Rgt1438. Reaction mixtures were incubated at room temperature, and, at specific time intervals, aliquots were quenched with equal volumes of cold 100% methanol with 0.05% trifluoroacetic acid (TFA). For analysis using thin-layer chromatography (TLC), RIF and UDP-glucose were used at 1 mM and 10 mM, respectively, and the reactions were initiated with the addition of Rgt1438 at 1 μM. At specified time intervals, a 10-μl aliquot of the reaction mixture was quenched with 10 μl of methanol and 5 μl was applied to a silica gel TLC plate and resolved using a 4:1 CHCl3:MeOH solvent system (51). Steady-state kinetics analysis was performed using 2.5 nM enzyme, and reactions were initiated with the addition of 50 μl UDP-glucose in a final reaction volume of 250 μl. To determine antibiotic Km values, the UDP-glucose concentration was held constant at 1 mM and the antibiotic concentrations were adjusted over a range of 1 to 100 μM. For the determination of Km values for UDP-glucose, the rifampin concentration was held constant at 100 μM and the UDP-glucose concentration was adjusted over a range of 5 to 400 μM. Progress curves were generated by analyzing 50-μl aliquots that were taken at several time points and quenched with equal volumes of cold methanol. Quenched reaction mixtures were stored on ice for 10 min and clarified by centrifugation at 4°C. Samples were then subjected to analytical HPLC using a Waters e2695 system (Milford, MA) with an XSelect CSH C18 5-μm column (4.6 by 100 mm) and a linear gradient of 25% to 97.5% acetonitrile in water with 0.05% TFA over 11 min at a flow rate of 1 ml/min. Product peaks were integrated using Waters Empower 3 software to determine the quantity of product generated. Initial rates were determined using the linear portion of progress curves. Reactions were performed in triplicate, and the results were averaged. Initial rates were analyzed by nonlinear least-squares fitting (equation 1) as follows:
Production and purification of 23-O-gluc-RIF.
A large-scale reaction to generate 23-(O-[β-d-glucopyranosyl])rifampin (23-O-gluc-RIF) consisted of 50 mM TAPS (pH 8.4), 1 mM rifampin, 10 mM UDP-glucose, 10 mM MgCl2, 2 mM DTT, and 5 μM Rgt1438 in a total of 10 ml. The 23-O-gluc-RIF product was purified using a Waters Sep-Pak C18 cartridge preequilibrated with distilled water (dH2O). Following the removal of water-soluble components, 23-O-gluc-RIF was eluted with 50% acetonitrile in water and lyophilized. The final product was resuspended in dimethyl sulfoxide (DMSO), and purity was assessed using LC-MS.
Physiochemical analysis of glycosylated RIF products.
High-resolution mass spectra (HRMS) of glycosylated products were obtained using a ThermoFisher LTQ-XL-Orbitrap Hybrid mass spectrometer (ThermoFisher, Bremen, Germany) equipped with an electrospray interface operated in positive-ion and negative-ion modes. The 23-O-gluc-RIF product was identified by comparison of one-dimensional (1D) nuclear magnetic resonance (NMR) spectral data with data previously reported in the literature (51) along with additional 2D NMR analysis performed using a Bruker AVIII 700 MHz instrument and DMSO-d6. Chemical shifts are reported in parts per million relative to tetramethylsilane results using the residual solvent signals at 2.50 and 39.50 ppm as internal references for the 1H and 13C spectra, respectively. The presence of the d-glucose moiety was confirmed by NMR by the signals at δ 102.9 (C-1′), 73.8 (C-2′), 76.6 (C-3′), 70.1 (C-4′), 76.8 (C-5′), and 62.1 (C-6′). The site of glycosylation was determined by a heteronuclear multiple-bond correlation (HMBC) experiment, which showed the couplings between H-23 (signal at 3.33) and C-1′, and the comparison of the 1H and 13C NMR spectra of RIF and 23-O-gluc-RIF. The signals for H-23 and C-23 had shifted downfield, with 0.5 and 10.6 ppm, respectively. As previously reported, the coupling constant of the anomeric proton at 4.42 ppm (d; J = 7.4Hz) was consistent with a β configuration of the glucose moiety (51). No other significant differences were observed.
Phylogenetic analysis of Rgt1438.
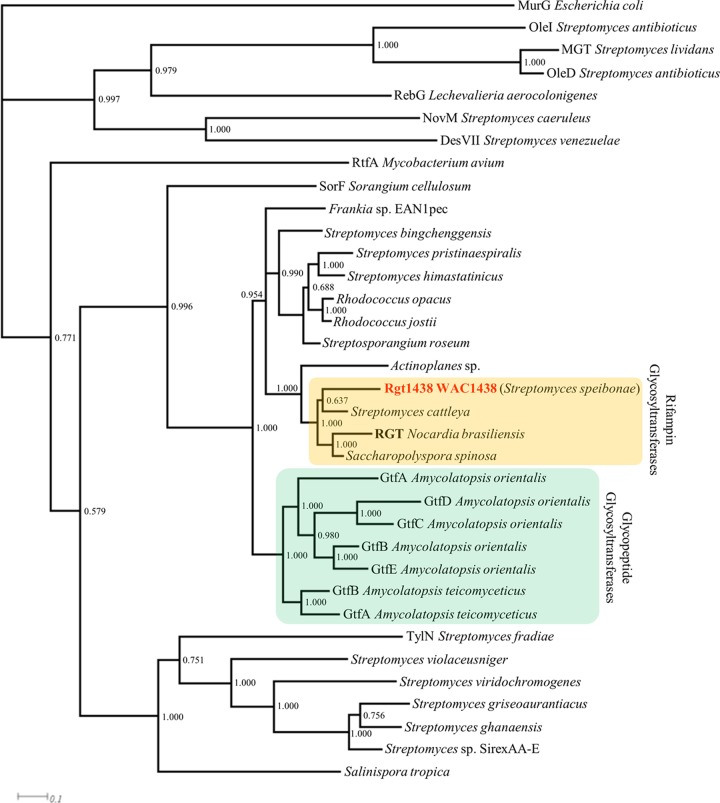
The sequences listed in Table S4 in the supplemental material were obtained from GenBank along with the WAC1438 glycosyltransferase and aligned using MUSCLE (14). The resulting alignment was inspected and manually corrected to remove excessively gapped regions, with a final total of 396 sites. A Bayesian phylogenetic analysis was performed using MrBayes software and a mixed amino acid model on two runs of four chains for 5 million generations (36). Samples were taken every 1,000 generations. Figure 2 shows the resulting consensus tree summarized using the 50% majority rule after removing a burn-in of 1,000 samples.
Fig 2.
Phylogenetic analysis of Rgt1438 with proteins highly homologous to Rgt and natural product glycosyltransferases. Protein sequences were aligned using MUSCLE (14) and a Bayesian phylogenetic analysis was conducted using MrBayes (36) as described in Materials and Methods. Complete lists of proteins (including accession numbers) are located in Table S4 in the supplemental material.
Nucleotide sequence accession number.
The nucleotide sequence for rgt1438 has been submitted to GenBank under accession number JX028276.
RESULTS
Multiple mechanisms of RIF inactivation.
A screen performed by our group identified soil-dwelling bacteria as a reservoir of diverse antibiotic resistance determinants (12). This survey identified a collection of environmental Streptomyces spp. with the ability to inactivate RIF. We explored the various modes of inactivation by analyzing culture filtrates of isolates grown in the presence of RIF. Two different modified antibiotic peaks were identified by LC-MS, with each resulting in a shift in retention time while maintaining a UV-visible light (UV-Vis) absorption spectrum similar to that seen with RIF. Mass spectrometry of the modified RIF products confirmed an increase in the mass-to-charge ratio (m/z) consistent with the glycosylation and phosphorylation of RIF (Table 1). Additionally, a number of isolates appeared to decompose RIF where no identifiable product could be detected. We sequenced the 16S rRNA gene of each isolate and confirmed that, with the exception of one Amycolatopsis species, they all belonged to the Streptomyces genus (Table 1). This demonstrates that soil-dwelling actinomycetes utilize a variety of mechanisms to inactivate RIF.
Table 1.
Rifampin-inactivating isolates and mechanisms of inactivation
| Isolate | 16S rRNA relatedness | Rifampin product (m/z [M+H]+)a | Mechanism of inactivation |
|---|---|---|---|
| WAC1438 | Streptomyces speibonae | 985.6 | Glycosylation |
| WAC4839 | Streptomyces olivoverticillatus | 985.1 | Glycosylation |
| WAC5038 | Streptomyces pseudogriseolus | 985.2 | Glycosylation |
| WAC4747 | Streptomyces viridochromogenes | 900.7 | Phosphorylation |
| WAC5090 | Streptomyces chartreusis | 900.7 | Phosphorylation |
| WAC5242 | Streptomyces flavovariabilis | 900.8 | Phosphorylation |
| WAC4791 | Streptomyces chartreusis | Decomposition | |
| WAC4782 | Streptomyces flavovariabilis | Decomposition | |
| WAC5041 | Amycolatopsis echigonensis | Decomposition | |
| WAC5122 | Streptomyces castaneus | Decomposition | |
| WAC5188 | Streptomyces umbrinus | Decomposition |
Rifampin m/z [M+H]+, 823.6.
WAC1438 inactivates RIF via a RIF glycosyltransferase.
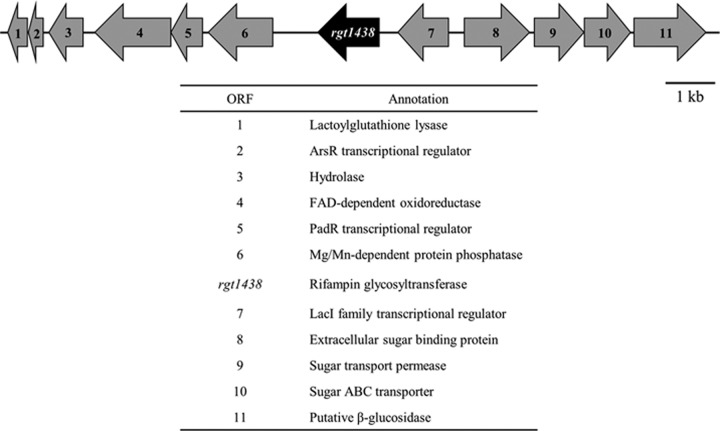
The glycosylation of RIF has been previously reported as a resistance mechanism shown by the pathogenic actinomycete Nocardia brasiliensis (51). We identified a total of 3 Streptomyces isolates that inactivated RIF by glycosylation and selected one of these, WAC1438, for follow-up (Table 1). To identify the gene associated with the RIF glycosyltransferase, we prepared a draft genome sequence of WAC1438 using the Roche 454 platform. Using the unpublished rgt sequence from N. brasiliensis as a reference (accession number AF394943.1), we identified an ORF, rgt1438, located on a 43-kb contig (accession number JX028276) which showed 72% nucleotide and 69% protein identity to the N. brasiliensis element. A blastp analysis of Rgt1438 searching the nonredundant protein sequence database revealed a similarity to members of the inverting GT1 family of glycosyltransferases, which are commonly involved in glycosylation of natural product scaffolds (Fig. 2) (28). In particular, Rgt1438 demonstrates strong similarity to members of the family of glycopeptide glycosyltransferases and shows 56% identity and 66% similarity to GtfB belonging to Amycolatopsis orientalis, responsible for transferring glucose from UDP-glucose to the 4-OH of PheGly4 of the vancomycin core (32). Within the WAC1438 genome, rgt1438 is localized with ORFs associated with sugar metabolism (Fig. 3).
Fig 3.
Genetic organization of rgt1438 and surrounding open reading frames in WAC1438.
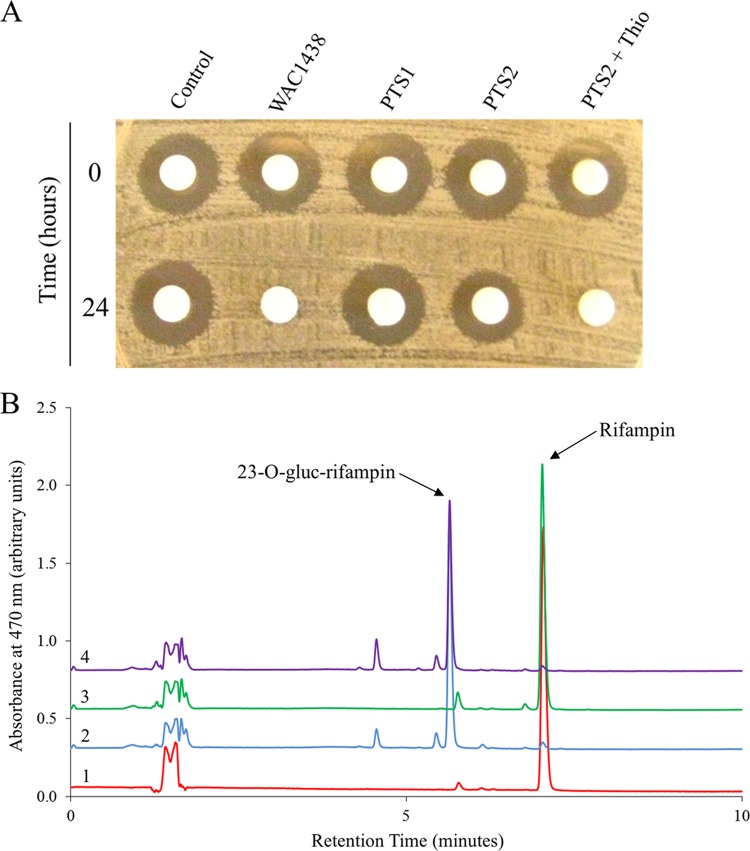
We next constructed a mutant strain of WAC1438 with a disrupted rgt1438 (strain PTS1) and examined its ability to inactivate RIF. In this experiment, strains were grown in the presence of RIF and culture filtrates were analyzed for their residual RIF concentration using the Kirby-Bauer disk diffusion assay. As shown in Fig. 4A, WAC1438 is capable of completely inactivating RIF within 24 h. In comparison, mutant PTS1 is incapable of inactivating RIF. We then complemented mutant PTS1 with an intact rgt1438 on an inducible integrating plasmid (strain PTS2). Under noninducing conditions, strain PTS2 appeared to inactivate a fraction of the RIF, as demonstrated by a slightly smaller zone of inhibition compared to the control (Fig. 4A), most likely attributable to leaky expression from the promoter (ptipA). With the addition of the inducer, strain PTS2 completely inactivated RIF (Fig. 4A), corroborating a role for rgt1438 in drug modification. These results were also confirmed using HPLC methods (Fig. 4B).
Fig 4.
The rgt1438 gene product is responsible for the glycosylative inactivation of rifampin. (A) WAC1438 strains were grown for 3 days at 30°C in liquid SIM, at which point rifampin was added at a concentration of 20 μg/ml, representing time 0 h. Cultures were grown for an additional 24 h, and supernatants were sampled. Supernatants (10 μl) were applied on filter paper disks overlaid on a lawn of B. subtilis 168 and incubated overnight. A control sample included SIM supplemented with rifampin (20 μg/ml). Thiostrepton (Thio [inducer]) was added to achieve a final concentration of 1 μg/ml. (B) Supernatant samples mentioned above were diluted with equal volumes of methanol, centrifuged, and analyzed by HPLC. 1, control (SIM with rifampin); 2, WAC1438; 3, PTS1; 4, PTS2 plus thiostrepton.
rgt1438 confers RIF resistance.
WAC1438 exhibits a MIC of 64 μg/ml for RIF (Table 2). Mutant PTS1, on the other hand, demonstrated a 4-fold-higher sensitivity to RIF (MIC of 16 μg/ml). This increase in sensitivity was reversed when PTS1 was complemented with rgt1438 (strain PTS2) (Table 2). Interestingly, mutant PTS1 displayed a relatively high RIF MIC, suggesting there may be an additional mechanism of resistance to RIF in this organism. Recently, Kim et al. explored the natural RIF resistance of Streptomyces spp. (26). They attributed resistance to a single amino acid polymorphism at position 531 of RpoB (E. coli numbering). In particular, Asn substitution at position 531 (Asn531) correlates with a higher RIF MIC level (≥16 μg/ml) compared to the normal Ser counterpart. This finding is consistent with biochemical and crystallographic studies that showed that Ser531 is important in forming hydrogen bond interactions with RIF (7, 19). Additionally, this amino acid position has been linked to natural RIF resistance in various bacterial genera (1, 18, 42). Investigation of the draft genome sequence of WAC1438 identified a single candidate rpoB gene predicted to encode an enzyme with the Asn531 substitution, thereby accounting for the high RIF MIC in mutant PTS1. Our initial survey identified an additional 2 isolates capable of glycosylating RIF (Table 1). We therefore amplified and sequenced the resistance-determining region of rpoB, which confirmed that those 2 isolates also possessed a RIF-resistant rpoB (data not shown). We conclude that WAC1438 and other Streptomyces strains possess redundant mechanisms of RIF resistance.
Table 2.
Rifampin MIC determinations
| Strain | Thiostreptona | Rifampin MIC (μg ml−1) |
|---|---|---|
| WAC1438 | − | 64 |
| PTS1 (WAC1438 rgt1438::pSET152) | − | 16 |
| PTS1 (WAC1438 rgt1438::pSET152 + pIJ8600K-rgt1438) | − | 64 |
| + | 64 | |
| S. albus J1074 | − | 2 |
| S. albus J1074 + pIJ8600K-rgt1438 | − | 64 |
| + | 128 |
+, thiostrepton added to achieve a final concentration of 1 μg ml−1; −, no thiostrepton added.
To further validate the role of rgt1438 as a resistance determinant, we introduced the associated gene into Streptomyces albus J1074, which has previously been shown to be RIF sensitive (38). S. albus J1074 contains a RIF-sensitive (Ser531) RpoB and demonstrated a RIF MIC of 2 μg/m. S. albus pIJ8600K-rgt1438, on the other hand, showed a 32- to 64-fold increase in RIF MIC (Table 2). This result confirmed the role of rgt1438 in high-level RIF resistance.
Steady-state kinetics analysis of Rgt1438.
Efforts to overexpress Rgt1438 in E. coli failed despite numerous attempts. We therefore turned to protein expression in the actinomycete Rhodococcus erythropolis, which we had previously found useful in generating soluble and active protein expressed from high-GC-content genes (30, 34, 40). Using this system, soluble and active N-terminal His6-tagged Rgt1438 was successfully overexpressed and purified to homogeneity using a two-step chromatographic protocol (see Fig. S1 in the supplemental material). Rgt1438 was assayed using UDP-glucose as a substrate and resolved by TLC, which showed complete turnover of RIF within 60 min with 1 μM enzyme (see Fig. S1 in the supplemental material). The RIF product was then purified and analyzed by high-resolution mass spectrometry and NMR, which verified the regiospecificity of glucose transfer and identified the product as 23-(O-[β-d-glucopyranosyl])rifampin or 23-O-gluc-RIF (Fig. 1B; see also Table 4 and Table S2 and Table S3 in the supplemental material) as previously described (51).
Table 4.
High-resolution mass spectrometry analysis of various glycosylated rifamycin antibiotics
| Antibiotic | Glycosylated product |
||||
|---|---|---|---|---|---|
| Exact mass (Da) | Expected m/z (Da) |
Observed m/z (Da) |
|||
| [M+H]+ | [M−H]− | [M+H]+ | [M−H]− | ||
| Rifampin | 984.4580 | 985.4659 | 983.4509 | 985.4696 | NDa |
| Rifamycin B | 917.3681 | 918.3760 | 916.3602 | ND | 916.3638 |
| Rifamycin SV | 857.3470 | 858.3549 | 856.3391 | ND | 856.3436 |
| Rifabutin | 1,008.4943 | 1,009.5022 | 1,007.4864 | 1,009.5054 | ND |
| Rifaximin | 947.4052 | 948.4131 | 946.3973 | 948.4166 | ND |
ND, not determined.
We then developed a quantitative HPLC method to assess the steady-state kinetics properties of Rgt1438. The enzyme demonstrated normal rectangular hyperbolic saturation with both RIF and UDP-glucose as variable substrates with Km values of 3.74 μM and 24.1 μM, respectively, and a kcat of 0.20 per second per enzyme molecule. We next explored the substrate specificity of Rgt1438 with a number of semisynthetic rifamycin antibiotics and the natural product rifamycin B. All the rifamycins tested were substrates for Rgt1438, with comparable kinetics constants (Table 3). The various glycosylated products were confirmed using high-resolution mass spectrometry (Table 4). A number of nonrifamycin antibiotics (glycopeptides, macrolides, etc.) were also evaluated as the substrates for Rgt1438; however, no detectable products could be identified (data not shown).
Table 3.
Kinetic constants of Rgt1438
| Substrate | Km (μM) | kcat (s−1) | kcat/Km (s−1 M−1) |
|---|---|---|---|
| Rifampin | 3.74 ± 0.04 | 0.200 | 5.35 × 104 |
| Rifamycin B | 6.41 ± 1.02 | 1.123 | 1.75 × 105 |
| Rifamycin SV | 1.43 ± 0.50 | 0.153 | 1.07× 105 |
| Rifabutin | 1.42 ± 0.39 | 0.151 | 1.06 × 105 |
| Rifaximin | 6.09 ± 0.14 | 0.163 | 2.68 × 104 |
| UDP-glucose | 24.1 ± 2.06 | 8.30 × 103 |
DISCUSSION
The discovery of rifamycin in extracts from A. mediterranei, a bacterium isolated from a soil sample collected in the French Riviera, was first reported in 1959 (39). It has been hypothesized that microorganisms may produce these sophisticated secondary metabolites to deter competitors in their respective niches, providing the producing organism a selective advantage. Consequently, microorganisms have also adapted to cope with this noxious chemical landscape and have coevolved specialized resistance mechanisms. The environment should thus serve as a reservoir of the most ancient and diverse antibiotic resistance determinants. There is now growing evidence that antibiotic resistance is ancient and that environmental microorganisms, including the antibiotic producers, are sources of resistant determinants found in pathogens (11, 49). The group transfer mode of antibiotic resistance is the largest and most diverse mechanism of resistance (50). This mode requires highly specialized and selective enzymes, which modify antibiotics in a manner that attenuates their ability to bind their target.
In this study, we demonstrated that environmental bacteria utilize an assortment of mechanisms to inactivate RIF. We have identified a subset of actinomycetes that inactivate RIF by glycosylation, phosphorylation, and decomposition (Table 1). Although these modes of inactivation have been previously recognized in other bacteria, primarily in pathogenic Rhodococcus and Nocardia spp., this is the first report identifying environmental Streptomyces and Amycolatopsis spp. as a reservoir of these elements as well (2, 43, 51, 52).
Glycosylation is a common mechanism in the elaboration of natural product chemical diversity. The associated genes are often colocated within the biosynthetic gene cluster and act late in the assembly of the natural product. Resistance by glycosylation is a strategy utilized by the macrolide producers Streptomyces antibioticus (oleandomycin) and Streptomyces venezuelae (methymycin-neomethymycin) as a mode of self-immunity (6, 47, 53). In these examples, the inactive glycosylated product is assembled intracellularly in a nontoxic state and then excreted by specialized transporters. Specific β-glucosidases on the exterior of the cell catalyze the removal of sugar moieties, revealing and activating the natural product.
In total, three group transfer mechanisms responsible for RIF inactivation have been reported: ADP-ribosylation, phosphorylation, and glycosylation. These mechanisms target one of two critical hydroxyl residues of the ansa chain required for hydrogen bond interactions with the RpoB target (7). The glycosylative inactivation of RIF was first described by Yazawa et al. as a mechanism of resistance by the pathogenic Nocardia brasiliensis (51). Outside the Nocardia genus, this mechanism of RIF inactivation has not been reported. Our analysis of Rgt1438 from WAC1438, described here, confirms that this enzyme is an efficient inactivator of a broad spectrum of natural and semisynthetic rifamycins. The strain does not encode an obvious rifamycin biosynthetic gene cluster, but the rgt1438 ORF is colocated with a number of genes encoding carbohydrate-metabolizing enzymes, which may suggest a broad substrate profile (Fig. 3).
N. farcinica has been shown to be resistant to RIF as a consequence of possessing an alternative RIF-resistant RpoB (target duplication with resistance-associated site mutation) as well as possessing the Rox enzyme which catalyzes the decomposition of RIF (21, 23). Dual modes of resistance are also shared by WAC1438 where, in addition to the enzyme-mediated inactivation of RIF by glycosylation, it also harbors a resistant rpoB allele. A blastp analysis of the nonredundant protein sequence database also identified rgt orthologs in the genomes of Saccharopolyspora spinosa NRRL 18395 and Streptomyces cattleya NRRL 8057 (Fig. 2). Examination of the rpoB sequences from these two organisms also reveals an Asn531 RpoB type. Interestingly, all Streptomyces spp. that harbor an orthologue of rgt appear to possess a resistant RpoB (Asn531). This highlights that, in addition to the multidrug phenotypes of environmental bacteria, they may also possess multiple modes of resistance to the same antibiotic.
The three group transfer modes of RIF resistance all target critical hydroxyl residues located on the ansa chain. As demonstrated biochemically using the ADP-ribosyltransferase Arr, and reiterated with our work presented here with Rgt1438, the available rifamycins in the arsenal developed thus far are susceptible to the group transfer inactivation mechanisms (5). Semisynthetic modifications to regions of the rifamycin scaffold other than the C3 and/or C3-C4 positions, where most of the semisynthesis has been focused (Fig. 1), may prevent these enzymes from inactivating RIF. The possibility of this has been highlighted with the generation of new rifamycins modified at the C25 position that retain activity against bacteria with Arr (9). While these group transfer resistance mechanisms have yet to emerge in pathogens, next-generation rifamycins would benefit by avoiding the known mechanisms of resistance prior to being launched into the clinic.
With the continued development of new rifamycin antibiotics along with their utility to now combat a much broader range of bacterial infections, the rifamycin family of antibiotics will likely experience an increase in use. This should apply a strong pressure for the selection of RIF resistance determinants that already exist in the environment. For example, Arr, which was originally identified from the nonpathogen Mycobacterium smegmatis, is now mobilized and has been found in a number of clinically relevant bacteria (4, 5, 44). Characterizing the RIF resistome should provide crucial insight for future drug development programs where resistance-proof antibiotics may be strategically designed.
Supplementary Material
ACKNOWLEDGMENTS
We thank Tomohiro Tamura for providing Rhodococcus erythropolis L-88 and the pTIP-QC1 used for overexpression of Rgt1438 and Michael Fischbach for the gift of Streptomyces albus J1074. We thank Inga Kireeva for assistance with the high-resolution mass spectrometry and Grace Yim for edits.
This research was supported by the Canadian Institutes of Health Research (MT-13536) and the Canada Research Chairs program.
Footnotes
Published ahead of print 16 July 2012
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Alekshun M, Kashlev M, Schwartz I. 1997. Molecular cloning and characterization of Borrelia burgdorferi rpoB. Gene 186:227–235 [DOI] [PubMed] [Google Scholar]
- 2. Andersen SJ, Quan S, Gowan B, Dabbs ER. 1997. Monooxygenase-like sequence of a Rhodococcus equi gene conferring increased resistance to rifampin by inactivating this antibiotic. Antimicrob. Agents Chemother. 41:218–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aristoff PA, Garcia GA, Kirchhoff PD, Hollis Showalter HD. 2010. Rifamycins—obstacles and opportunities. Tuberculosis (Edinb.) 90:94–118 [DOI] [PubMed] [Google Scholar]
- 4. Arlet G, et al. 2001. Plasmid-mediated rifampin resistance encoded by an arr-2-like gene cassette in Klebsiella pneumoniae producing an ACC-1 class C beta-lactamase. Antimicrob. Agents Chemother. 45:2971–2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baysarowich J, et al. 2008. Rifamycin antibiotic resistance by ADP-ribosylation: structure and diversity of Arr. Proc. Natl. Acad. Sci. U. S. A. 105:4886–4891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bolam DN, et al. 2007. The crystal structure of two macrolide glycosyltransferases provides a blueprint for host cell antibiotic immunity. Proc. Natl. Acad. Sci. U. S. A. 104:5336–5341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Campbell EA, et al. 2001. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 104:901–912 [DOI] [PubMed] [Google Scholar]
- 8. Chevreux B, Wetter T, Suhai S. 1999. Genome sequence assembly using trace signals and additional sequence information, p 45–56 Proceedings of the German Conference on Bioinformatics (GCB) Universität Trier, Hanover, Germany [Google Scholar]
- 9. Combrink KD, et al. 2007. New C25 carbamate rifamycin derivatives are resistant to inactivation by ADP-ribosyl transferases. Bioorg. Med. Chem. Lett. 17:522–526 [DOI] [PubMed] [Google Scholar]
- 10. Cremonini F, Lembo A. 2012. Rifaximin for the treatment of irritable bowel syndrome. Expert Opin. Pharmacother. 13:433–440 [DOI] [PubMed] [Google Scholar]
- 11. D'Costa VM, et al. 2011. Antibiotic resistance is ancient. Nature 477:457–461 [DOI] [PubMed] [Google Scholar]
- 12. D'Costa VM, McGrann KM, Hughes DW, Wright GD. 2006. Sampling the antibiotic resistome. Science 311:374–377 [DOI] [PubMed] [Google Scholar]
- 13. Desomer J, Dhaese P, Montagu MV. 1990. Transformation of Rhodococcus fascians by high-voltage electroporation and development of R. fascians cloning vectors. Appl. Environ. Microbiol. 56:2818–2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fischbach MA, Walsh CT. 2009. Antibiotics for emerging pathogens. Science 325:1089–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Floss HG, Yu TW. 2005. Rifamycin—mode of action, resistance, and biosynthesis. Chem. Rev. 105:621–632 [DOI] [PubMed] [Google Scholar]
- 17. Forrest GN, Tamura K. 2010. Rifampin combination therapy for nonmycobacterial infections. Clin. Microbiol. Rev. 23:14–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gaurivaud P, Laigret F, Bove JM. 1996. Insusceptibility of members of the class Mollicutes to rifampin: studies of the Spiroplasma citri RNA polymerase beta-subunit gene. Antimicrob. Agents Chemother. 40:858–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gill SK, Garcia GA. 2011. Rifamycin inhibition of WT and Rif-resistant Mycobacterium tuberculosis and Escherichia coli RNA polymerases in vitro. Tuberculosis (Edinb.) 91:361–369 [DOI] [PubMed] [Google Scholar]
- 20. Heuer H, Krsek M, Baker P, Smalla K, Wellington EM. 1997. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 63:3233–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hoshino Y, et al. 2010. Monooxygenation of rifampicin catalyzed by the rox gene product of Nocardia farcinica: structure elucidation, gene identification and role in drug resistance. J. Antibiot. (Tokyo) 63:23–28 [DOI] [PubMed] [Google Scholar]
- 22. Huang DB, DuPont HL. 2005. Rifaximin—a novel antimicrobial for enteric infections. J. Infect. 50:97–106 [DOI] [PubMed] [Google Scholar]
- 23. Ishikawa J, Chiba K, Kurita H, Satoh H. 2006. Contribution of rpoB2 RNA polymerase beta subunit gene to rifampin resistance in Nocardia species. Antimicrob. Agents Chemother. 50:1342–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jin DJ, Gross CA. 1988. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J. Mol. Biol. 202:45–58 [DOI] [PubMed] [Google Scholar]
- 25. Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. 2000. Practical streptomyces genetics. John Innes Foundation Press, Norwich, United Kingdom [Google Scholar]
- 26. Kim H, et al. 2005. Mechanism of natural rifampin resistance of Streptomyces spp. Syst. Appl. Microbiol. 28:398–404 [DOI] [PubMed] [Google Scholar]
- 27. Kim TK, Hewavitharana AK, Shaw PN, Fuerst JA. 2006. Discovery of a new source of rifamycin antibiotics in marine sponge actinobacteria by phylogenetic prediction. Appl. Environ. Microbiol. 72:2118–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lairson LL, Henrissat B, Davies GJ, Withers SG. 2008. Glycosyltransferases: structures, functions, and mechanisms. Annu. Rev. Biochem. 77:521–555 [DOI] [PubMed] [Google Scholar]
- 29. Li J, et al. 2007. Preparation and in vitro anti-staphylococcal activity of novel 11-deoxy-11-hydroxyiminorifamycins. Bioorg. Med. Chem. Lett. 17:5510–5513 [DOI] [PubMed] [Google Scholar]
- 30. Mitani Y, Meng X, Kamagata Y, Tamura T. 2005. Characterization of LtsA from Rhodococcus erythropolis, an enzyme with glutamine amidotransferase activity. J. Bacteriol. 187:2582–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Morisaki N, et al. 2000. Structures of ADP-ribosylated rifampicin and its metabolite: intermediates of rifampicin-ribosylation by Mycobacterium smegmatis DSM43756. J. Antibiot. (Tokyo) 53:269–275 [DOI] [PubMed] [Google Scholar]
- 32. Mulichak AM, Losey HC, Walsh CT, Garavito RM. 2001. Structure of the UDP-glucosyltransferase GtfB that modifies the heptapeptide aglycone in the biosynthesis of vancomycin group antibiotics. Structure 9:547–557 [DOI] [PubMed] [Google Scholar]
- 33. Murphy CK, et al. 2006. In vitro activity of novel rifamycins against rifamycin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 50:827–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nakashima N, Tamura T. 2004. Isolation and characterization of a rolling-circle-type plasmid from Rhodococcus erythropolis and application of the plasmid to multiple-recombinant-protein expression. Appl. Environ. Microbiol. 70:5557–5568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ramaswamy S, Musser JM. 1998. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber. Lung Dis. 79:3–29 [DOI] [PubMed] [Google Scholar]
- 36. Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61:539–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rothstein DM, van Duzer J, Sternlicht A, Gilman SC. 2007. Rifalazil and other benzoxazinorifamycins in the treatment of chlamydia-based persistent infections. Arch. Pharm. (Weinheim) 340:517–529 [DOI] [PubMed] [Google Scholar]
- 38. Sánchez-Hidalgo M, Nunez LE, Mendez C, Salas JA. 2010. Involvement of the beta subunit of RNA polymerase in resistance to streptolydigin and streptovaricin in the producer organisms Streptomyces lydicus and Streptomyces spectabilis. Antimicrob. Agents Chemother. 54:1684–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sensi P, Margalith P, Timbal MT. 1959. Rifomycin, a new antibiotic; preliminary report. Farmaco Sci. 14:146–147 [PubMed] [Google Scholar]
- 40. Shakya T, et al. 2011. A small molecule discrimination map of the antibiotic resistance kinome. Chem. Biol. 18:1591–1601 [DOI] [PubMed] [Google Scholar]
- 41. Shao Z, Dick WA, Behki RM. 1995. An improved Escherichia coli-Rhodococcus shuttle vector and plasmid transformation in Rhodococcus spp. using electroporation. Lett. Appl. Microbiol. 21:261–266 [DOI] [PubMed] [Google Scholar]
- 42. Stamm LV, Bergen HL, Shangraw KA. 2001. Natural rifampin resistance in Treponema spp. correlates with presence of N531 in RpoB Rif cluster I. Antimicrob. Agents Chemother. 45:2973–2974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tanaka Y, et al. 1996. Different rifampicin inactivation mechanisms in Nocardia and related taxa. Microbiol. Immunol. 40:1–4 [DOI] [PubMed] [Google Scholar]
- 44. Tribuddharat C, Fennewald M. 1999. Integron-mediated rifampin resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:960–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tupin A, et al. 2010. Resistance to rifampicin: at the crossroads between ecological, genomic and medical concerns. Int. J. Antimicrob. Agents 35:519–523 [DOI] [PubMed] [Google Scholar]
- 46. Van Scoy RE, Wilkowske CJ. 1999. Antimycobacterial therapy. Mayo. Clin. Proc. 74:1038–1048 [DOI] [PubMed] [Google Scholar]
- 47. Vilches C, Hernandez C, Mendez C, Salas JA. 1992. Role of glycosylation and deglycosylation in biosynthesis of and resistance to oleandomycin in the producer organism, Streptomyces antibioticus. J. Bacteriol. 174:161–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wehrli W. 1977. Ansamycins. Chemistry, biosynthesis and biological activity. Top. Curr. Chem. 72:21–49 [DOI] [PubMed] [Google Scholar]
- 49. Wright GD. 2007. The antibiotic resistome: the nexus of chemical and genetic diversity. Nat. Rev. Microbiol. 5:175–186 [DOI] [PubMed] [Google Scholar]
- 50. Wright GD. 2005. Bacterial resistance to antibiotics: enzymatic degradation and modification. Adv. Drug Deliv. Rev. 57:1451–1470 [DOI] [PubMed] [Google Scholar]
- 51. Yazawa K, et al. 1993. Inactivation of rifampin by Nocardia brasiliensis. Antimicrob. Agents Chemother. 37:1313–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yazawa K, Mikami Y, Maeda A, Morisaki N, Iwasaki S. 1994. Phosphorylative inactivation of rifampicin by Nocardia otitidiscaviarum. J. Antimicrob. Chemother. 33:1127–1135 [DOI] [PubMed] [Google Scholar]
- 53. Zhao L, Beyer NJ, Borisova SA, Liu HW. 2003. Beta-glucosylation as a part of self-resistance mechanism in methymycin/pikromycin producing strain Streptomyces venezuelae. Biochemistry 42:14794–14804 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.