Abstract
The heterogeneity and the increasing clinical importance of the Enterobacter cloacae complex have often been discussed. However, little is known about molecular factors causing pathogenicity within this nomenspecies. Here, we analyzed the genetic differences between an avirulent plant isolate and a pathogenic strain causing an outbreak with septicemia in three patients. We identified an IncHI-2 plasmid as a major difference between these two strains. Besides resistance to several antibiotics, this plasmid encoded a silver resistance determinant. We further showed that this sil determinant was present not only in the analyzed outbreak strain but also in the vast majority of clinical isolates of the E. cloacae complex, predominantly in (sub)species that frequently cause nosocomial infections. The identified sil determinant was highly conserved within the E. cloacae complex and mediated resistance to up to 600 μM silver nitrate. As silver is often used as a disinfectant and treatment for burn wounds, we present here an important fitness factor within the clinically most prevalent subspecies of the E. cloacae complex. This provides a possible explanation for their unequal involvement in nosocomial and especially burn wound infections.
INTRODUCTION
The term “Enterobacter cloacae” is often used for a complex consisting of nine species and subspecies and at least three additional genovars, which are all genetically and phenotypically closely related to each other. Members of this complex have been isolated from plants (38), insects (11), and open bodies of water (19), reflecting the wide distribution of this intestinal commensal in the environment. However, the increasing number of epidemiological studies highlighting severe cases of bacteremia caused by the E. cloacae complex reflects the particular importance of these microorganisms as clinical pathogens (4, 21, 36). Especially at (neonatal) intensive care (2, 10, 12, 24, 25, 29, 34, 44, 45) and burn units (31, 32), (sub)species of the E. cloacae complex often emerge as the cause of severe outbreaks, with several patients being infected by the same clone.
Despite this clinical importance, few pathogenicity factors have so far been characterized within the E. cloacae complex, and most of them have been found by screening for known virulence-associated properties. Keller et al. (26) demonstrated broad distribution of serum resistance, aerobactin production, and hemagglutinin production, whereas Simi et al. (42) found a low-molecular-weight enterotoxic hemolysin in seven of 50 E. cloacae strains analyzed. Sevcik et al. (39) described a paralytic toxin called saxitoxin found in two species of the E. cloacae complex, and the level of the outer membrane protein OmpX has been directly linked to invasiveness of E. cloacae in rabbit ileal tissue (9). Unfortunately, these studies did not account for the diversity of the E. cloacae complex. This might be of importance, as some genovars were repeatedly overrepresented in clinical isolates (15, 22, 30), while others were significantly underrepresented. Furthermore, our recent data show that some genotypes are significantly associated with certain infection sites (27).
Paauw et al. (36) studied the pathogenicity of the E. cloacae complex by performing comparative genomic hybridization to identify genes specific for an Enterobacter hormaechei outbreak strain. They thereby found the large IncHI-2 plasmid pQC, which harbored many resistance determinants, probably leading to a greater likelihood of survival in the hospital environment.
In the present study, we sought to identify virulence-associated genes within the E. cloacae complex by direct whole-genome comparison of an outbreak strain and an avirulent plant isolate using a subtractive hybridization assay (1). With this approach, we also found a large IncHI-2 plasmid as a major difference between the two strains. Besides several antibiotic resistance determinants, this plasmid harbored a silver resistance determinant (sil). We could demonstrate that the sil determinant was phenotypically functional and was present significantly more often in genotypes that frequently cause nosocomial infections. This resistance to silver ions represents a potent fitness factor and could therefore partly explain clonal outbreaks as well as the general predominance of members of the E. cloacae complex with certain genotypes as nosocomial pathogens.
MATERIALS AND METHODS
Bacterial strain collection.
The strains used in the subtraction studies belonged to the species Enterobacter asburiae. The presumed virulent strain EN-373 (= M40, tester) had infected three infants nursed at a neonatal intensive care unit (12) and was kindly provided by Victoria Fernandez-Baca, Palma de Mallorca, Spain. This outbreak strain (tester) was compared with strains 501R3 and E6 (driver), both plant-beneficial bacteria that have been recovered independently from cucumber (38) and were considered avirulent. These strains were kindly provided by Daniel P. Roberts, Beltsville, MD.
All clinical isolates were recovered from specimens sent from the University Hospital Großhadern, Munich, Germany, to its diagnostic bacteriology department (Max von Pettenkofer Institute). One hundred sixty-four isolates were enrolled, including all blood culture isolates (n = 67) identified as E. cloacae complex between 1997 and 2002. Furthermore, study isolates included 97 isolates randomly taken from our E. cloacae strain bank collected from 2000 to 2002. This included isolates recovered from respiratory tract (n = 39), gastrointestinal tract (n = 13), genitourinary tract (n = 16), wound swabs (n = 12), central venous lines and Foley catheters (n = 9), and abdominal (n = 6) and pleural cavities (n = 2). All isolates were identified as members of the E. cloacae complex by API20E (bioMérieux, Marcy l'Étoile, France) and were assigned to their genovars by partial hsp60 sequence analysis as described previously (22). To our knowledge, none of the patients was treated with silver compounds for respective infections. However, silver-coated devices such as central venous lines and Foley catheters were widely used in the hospital. In all analyses, Escherichia coli DH5α served as negative control and E. coli DH5α pMG101 (kindly provided by Simon Silver, Chicago, IL) as positive control. All bacteria were grown in Luria-Bertani (LB) medium and cryopreserved at −80°C with 20% glycerol.
Preparation of chromosomal DNA.
For preparations of chromosomal DNA, strains were incubated overnight in 5 ml LB broth, washed and incubated in Tris-HCl buffer (pH 8.5), and lysed with 0.5% SDS and 100 μg/ml proteinase K for 1 h at 37°. After the addition of NaCl to a concentration of 0.7 M, cell debris was removed using cetyltrimethylammonium bromide (CTAB) and chloroform-isoamyl alcohol extraction. DNA was recovered by precipitation in 60% isopropanol and dissolved in 500 μl of Tris Cl buffer (pH 8.5). Small and medium-sized plasmids (up to 20 kb) of the clinical isolate EN-373 were prepared using a Nucleobond PC100 kit (Macherey & Nagel, Düren, Germany). Chromosomal DNA and plasmid DNA solutions of clinical strain EN-373 were mixed in a proportion of 3:1 (vol/vol). DNA (10 μg) of each of the clinical and the environmental strains was cleaved with RsaI (Fermentas GmbH, St. Leon-Roth, Germany).
Subtractive hybridization.
For DNA subtraction, a bacterial PCR-Select DNA subtraction kit (Clontech, La Jolla, CA) and GeneAmp PCR system 9700 (Applied Biosystems, Darmstadt, Germany) were used, following the manufacturer's instructions. After subtractive hybridization, DNA fragments unique to the clinical isolate EN-373 were PCR amplified, cloned into pCR2-1-TOPO vector, and transformed into E. coli TOP-10 One Shot chemically competent cells (Invitrogen BV, Groningen, The Netherlands). After beta-galactosidase read-out of the transformants, white colonies were separated twice on LB agar containing 100 μg/ml ampicillin. After quick heat lyses of single colonies in 100 μl H2O, the cloned inserts were PCR amplified with the vector-specific primers M13F and M13R. The primers bind 114 bp upstream and 95 bp downstream, respectively, from the insertion site of the TOPO cloning vector. Hence, PCR products obtained corresponded to the DNA fragment unique to the outbreak strain plus 209 bp of the cloning vector. PCR products were visualized in 2% agarose gel after ethidium bromide staining. After purification using a Nucleospin PCR cleaning kit (Macherey-Nagel, Düren, Germany), PCR products were sequenced with the dye terminator method (BigDye reaction solution, API Prism 277 sequencer).
PCR amplifications and sequencing.
Amplification reactions were performed with an Applied Biosystems GeneAmp PCR system 2700 and Taq Gold DNA polymerase (Perkin-Elmer, Branchburg, NJ) under standard conditions (500 pmol of each primer, 200 μM nucleotide mix, 10 mM Tris-HCl [pH 8.3], 5 mM KCl, 1,5 mM MgCl2). Primers are listed in Table 1. Reactions were carried out in a total volume of 50 μl and started after a preheating time of 10 min at 95°C. Samples were subjected to 30 cycles of 30 s denaturation at 95°C, 30 s annealing (annealing temperatures are listed in Table 1), and 2 min of extension at 72°C. Cycles were followed by a final elongation step of 15 min at 72°C. PCR products were visualized in 2% agarose gels after electrophoresis and ethidium bromide staining. DNA was cleaned using a Nucleospin extraction kit (Macherey-Nagel, Düren, Germany) for further processing, following the manufacturer's guidelines.
Table 1.
Primers and annealing temperatures used for PCR amplifications
| PCRa | Primer | Primer sequenceb | Size of PCR product (bp) and temp (°C) | Position on plasmid (accession no) |
|---|---|---|---|---|
| silE-silS | sil 1105 | GTT CGT CAT GGT TTC ATG AGCc | 1,915; 58.0 | SATY pMG101, silE 1105–1125 (AF067954) |
| sil 3019 | CTG ACC TTT TTT ATC AGC CTGc | SATY pMG101, silS 3019–2999 (AF067954) | ||
| silS | sil 3019 | CTG ACC TTT TTT ATC AGC CTGc | 741; 58.0 | SATY pMG101, silS 3019–2999 (AF067954) |
| sil 2279 | GCG GGT AAA RAC ATC CTC AATc | SATY pMG101, silS 2279–2299 (AF067954) | ||
| silS-silR | sil 2999 | CAG GCT GAT AAA AAA GGT CAGc | 574; 58.0 | SATY pMG101, silS 2999–3019 (AF067954) |
| sil 3572 | AAC GGC TGG GAT ATC RTS CGc | SATY pMG101, silR 3572–3553 (AF067954) | ||
| silR | sil 3244 | CCC TGA TGG CGA AKG AAG AAc | 329; 58.0 | SATY pMG101, silR 3244–3263 (AF067954) |
| sil 3572 | AAC GGC TGG GAT ATC RTS CGc | SATY pMG101, silR 3572–3553 (AF067954) | ||
| silS-silC | sil 2999 | CAG GCT GAT AAA AAA GGT CAGc | 1,472; 58.0 | SATY pMG101, silS 2999–3019 (AF067954) |
| sil 4470 | CCA GTT GCT GRY TGA AAT AGCc | SATY pMG101, silC 4470–4449 (AF067954) | ||
| silC | sil 4090 | CGG GCT GGC GHA MCT TTT TTGc | 381; 58.0 | SATY pMG101, silC 4090–4111 (AF067954) |
| sil 4470 | CCA GTT GCT GRY TGA AAT AGCc | SATY pMG101, silC 4470–4449 (AF067954) | ||
| silC-silB | sil 4090 | CGG GCT GGC GHA MCT TTT TTGc | 1,942; 58.0 | SATY pMG101, silC 4090–4111 (AF067954) |
| sil 6031 | TCC ATA TCC ATA AAV GGC GATc | SATY pMG101, silB 6031–6011 (AF067954) | ||
| silB-silA | sil 6549 | ATT CAG GGD ATG GAC CCG GTc | 732; 58.0 | SATY pMG101, silB 6549–6568 (AF067954) |
| sil 7280 | CCG GGA TAG CTG GTT TTR ATAc | SATY; pMG101, silA 7280–7260 (AF067954) | ||
| silA-silP | sil 9936 | TGG CGT GGT SAT GCT GAT GTAc | 1,228; 58.0 | SATY pMG101, silA 9936–9956 (AF067954) |
| sil 11163 | TGT TCA CTG GCA TCT TCG GTAc | SATY pMG101, silP 11163–11143 (AF067954) | ||
| silP | sil 11882 | GGC GAT AAG CTC CGC ATC AGAc | 524; 58.0 | SATY pMG101, silP 11882–11902 (AF067954) |
| sil 12405 | TCC ACT TTT TCA AGA CGC TCAc | SATY pMG101, silP 12405–12385 (AF067954) | ||
| RepHIA | RepHIA-F | ACC TTT CTT AAC YTC TTG ATAd | 450; 51.1 | SEMA pR478, 813–793 (BX664015) |
| RepHIA-R | TGT GGT ATC AGT TCW ACA AAC ATd | SEMA pR478, 364–386 (BX664015) | ||
| M13 | M13-F | GTA AAA CGA CGG CCA G | Depending on insert; 50.0 | pCR2.1-TOPO, 391–406 |
| M13-R | CAG GAA ACA GCT ATG AC | pCR2.1-TOPO, 205–221 |
The structure of the complete sil operon is depicted in Fig. 2.
A = adenine, C = cytosine, G = guanosine, T = thymine, I = inosine; D = G, T or A; H = A, C or T; K = G or T; M = A or C; R = A or G; S = G or C; V = G, C or A; W = A or T; Y = C or T.
PCR-amplified and cleaned DNA was sequenced using the dye terminator method. To prepare cycle sequencing reaction mixtures, 0.2 μg of purified DNA was added to a 20-μl reaction solution containing 4 μl of BigDye dye terminator sequencing mix and 0.7 μl of a 5-pmol/μl primer solution. Cycle sequencing was performed in an Applied Biosystems GeneAmp 2700 PCR system. Cycle parameters were 25 cycles with an initial 96°C denaturation step of 10 s, followed by an annealing step at 55°C of 10 s and an extension step of 4 min at 60°C. Sequences were determined by electrophoresis with the ABI Prism 377 DNA sequencer.
Phenotypic tests.
Inhibition of bacterial growth was assessed by measuring the OD600 after incubation (16 h, 36 ± 1°C, room air) of five representative isolates in liquid LB medium supplemented with different concentrations of AgNO3 (dissolved to a stock solution of 40 mM in H2O; Sigma-Aldrich). Three PCR-negative isolates (114, 501R3, and E6), two PCR-positive isolates (244 and 373), and E. coli strains DH5α and DH5α(pMG101) (negative and positive controls, respectively) were included. To measure growth inhibition in the absence of NaCl ions, LB agar plates were prepared without addition of NaCl but supplemented with different concentrations of AgNO3 medium as previously described (17). After incubation (16 h, 36 ± 1°C, room air), growth of strains was assessed and documented by photography.
RESULTS
Subtractive hybridization of EN-373 and 501R3.
One hundred seventy-eight white colonies were obtained by subtraction of strains 373 and 501R3. Fifty of them were randomly selected for further analysis. Twenty-one of the colonies analyzed (42%) yielded PCR products of 210 bp, meaning that they did not contain a DNA fragment. The PCR products of the other 29 clones were longer than 210 bp.
BLAST search for homologies yielded the results listed in Table 2. Four sequences were not usable. Ten clones contained novel sequences with no homology to any gene previously identified. Fifteen inserts gave homologies to known sequences. Their GenBank accession numbers are listed in Table 2. Six sequences (A-051, D-007, C-005, C-007, C-006, and A-000) showed homology to housekeeping genes of various members of the Enterobacteriaceae, one (A-053) to a two-component regulatory system, one (A-088) to a hypothetical protein, and one (B-031) to a putative kinase, both of unknown functions. Two clones (A-007 and B-007) showed homology to nirAB, nreAB-like genes, and ncrABC, genes of the nickel/cobalt resistance determinants present in Klebsiella oxytoca, Legionella pneumophila, and the Hafnia alvei plasmid NRS148.
Table 2.
BLAST GenBank search with sequences of DNA fragments found in 28 clones after subtractive hybridization of clinical outbreak strain EN-373 and plant isolates 501R3 and E6
| Subtractive hybridization | Clone |
Database information about homologous genes |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Namea | % C+G | Segment (bp) with homologyb | Accession no. | Homologous gene(s) | Species, serovar, and plasmidc | Homologous regione | % IDd | Scoref | Protein function | |
| EN-373 and 501R3 | A-114 | 56 | 1–288 | BX664015 | Hypothetical | SEMA pR478 | 71129–70840 | 97 | 511 | Putative ATP/GTP-binding protein integrated in tellurium resistance operon |
| A-073 | 56 | 1–275 | BX664015 | Hypothetical | SEMA pR478 | 71123–70848 | 96 | 468 | Putative ATP/GTP-binding protein integrated in tellurium resistance operon | |
| B-036 | 56 | 21–214 | BX664015 | Hypothetical | SEMA pR478 | 71169–70976 | 99 | 377 | Putative ATP/GTP-binding protein integrated in tellurium resistance operon | |
| B-007 | 48 | 87–321 | AY492000 | nirA and -B and nreA and -B homologues | KLOX | 1318–1553 | 92 | 325 | Nickel/cobalt resistance determinant; CCUG15788 integral membrane protein | |
| AY030282 | LEPN | |||||||||
| A-104 | 55 | 19–447 | AF067954 | silS | SATM pMG101 | 2624–2196 | 82 | 242 | sil operon (plasmidal silver resistance genes) | |
| BX664015 | SEMA pR478 | 127176–126824 | 83 | 226 | ||||||
| A-007 | 49 | 87–267 | AY492000 | nirAB, nreAB, and ncrABC homologues | KLOX | 1318–1499 | 91 | 226 | Nickel/cobalt resistance determinant CCUG15788 integral membrane protein | |
| AY030282 | LEPN | 1690–1875 | 226 | |||||||
| AF322866 | HAAL pNRS148 | 143 | ||||||||
| A-051 | 52 | 245–415 | AE008877 | ibpB and ibpA | SATM LT2 | 12266–12437 | 88 | 174 | Small heat shock protein | |
| D-007 | 63 | 32–170 | AE016840 | hcr | SATY Ty2 | 255337–255475 | 89 | 165 | NADH oxidoreductase | |
| C-005 | 58 | 54–212 | AE016762 | dacD | ESCO CFT073 | 261785–261943 | 84 | 117 | Putative enzyme d-alanyl-d-alanine carboxypeptidase | |
| B-031 | 57 | 26–188 | AE005674 | ycfW | SHFL 2a | 1166030–1166192 | 82 | 101 | Putative kinase | |
| A-000 | 55 | 23–165 | AE008886 | trkH | SATM LT2 | 6464–6322 | 83 | 101 | Trk family potassium transport protein | |
| A-053 | 59 | 174–244 | AE008803 | yfbE | SATM LT2 | 4172–4242 | 88 | 78 | Putative DegT/DnrJ/EryC1/StrS family regulatory system | |
| C-007 | 48 | 13–48 | AE008706 | yafB | SATM LT2 | 15741–15706 | 94 | 56 | 2,5-Diketo-d-gluconate reductase B | |
| A-088 | 61 | 139–225 | BA000007 | Hypothetical | ESCO O157:H7 | 3370219–3370306 | 82 | 48 | Ecs3386, unknown function | |
| C-006 | 60 | 1–24 | KPN293850 | EVGA | KLPN | 56–79 | 95 | 40 | Putative positive transcription regulator EVGA | |
| EN-373 and E6 | E-013 | 48 | 1–624 | AF550679 | orf18 | ESCO p1658/97 | 54236–54860 | 99 | 1199 | Unknown |
| E-001 and E-007 | 47 | 1–571 | SMU60283 | trag1 | SEMA pR478 | 386–958 | 99 | 1098 | Putative restriction methylase | |
| 19–125, 491–510 | AF250878 | SATY pR27 | 47386–47468 | 86 | 101 | |||||
| E-022 | 42 | 1–451 | SMU62007 | PR478 intergenic region between RepHI1A and tra2 | SEMA pR478 | 2288–2738 | 99 | 886 | None; RepHI1A, putative Rep protein Tra2; putative pilin precursor; similar to TrhA from SATY plasmid HCM1 | |
| E-010 | 42 | 1–386 | SMU62007 | Like traE of F plasmid | SEMA pR478 | 4201–4586 | 95 | 632 | Plasmid transfer protein | |
| 22–204 | AF250878.1 | SATY pR27 | 36704–36522 | 80 | 100 | |||||
| E-009 | 42 | 153–457 | STYFIMCLUS | Putative tnpB | SHFL 2a | 1356–1052 | 97 | 541 | Putative transposase B | |
| E-011 | 44 | 1–275 | AE000134 | insA-2 | ESCO | 4764–5038 | 96 | 472 | IS element 30 | |
| E-034 | 44 | 8–188 | EAU67194 | IS4321R | ENAE pR751 | 14134–13954 | 100 | 359 | Putative transposase binding site | |
| E-037 | 31 | 1–169 | SMU62007 | htdV | SEMA IncHI2 plasmid R478 | 7640–7472 | 84 | 183 | Putative transfer-associated protein | |
| E-002 | 42 | 531–612 | AF550679 | tnpA | ESCO p1658/97 | 68019–68100 | 92 | 115 | Transposase | |
| AF342826 | 7419–7338 | 92 | 115 | |||||||
| E-023 | 43 | 83–330 | AF250878 | R0009, putative gene | SATY pR27 | 12627–12380 | 80 | 103 | Hypothetical cell surface glycoprotein (S-layer protein) | |
| 83–330 | STYPPHCM1 | SATY CT18 pHCM1 | 69452–69699 | 80 | 103 | |||||
| E-029 | 48 | 1–247 | AF250878 | R0135, putative gene | SATY pR27 | 121746–121500 | 79 | 86 | Cell envelope; putative outer membrane constituents | |
| 1–247 | STYPPHCM1 | SATY CT18 pHCM1 | 187835–188081 | 79 | 86 | |||||
| E-033 | 50 | 51–139, 216–268, 330–450 | AF250878 | R0156 | SATY pR27 | 142955–143354 | 83 | 68 | Unknown | |
| STYPPHCM1 | Putative gene | SATY pHCM1 | 137009–137129 | 83 | 68 | |||||
Clones A-117, A-008, A-018, A-056, A-087, B-008, B-018, C-001, C-004, C-011, E-004, E-005, E-008, E-014, E-015, E-017, E-019, E-020, E-021, E-024, E-025, E-030, E-031, E-032, and E-035 are novel sequences.
Sequence section of insert of respective clone with homology to the genes with the indicated accession numbers.
ENAE, Enterobacter aerogenes; ENCL, Enterobacter cloacae; ESCO, Escherichia coli; HAAL, Hafnia alvei; LEPN, Legionella pneumophila; KLOX, Klebsiella oxytoca; KLPN, Klebsiella pneumoniae; SAEN, Salmonella enterica serotype Enteritidis; SATY, Salmonella enterica serotype Typhi; SATM, Salmonella enterica serotype Typhimurium; SHFL, Shigella flexneri.
Percent nucleotide identity.
Sequence section of accession number with homology to insert of respective clone.
BLASTN 2.2.5 score (bits).
Four (A-114, A-073, B-036, and A-104) of the five clones with the best BLAST scores contained DNA fragments with high-level identity to genes on plasmid R478, found in Serratia marcescens. pR478 is a large group H incompatibility (IncHI) conjugative plasmid with a length of 274.762 bp which harbors determinants of resistance to several heavy metals (ter operon for tellurium resistance, mer operon for mercuric resistance, sil operon for silver resistance, cop operon for copper resistance, and ars operon for arsenic resistance) and antibiotics (tetA gene for tetracycline resistance and aphA gene for kanamycin resistance). While three clones (A-114, A-073, and B-036) showed homology to plasmidic genes with unknown functions, one clone (A-104) showed homology to the silS gene. silS is one of seven genes of a silver resistance determinant characterized by Gupta et al. (16). It encodes a membrane sensor functioning as part of a regulatory system of the resistance operon. The silPABCRSE silver resistance determinant has been identified on a variety of group H incompatibility plasmids, i.e., MG101, MIP233, WR23, and MIP235 from Salmonella enterica serovars and R476b and R478 from Serratia marcescens. Two accession numbers for sequences with homologies are listed in Table 2, i.e., AF067954 for pMG101 and BX664015 for pR478. Both are members of IncHI subgroup 2 (IncHI-2). The DNA sections with homology to IncHI plasmids were the most important genomic differences found between clinical outbreak strain EN-373 and plant isolate 501R3.
Subtractive hybridization of EN-373 and E6.
A second subtractive hybridization experiment was performed with another plant isolate as a driver (E6), in order to confirm the results of the first experiment. After subtractive hybridization, about 220 white clones were isolated, of which 40 were randomly selected for further analysis. Eight of them did not contain an insert. Four clones yielded nonsense sequences with large clusters of repeats. The remaining 28 sequences were compared to sequences in the GenBank database. BLAST search results of this experiment are listed in Table 2. Fifteen clones yielded unknown sequences. Thirteen sequences had homology to transferable DNA, i.e., transposons (E-002 and E-009), IS elements (E-011) or plasmid DNA. Again, five (E-001, E-007, E-022, E-010, and E-037) of the clones with the best BLAST scores showed homologies to the IncHI-2 plasmid R478 of S. marcescens. Three clones (E-023, E-029, and E-033) had homology to IncHI-1 plasmids of Salmonella enterica serovar Typhi. Two further clones were homologous to plasmids of Escherichia coli (E-013) and Enterobacter aerogenes (E-034), respectively.
RepHIA PCR.
In both subtractive hybridizations, the main differences between the two strains were homologies to IncHI plasmids. Beside multiple resistance genes, few functional genes have been described as being located on IncHI-1 as well as on IncHI-2 plasmids, which carry, for example, silver resistance genes of the sil type found in clone A-104. Homologues of the autoreplicative region RepHIA were found on both IncHI-1 and IncHI-2 plasmids (37, 40). In order to verify whether EN-373 truly harbored an IncHI plasmid and the plant isolates did not, a PCR amplifying 450 bp of RepHIA was performed. The clinical outbreak strain EN-373 was PCR positive, while plant isolates 501R3 and E6 were negative, suggesting that EN-373 harbored an IncHI plasmid of subgroups IncHI-1 or IncHI-2 while the plant isolates did not (Fig. 1B). This experiment confirmed the specificity of the second subtractive hybridization in which clone E-022 showed 99% nucleotide identity to the pR478 intergenic region of RepHI1A and tra2. tra2 is one of two clusters encoding transfer genes in IncH plasmids. This transfer region varies widely between the subgroups IncHI-1 and IncHI-2, and clone E-022 has shown homology only to tra2 of IncHI-2 plasmid R478. This suggests that the plasmid of our clinical outbreak strain EN-373 belongs to IncHI subgroup 2 rather than to subgroup 1. This also fits with reports of other authors, who found the majority of sil genes on IncHI-2 plasmids rather than on IncHI-1 plasmids (18).
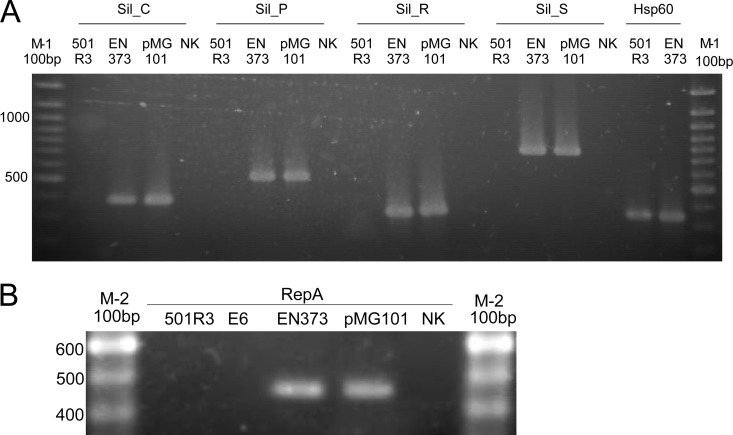
Fig 1.
Silver resistance genes and IncHI plasmids in pathogenic isolate EN-373. (A) PCRs of four genes of the sil resistance determinant in strains EN-373, a clinical outbreak strain, and 501R3, a plant isolate. Plasmid MG101 was used as a positive control; E. coli strain DH5α was the negative control (NK). The hsp60 PCR was used as control PCR. (B) PCR of repA, a marker gene for IncHI-1 and IncHI-2 plasmids. 501R3 and E6, two plant isolates, were negative in the repA PCR, suggesting that they did not carry IncHI-1 or -2 plasmids. EN-373 was PCR positive, confirming the presence of an IncHI-1 or -2 plasmid, presumably carrying the silver resistance genes.
sil PCRs.
To confirm existence of the sil operon in the tester strain, PCRs of four of the sil genes (silS, silR, silC, and silP) were performed. PCR results are displayed in Fig. 1A. 501R3 was negative and EN-373 positive in all sil PCRs, suggesting that the clinical outbreak strain did harbor the complete silver resistance determinant while the plant isolate did not. hsp60 PCR, amplifying 341 bp of the gene encoding the 60-kDa heat shock protein, was performed as a positive-control PCR as previously described (22).
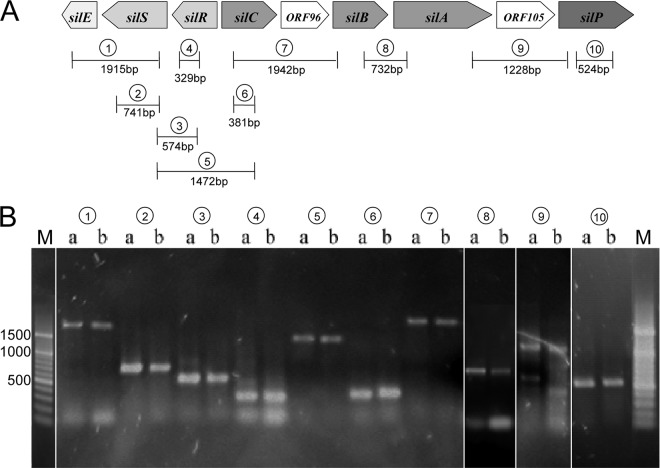
To verify whether the sil operon has the same configuration as on Salmonella plasmid pMG101, PCRs overlapping from one gene to the other were performed (silE to silS, silS to silR, silS to silC, silC to silB, and silA to silP). All amplification products were comparable to those of pMG101 (Fig. 2).
Fig 2.
The sil operon in Enterobacter cloacae. (A) Graphic illustration of the sil operon with marked PCR amplification sites. (B) PCRs of the sil operon; Lanes a, positive control DH5α pMG101; lanes b, tester strain EN-373. Lane pairs: 1, silE to silS; 2, silS; 3, silS to silR; 4, silR; 5, silS to silC; 6, silC; 7, silC to silB; 8, silB to silA; 9, silA to silP; 10, silP. M, 100-bp DNA marker.
Sequencing of sil genes.
Partial sil gene sequencing revealed that sil genes in the E. cloacae complex were highly conserved (Table 3). ORF105 was the least conserved gene (mean nucleotide identity, 88%). Partial sequences of silS, silR, silC, silB, silA, and silP revealed nucleotide identities of 99%, 98%, 99%, 97%, 94%, and 95%, respectively, confirming their higher degree of conservation between species. Deduced amino acid sequences were all more than 90% identical to that of pMG101 (Table 3).
Table 3.
Identity of sil nucleotide and amino acid sequences of Enterobacter species with pMG101
| Type of identitya | % Identity with pMG101b |
||||||
|---|---|---|---|---|---|---|---|
| SilS | SilR | SilC | SilB | SilA | ORF105 | SilP | |
| Nucleotide | 99 | 98 | 99 | 97 | 94 | 88 | 95 |
| Amino acid | |||||||
| Identical | 100 | 97 | 99 | 94 | 97 | 96 | 97 |
| Positive | 100 | 98 | 100 | 96 | 98 | 98 | 98 |
“Positive” amino acids are those that are different but have the same characteristics.
PCR products resulting in partial sequences of 818 bp for silS, 525 bp for silR, 381 bp for silC, 243 bp for silB, 277 bp for silA, 317 bp for OFR105, and 720 bp for silP.
Distribution of the sil operon within the E. cloacae complex.
One hundred sixty-four clinical isolates of all genotypes of the E. cloacae complex were screened for silS, silR, silC, and silP. Of these isolates, 63% were positive in all sil PCRs, suggesting that about two-thirds of clinical isolates of the E. cloacae complex harbor the complete silver resistance determinant. The genovars most prevalent in the population structure of the E. cloacae complex, i.e., clusters III, VI (Enterobacter hormaechei subsp. oharae) and VIII (Enterobacter hormaechei subsp. hormaechei) (22), were also most frequently positive in the sil PCRs (80% were positive in all four PCRs performed). Of clusters IX and xiii, which were rarely recovered from clinical specimens, no isolate was positive in all PCRs, although most isolates of cluster xiii seemed to harbor at least one or two sil genes. Similarly, most isolates of cluster V (Enterobacter ludwigii) were positive in the silS PCR, and half were positive in the silC or silR PCRs, but only two were positive in all PCRs performed, suggesting that isolates of this cluster could harbor either partial sil determinants or sil genes too divergent to be amplified by the PCRs employed. Cluster V represents about 6% of strains of the recently presented population structure (22) and about 11% of isolates in the present study. Clusters I, II, and IV represented 5 to 10% of our clinical isolates (27) and had an intermediate rate of sil PCR positivity (60 to 63%). It is noteworthy that isolates from these clusters yielded 100% positive results in the silC PCR. To summarize, genovars III, VI, and VIII had the complete silver resistance determinant significantly more often and were more frequently recovered from clinical specimens than genovars V, IX, and xiii (chi-square test, P < 0.0001). Data are shown in Table 4.
Table 4.
sil PCRs of clinical isolates from different genetic clusters
| Genetic clustera | n | No. (%) positive in PCR |
||||
|---|---|---|---|---|---|---|
| SilC | SilP | SilR | SilS | Allb | ||
| I | 9 | 9 (100) | 6 (67) | 6 (67) | 9 (100) | 6 (67) |
| II | 11 | 11 (100) | 9 (82) | 11 (100) | 10 (91) | 8 (73) |
| III | 33 | 27 (82) | 27 (82) | 27 (82) | 27 (82) | 27 (82) |
| IV | 15 | 15 (100) | 9 (60) | 14 (93) | 15 (100) | 9 (60) |
| V | 18 | 9 (50) | 11 (61) | 5 (28) | 17 (94) | 2 (11) |
| VI + VII | 19 | 17 (89) | 16 (84) | 17 (89) | 16 (84) | 16 (84) |
| VIII | 40 | 32 (80) | 32 (80) | 32 (80) | 32 (80) | 32 (80) |
| IX | 6 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| XI + XIIc | 7 | 6 (86) | 5 (71) | 6 (86) | 4 (57) | 4 (57) |
| xiii | 6 | 5 (83) | 3 (50) | 3 (50) | 3 (50) | 0 (0) |
| Total | 164 | 131 (80) | 118 (71) | 121 (74) | 133 (81) | 104 (63) |
Genetic clusters according to Hoffmann and Roggenkamp (22).
Distribution of isolates positive for all PCRs in genetic clusters; chi-square test, P < 0.0001.
Includes E. cloacae type strain ATCC 13047.
Phenotypic tests.
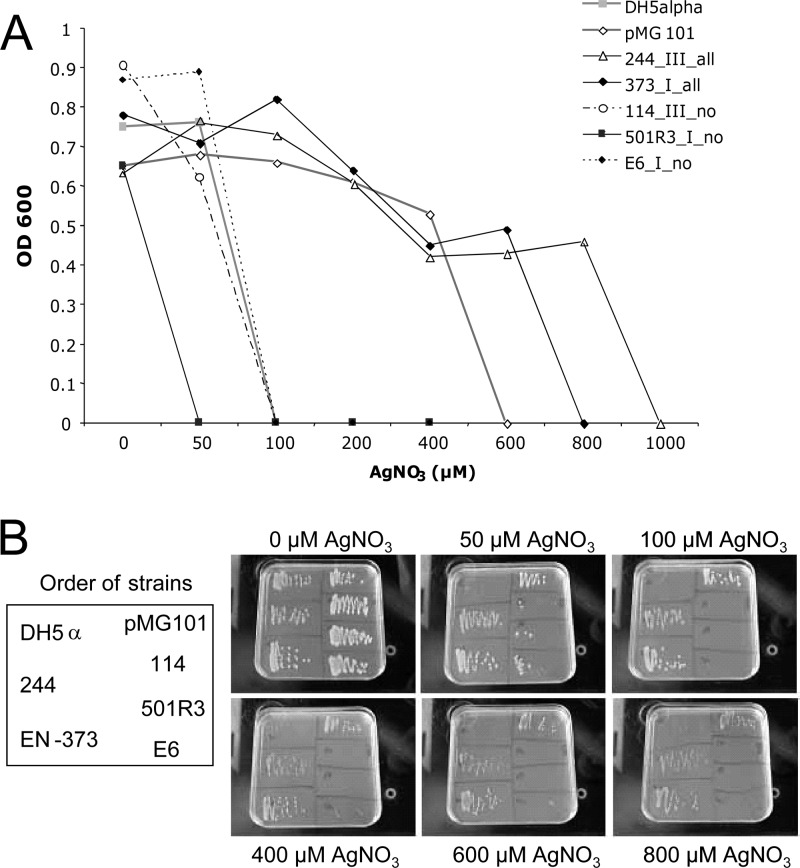
Plasmids harboring the sil determinant were reported to transfer resistance to up to 600 μM AgNO3 (17). In order to verify a potential correlation between the presence of silver resistance genes and phenotypic behavior, the extent of silver resistance was determined for five representative isolates of the E. cloacae complex. Inhibition of bacterial growth in the presence of increasing amounts of AgNO3 was assessed by measuring the optical density at 600 nm (OD600) after overnight incubation with different concentrations of AgNO3 (Fig. 3A). PCR-negative isolates 114, 501R3, and E6 and the negative control DH5α did not grow in AgNO3 concentrations of more than 100 μM. In contrast, PCR-positive isolates 244 and 373 and the positive control DH5α(pMG101) sustained concentrations of up to 800 μM. This finding was confirmed on solid Luria-Bertani (LB) medium without added NaCl (Fig. 3B).
Fig 3.
Phenotypic silver resistance. (A) Killing of bacteria by AgNO3. OD600 was measured after incubation (16 h, 36 ± 1°C, room air) of the indicated strains in LB medium supplemented with increasing amounts of AgNO3. E. coli strains DH5α and DH5α(pMG101) served as negative (AgNO3 MIC, 50 μM) and positive (AgNO3 MIC, 600 μM) controls, respectively. Two sil PCR-positive strains (244 and 373) as well as three sil PCR-negative strains (114, 501R3, and E6) were included. Symbols in the key are labeled with the strain name, genetic cluster, and PCR results. Clinical outbreak strain 373 of genetic cluster I (22) was positive in all sil PCRs and showed an AgNO3 MIC of 600 μM. Both strains 501R3 and E6 yielded negative PCRs with all sil genes and had AgNO3 MICs equal to or below 50 μM. (B) Growth of strains on LB agar containing rising concentration of AgNO3. The positions of the strains on the plates are depicted on the left.
DISCUSSION
In this study, we assessed the molecular background of the imbalanced distribution of genotypes within the E. cloacae complex as nosocomial pathogens by direct whole-genome comparison of an outbreak strain and an avirulent strain. Using subtractive hybridization, we found an IncHI plasmid harboring a silver resistance determinant as a major difference between pathogenic and nonpathogenic isolates. Serving as a hygienic fitness factor, the presence of silver resistance determinants could improve survival in hospital environments and might be an explanation for the rising numbers of nosocomial infections caused by the E. cloacae complex. Moreover, the distinct distribution of the sil genes among different (sub)species of the E. cloacae complex could partially explain their unequal prevalence as nosocomial pathogens (22, 27, 28).
Silver ions are frequently used as antiseptics in medicine and health (41). The best known uses of silver preparations are Crédé's prophylaxis and treatment of burn wounds, e.g., in topical creams containing 1% silver sulfadiazine (14). Ag-coated bandages are used to cover burns and traumatic injuries. Silver-impregnated polymers are commonly used in medical devices such as catheters to prevent the formation of bacterial biofilms (7, 43). Besides these and other medical applications, silver compounds are employed in a variety of nonmedical products. For example, silver ions are used in disinfectants (5), water purifiers, and sterilizers in hospitals and hotels, e.g., to control Legionella growth in water pipe systems (41). Hence, a resistance mechanism against silver ions would serve as a potent hygienic fitness factor for bacteria, facilitating their survival in hospital environments and creating a potential infection risk for patients. This has directed our attention to the silS gene, a silver resistance regulator gene, which was found to be unique to the clinical outbreak strain by subtractive hybridization. We showed that the complete silver resistance determinant was present and functional in the majority of isolates of the E. cloacae complex. The distribution of the sil operon among the different genotypes of the E. cloacae complex was significantly shifted in favor of genotypes frequently isolated in hospital environments (Table 4). The prevalence of 63% found in our cohort was surprisingly high; however, the study set consisted of clinical isolates collected over several years without any specific selection criteria and without knowledge of their susceptibility or resistance to silver. Woods et al. (46) screened 172 bacterial strains isolated from human and equine wounds for the presence of the sil determinant. They found only six positive strains; however, all of them were E. cloacae, representing 60% of all investigated E. cloacae strains. In our study cohort, we observed that a number of isolates were positive for some but not all sil genes. This might have been partly due to amplifications being missed by the PCR assays designed from reference sequences. However, it is interesting to speculate whether these genes also might have additional functions independent of the context of the sil operon. This would be of particular interest for silC, which is consistently present in all isolates of genotypes I, II, and IV.
In the 1960s, the advent of silver compounds as therapeutics in medicine revolutionized the care of surgical and especially burn patients. Before topical silver nitrate was available, burn wound sepsis accounted for about two-thirds of mortality in burn patients. Since the late 1960s, when silver compounds were routinely applied to burn wounds (e.g., silver sulfadiazine in Flamazine), mortality rates rapidly reduced to about 1/4 (35). A decade later, a series of studies reported silver resistance in Enterobacteriaceae and nonfermenters (3, 6, 20, 33). Since that time, an increasing number of outbreaks due to silver-resistant strains of Enterobacteriaceae have been reported. In 1983, Markowitz et al. (31) reported a strong correlation between silver resistance and the presence of a typical “epidemic” plasmid pattern in E. cloacae outbreak strains causing infections in burn patients. These silver-resistant E. cloacae strains contained a 55-MDa plasmid which was not found in strains susceptible to silver sulfadiazine.
Irrespective of the medical impact of silver resistance in bacteria, the molecular genetics of silver resistance were not deciphered until the late nineties (16). Gupta et al. described a silver resistance determinant composed of nine open reading frames (ORFs), of which seven are named and two are still called ORFs. The unique feature of this determinant is the composition of two parallel membrane Ag(I) efflux pumps (silCBA and silP) and a periplasmic metal-binding protein (silE). Furthermore, Gupta et al. (18) described that sil genes were highly conserved in different IncHI plasmids of Salmonella serovars (MIP233, MIP235, and WR23) and Serratia marcescens (R476b and R478). Also, in the present study, partial sil sequences proved to be highly conserved compared to the prototype of the sil determinant in plasmid MG101 (16). Nucleotide identities ranged from 94% for silA to 99% for silS and silC (Table 3).
These results, together with the other plasmid-derived sequences found in the subtractive hybridization and the verification with the RepHIA PCR, clearly favor a plasmidic origin of the silver resistance in our E. cloacae strains. An extrachromosomal origin was also suggested by Davis et al. (8), who isolated two highly silver-resistant E. cloacae strains from teeth. This is also in line with the recent findings by Paauw et al. (36), who identified a large plasmid, pQC, in an E. hormaechei strain that caused a hospital-wide outbreak in the Netherlands. This plasmid also belonged to the IncHI-2 group and showed the highest similarity to plasmid R478. Besides resistance to several antibiotics, it was reported to carry genes for resistance to heavy metals, including the sil operon. The presence and absence of the putative IncHI-2 plasmid are then the major genomic differences between clinical outbreak strain EN-373 and two independent plant isolates. However, as the large plasmid was isolated in the chromosomal DNA fraction, it might have been overrepresented in the tester DNA and—depending on the number of plasmidic copies—may have obscured other genomic differences between the pathogenic and nonpathogenic strains.
In conclusion, by subtractive hybridization we identified a silver resistance determinant selectively present in a pathogenic Enterobacter strain. Furthermore, this sil operon was found in 63% of all isolates of the E. cloacae complex and was predominantly present in genotypes with high clinical prevalence. This distribution could partially explain the unequal prevalence rates of the (sub)species of the E. cloacae complex in the hospital setting and might also provide some explanation for the high infection rates in burn patients; however, the relatively high frequency of infections caused by Enterobacter in newborns is still not sufficiently elucidated by our findings. The association of such a plasmid with certain genotypes, however, also raises the question of why it is not more extensively distributed among other genotypes. This again suggests further differences between the pathogenic and nonpathogenic variants. Furthermore, the question of whether silver resistance is only a fitness factor promoting survival in hospital environment or whether the sil determinant itself confers pathogenicity-associated functions needs to be investigated. The amino acid sequence of the product of silC, for instance, shows 70% identity with that of ibeB, a gene which has been described as being required in E. coli for the penetration of brain microvascular endothelial cells (23). This speculation is supported by a recent report demonstrating that isolates of the E. cloacae complex are able to adhere to and penetrate epithelial cells (28). Other studies have shown that mutations in the copper transport ATPase reduce the virulence in Listeria monocytogenes (13). In line with those results, it is intriguing to speculate that silver resistance might not only promote survival but also increase bacterial pathogenicity. In this context, therapeutic usage of silver compounds would not only be ineffective but could even be harmful, as they would select for more pathogenic strains. Whether there are indeed such associations is currently unknown, and further studies are needed to focus on this issue.
Footnotes
Published ahead of print 25 July 2012
REFERENCES
- 1. Akopyants NS, et al. 1998. PCR-based subtractive hybridization and differences in gene content among strains of Helicobacter pylori. Proc. Natl. Acad. Sci. U. S. A. 95:13108–13113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Andersen BM, et al. 1989. Multiply beta-lactam resistant Enterobacter cloacae infections linked to the environmental flora in a unit for cardiothoracic and vascular surgery. Scand. J. Infect. Dis. 21:181–191 [DOI] [PubMed] [Google Scholar]
- 3. Annear DI, Mee BJ, Bailey M. 1976. Instability and linkage of silver resistance, lactose fermentation, and colony structure in Enterobacter cloacae from burn wounds. J. Clin. Pathol. 29:441–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bodey GP, Elting LS, Rodriguez S. 1991. Bacteremia caused by Enterobacter: 15 years of experience in a cancer hospital. Rev. Infect. Dis. 13:550–558 [DOI] [PubMed] [Google Scholar]
- 5. Brady MJ, Lisay CM, Yurkovetskiy AV, Sawan SP. 2003. Persistent silver disinfectant for the environmental control of pathogenic bacteria. Am. J. Infect. Control. 31:208–214 [DOI] [PubMed] [Google Scholar]
- 6. Bridges K, Kidson A, Lowbury EJ, Wilkins MD. 1979. Gentamicin- and silver-resistant pseudomonas in a burns unit. Br. Med. J. 1:446–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Choi L, et al. 1999. Development of an infection-resistant LVAD driveline: a novel approach to the prevention of device-related infections. J. Heart Lung Transplant. 18:1103–1110 [DOI] [PubMed] [Google Scholar]
- 8. Davis IJ, Richards H, Mullany P. 2005. Isolation of silver- and antibiotic-resistant Enterobacter cloacae from teeth. Oral Microbiol. Immunol. 20:191–194 [DOI] [PubMed] [Google Scholar]
- 9. de Kort G, Bolton A, Martin G, Stephen J, van de Klundert JA. 1994. Invasion of rabbit ileal tissue by Enterobacter cloacae varies with the concentration of OmpX in the outer membrane. Infect. Immun. 62:4722–4726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dorsey G, et al. 2000. A heterogeneous outbreak of Enterobacter cloacae and Serratia marcescens infections in a surgical intensive care unit. Infect. Control Hosp. Epidemiol. 21:465–469 [DOI] [PubMed] [Google Scholar]
- 11. Dushay MS, Asling B, Hultmark D. 1996. Origins of immunity: Relish, a compound Rel-like gene in the antibacterial defense of Drosophila. Proc. Natl. Acad. Sci. U. S. A. 93:10343–10347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fernandez-Baca V, et al. 2001. Molecular epidemiological typing of Enterobacter cloacae isolates from a neonatal intensive care unit: three-year prospective study. J. Hosp. Infect. 49:173–182 [DOI] [PubMed] [Google Scholar]
- 13. Francis MS, Thomas CJ. 1997. Mutants in the CtpA copper transporting P-type ATPase reduce virulence of Listeria monocytogenes. Microb. Pathog. 22:67–78 [DOI] [PubMed] [Google Scholar]
- 14. Fraser JF, Bodman J, Sturgess R, Faoagali J, Kimble RM. 2004. An in vitro study of the anti-microbial efficacy of a 1% silver sulphadiazine and 0.2% chlorhexidine digluconate cream, 1% silver sulphadiazine cream and a silver coated dressing. Burns 30:35–41 [DOI] [PubMed] [Google Scholar]
- 15. Grimont F, Grimont PA. 1992. The genus Enterobacter, p 2797–2815 In Balows A, Tr̈uper HG, Dworkin M, Harder W, Schleifer KH. (ed), The prokaryotes, 2nd ed Springer Verlag, New York, NY [Google Scholar]
- 16. Gupta A, Matsui K, Lo JF, Silver S. 1999. Molecular basis for resistance to silver cations in Salmonella. Nat. Med. 5:183–188 [DOI] [PubMed] [Google Scholar]
- 17. Gupta A, Maynes M, Silver S. 1998. Effects of halides on plasmid-mediated silver resistance in Escherichia coli. Appl. Environ. Microbiol. 64:5042–5045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gupta A, Phung LT, Taylor DE, Silver S. 2001. Diversity of silver resistance genes in IncH incompatibility group plasmids. Microbiology 147:3393–3402 [DOI] [PubMed] [Google Scholar]
- 19. Halda-Alija L, Hendricks SP, Johnston TC. 2001. Spatial and temporal variation of Enterobacter genotypes in sediments and the underlying hyporheic zone of an agricultural stream. Microb. Ecol. 42:286–294 [DOI] [PubMed] [Google Scholar]
- 20. Hendry AT, Stewart IO. 1979. Silver-resistant Enterobacteriaceae from hospital patients. Can. J. Microbiol. 25:915–921 [DOI] [PubMed] [Google Scholar]
- 21. Hervas JA, et al. 2001. Increase of Enterobacter in neonatal sepsis: a twenty-two-year study. Pediatr. Infect. Dis. J. 20:134–140 [DOI] [PubMed] [Google Scholar]
- 22. Hoffmann H, Roggenkamp A. 2003. Population genetics of the nomenspecies Enterobacter cloacae. Appl. Environ. Microbiol. 69:5306–5318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang SH, et al. 1999. Identification and characterization of an Escherichia coli invasion gene locus, ibeB, required for penetration of brain microvascular endothelial cells. Infect. Immun. 67:2103–2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kaminska W, Patzer J, Dzierzanowska D. 2002. Urinary tract infections caused by endemic multi-resistant Enterobacter cloacae in a dialysis and transplantation unit. J. Hosp. Infect. 51:215–220 [DOI] [PubMed] [Google Scholar]
- 25. Kartali G, et al. 2002. Outbreak of infections caused by Enterobacter cloacae producing the integron-associated beta-lactamase IBC-1 in a neonatal intensive care unit of a Greek hospital. Antimicrob. Agents Chemother. 46:1577–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Keller R, Pedroso MZ, Ritchmann R, Silva RM. 1998. Occurrence of virulence-associated properties in Enterobacter cloacae. Infect. Immun. 66:645–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kremer A, Hoffmann H. 2012. Prevalences of the Enterobacter cloacae complex and its phylogenetic derivatives in the nosocomial environment. Eur. J. Clin. Microbiol. Infect. Dis. doi:10.1007s10096-012-1646-2 [DOI] [PubMed] [Google Scholar]
- 28. Krzyminska S, Koczura R, Mokracka J, Puton T, Kaznowski A. 2010. Isolates of the Enterobacter cloacae complex induce apoptosis of human intestinal epithelial cells. Microb. Pathog. 49:83–89 [DOI] [PubMed] [Google Scholar]
- 29. Kuboyama RH, de Oliveira HB, Moretti-Branchini ML. 2003. Molecular epidemiology of systemic infection caused by Enterobacter cloacae in a high-risk neonatal intensive care unit. Infect. Control Hosp. Epidemiol. 24:490–494 [DOI] [PubMed] [Google Scholar]
- 30. Lindh E, Ursing J. 1991. Genomic groups and biochemical profiles of clinical isolates of Enterobacter cloacae. APMIS 99:507–514 [DOI] [PubMed] [Google Scholar]
- 31. Markowitz SM, Smith SM, Williams DS. 1983. Retrospective analysis of plasmid patterns in a study of burn unit outbreaks of infection due to Enterobacter cloacae. J. Infect. Dis. 148:18–23 [DOI] [PubMed] [Google Scholar]
- 32. Mayhall CG, Lamb VA, Gayle WE, Jr, Haynes BW., Jr 1979. Enterobacter cloacae septicemia in a burn center: epidemiology and control of an outbreak. J. Infect. Dis. 139:166–171 [DOI] [PubMed] [Google Scholar]
- 33. McHugh GL, Moellering RC, Hopkins CC, Swartz MN. 1975. Salmonella typhimurium resistant to silver nitrate, chloramphenicol, and ampicillin. Lancet i:235–240 [DOI] [PubMed] [Google Scholar]
- 34. Modi N, Damjanovic V, Cooke RW. 1987. Outbreak of cephalosporin resistant Enterobacter cloacae infection in a neonatal intensive care unit. Arch. Dis. Child. 62:148–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moyer CA, Brentano L, Gravens DL, Margraf HW, Monafo WW., Jr 1965. Treatment of large human burns with 0.5 per cent silver nitrate solution. Arch. Surg. 90:812–867 [DOI] [PubMed] [Google Scholar]
- 36. Paauw A, et al. 2009. Identification of resistance and virulence factors in an epidemic Enterobacter hormaechei outbreak strain. Microbiology 155:1478–1488 [DOI] [PubMed] [Google Scholar]
- 37. Page DT, Whelan KF, Colleran E. 2001. Characterization of two autoreplicative regions of the IncHI2 plasmid R478: RepHI2A and RepHI1A((R478)). Microbiology 147:1591–1598 [DOI] [PubMed] [Google Scholar]
- 38. Roberts DP, et al. 1999. Role of pfkA and general carbohydrate catabolism in seed colonization by Enterobacter cloacae. Appl. Environ. Microbiol. 65:2513–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sevcik C, Noriega J, D'Suze G. 2003. Identification of Enterobacter bacteria as saxitoxin producers in cattle's rumen and surface water from Venezuelan savannahs. Toxicon 42:359–366 [DOI] [PubMed] [Google Scholar]
- 40. Sherburne CK, et al. 2000. The complete DNA sequence and analysis of R27, a large IncHI plasmid from Salmonella typhi that is temperature sensitive for transfer. Nucleic Acids Res. 28:2177–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Silver S. 2003. Bacterial silver resistance: molecular biology and uses and misuses of silver compounds. FEMS Microbiol. Rev. 27:341–353 [DOI] [PubMed] [Google Scholar]
- 42. Simi S, et al. 2003. A low molecular weight enterotoxic hemolysin from clinical Enterobacter cloacae. Can. J. Microbiol. 49:479–482 [DOI] [PubMed] [Google Scholar]
- 43. Sutterlin S, Tano E, Bergsten A, Tallberg AB, Melhus A. 2012. Effects of silver-based wound dressings on the bacterial flora in chronic leg ulcers and its susceptibility in vitro to silver. Acta Derm. Venereol. 92:34–39 [DOI] [PubMed] [Google Scholar]
- 44. van Nierop WH, Duse AG, Stewart RG, Bilgeri YR, Koornhof HJ. 1998. Molecular epidemiology of an outbreak of Enterobacter cloacae in the neonatal intensive care unit of a provincial hospital in Gauteng, South Africa. J. Clin. Microbiol. 36:3085–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wenger PN, et al. 1997. An outbreak of Enterobacter hormaechei infection and colonization in an intensive care nursery. Clin. Infect. Dis. 24:1243–1244 [DOI] [PubMed] [Google Scholar]
- 46. Woods EJ, Cochrane CA, Percival SL. 2009. Prevalence of silver resistance genes in bacteria isolated from human and horse wounds. Vet. Microbiol. 138:325–329 [DOI] [PubMed] [Google Scholar]