Abstract
We aimed to determine whether additional molecular and microbiological evaluations of methicillin-resistant Staphylococcus aureus (MRSA) isolated from patients newly identified as nasal carriers were useful for control strategies and whether longitudinal testing during the same or repeat hospitalization changed MRSA status. Nasal swabs from patients positive by Xpert MRSA PCR and not known to be colonized in the previous year were cultured for S. aureus. Isolates were tested for resistance to a variety of antibiotics, including high-level mupirocin resistance (HLMR) and low-level mupirocin resistance (LLMR) and the presence of genes mecA and mupA and those for Panton-Valentine leukocidin (PVL), USA300, and USA400. Repeat nasal screens during the 6-month study were tested for continued presence of MRSA. Among 130 patients, cultures revealed MRSA in 85 (65.4%), methicillin-susceptible S. aureus in 19 (14.6%), and no growth in 26 (20%). MRSA isolates were USA300 positive in 13/85 (15.3%) and LLMR in 8/85 (9.4%) patients. No isolates were HLMR or mupA positive. mecA dropout was detected in 9/130 (6.9%) patients. The rate of subsequent MRSA infections in USA300-positive versus -negative patients was not different. MRSA nasal status remained concordant in 69/70 (98.6%) patients who had follow-up testing. The findings do not support expanding MRSA surveillance to include routine detection of genes for USA300, PVL, or mupA, all of which were either of low frequency or not significantly associated with MRSA infection risk in our population of newly identified nasal carriers. Repeat nasal screening for MRSA during the same or subsequent hospitalizations over 6 months could also be deferred, reducing costs associated with screening.
INTRODUCTION
The VA has a successful MRSA bundle for prevention of health care-associated methicillin-resistant Staphylococcus aureus (MRSA) infections (9). Every veteran is eligible for nasal screening for MRSA carriage on admission to an acute care setting, transfer between units, and at discharge. Currently, the main assessment on admission is a dichotomous result of positive or negative for MRSA carriage. Additional molecular and microbiological characteristics of the colonizing isolates are not routinely evaluated, understandably, given the large scale of this national quality improvement program and use of a PCR-based assay rather than culture for bacterial isolates.
Although the assessment of MRSA nasal colonization provides useful information for infection prevention activities, such as implementation of special contact precautions, the information regarding clonal type (i.e., USA300 versus other) and susceptibility to other antimicrobials, such as mupirocin, could further inform clinicians regarding virulence and expected success of decolonization regimens. USA300 MRSA has been associated with severe community-associated skin and soft tissue infections, as well as severe sepsis and other invasive infections (11, 20, 23). Differentiating between USA300 and non-USA300 colonization may be useful for predicting risk of infection during hospitalization (11, 20).
Once colonized with MRSA, it is anticipated that most patients will continue to carry MRSA for months to years (5, 8, 17). However, changes in the colonizing strain may occur during acute hospitalization, where patients are exposed to antibiotics and the competing flora of other patients and health care workers (15, 16). Documenting persistence of carriage of the index strain or a change or loss of the index strain could be useful for evaluating the utility of repeated assessments in MRSA-positive patients. If the index strain tends to persist despite hospital-related exposures, continued assessment for MRSA nasal carriage during hospitalization and on subsequent hospitalizations may not be necessary. The aims of our study were to assess the molecular and microbiological characteristics of nasal MRSA isolated from patients who were newly identified as MRSA nasal carriers in our health care system and to evaluate their subsequent colonizing strains for concordance to the initial positive strain.
MATERIALS AND METHODS
Study population.
The study population consisted of patients admitted to acute care at VA Boston HCS between March 2011 and August 2011. All patients were screened for MRSA nasal carriage as part of the National VA MRSA Prevention Program (9). Patients were eligible for inclusion if they had a positive nasal Xpert MRSA PCR result either on admission or during hospitalization and did not have a recorded positive nasal screen or culture any time in the year prior. We cannot exclude the possibility of positive nasal screens or cultures for these patients at other institutions. There were no specific exclusion criteria. Patients were entered into the study based on availability of a partner swab from the original nasal swab (some were inadvertently discarded). Once patients were entered into the cohort, available swabs from subsequent nasal MRSA screens that were performed during the same hospitalization or subsequent hospitalizations during the 6-month study period were also evaluated.
The electronic medical record was reviewed for demographics, including gender and age, previous MRSA nasal screening results, and for associated MRSA cultures. National Hospital Surveillance Network (NHSN) definitions were used to identify MRSA infections in the 90 days after the index nasal MRSA date. All study procedures were approved by the Boston VA Institutional Review Board.
Laboratory methods.
The initial nasal screen for MRSA (the index isolate) was evaluated by using the Xpert MRSA kit (Cepheid, Sunnydale, CA) as per the standard protocol at the facility. This real-time PCR method targets the chromosomal junction of orfX-SCCmec, which serves as a surrogate for the identification of mecA and methicillin resistance. The second swab of the double-swab collection kit is then sent to the research laboratory for culture. The nasal swabs are packaged as one item, and both are inserted simultaneously into the nose; only one is used for the PCR test. Swabs from eligible patients were cultured onto either Columbia agar with colistin and nalidixic acid (CNA) or phenylethyl alcohol agar plates and incubated at 35°C for 48 h. S. aureus colonies were identified visually, and the identification was confirmed by a positive coagulase test.
Index isolates recovered from swabs representing the first positive Xpert MRSA result for the patient in 365 days, either from the admission swab or the first subsequent swab if the admission swab was negative, were recovered from frozen stocks. These were tested according to CLSI guidelines for disk diffusion susceptibility (4) to cefoxitin, trimethoprim-sulfamethoxazole, cefazolin, amoxicillin-clavulanic acid, erythromycin, rifampin, clindamycin, gentamicin, ciprofloxacin, and tetracycline. These isolates were also assessed for mupirocin susceptibility by disk diffusion using 5-μg and 20-μg disks. Isolates that were found to have low-level resistance (LLR) or high-level resistance (HLR) to mupirocin were then evaluated by Etest to determine the MIC. MIC breakpoints were defined as follows: susceptible, ≤1 μg/ml; intermediate, 2 to 256 μg/ml; resistant, >256 μg/ml (3). Mupirocin MICs of 2 to 256 μg/ml were designated LLR, while MICs of >256 μg/ml were considered HLR. PCR was also performed for the mupA gene, which confers high-level mupirocin resistance (25).
All isolates were characterized by a multiplex PCR assay to identify the presence of S. aureus-specific thermostable nuclease (nuc), Panton-Valentine leukocidin (PVL) genes (lukS-PV/lukF-PV), the genetic marker for USA300 pulsed-field gel electrophoresis (PFGE) type (the arcA gene on the arginine catabolic mobile element), the genetic marker for USA400 PFGE type (genomic island gene locus MW0756), and the gene responsible for methicillin resistance (mecA) (24). A subset of nasal swabs positive for MRSA by the Xpert MRSA screen yielded isolates by culture that were susceptible to cefoxitin by disk diffusion and lacked the mecA gene when tested in the multiplex PCR assay. These isolates were subcultured for isolation on CNA plates, incubated at 35°C overnight, and prepared for repeat testing on the Xpert MRSA system per the manufacturer's protocol (Cepheid, Sunnyvale, CA) (1). The isolates were classified as mecA dropout or empty cassette mutants if they were positive by Xpert MRSA, negative for mecA by PCR, and phenotypically methicillin sensitive to cefoxitin by disk diffusion susceptibility testing.
RESULTS
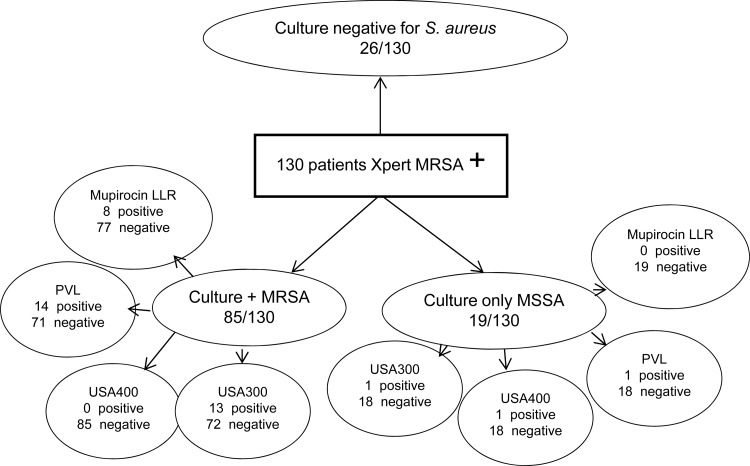
Over a 6-month period, 130 patients meeting our inclusion criteria had a nasal swab that was positive by Xpert MRSA. The mean age of the cohort was 69.3 years, and 94.6% were male. Culture of the Xpert PCR-positive nasal swab revealed MRSA in 85 patients, methicillin-susceptible S. aureus in 19, and no growth in 26 patients (Fig. 1). Cocolonization with MRSA and MSSA was present in 4 patients, and they are shown in the MRSA group in Fig. 1. False-positive Xpert MRSA results due to mecA dropout mutants occurred in 9/130 (6.9%). The patients with MSSA only, without evidence of a mecA dropout, and those with no growth on culture were considered to have low quantities of MRSA or nonviable organisms that were not retrieved by culture.
Fig 1.
Molecular and microbiological results for cultures from patients newly identified to be nasal carriers by using the Xpert MRSA nasal screen.
Among the 85 patients with MRSA, 13 (15.3%) had a strain positive for USA300, 14 (16.5%) had a strain positive for PVL, and none were positive for USA400. High-level mupirocin resistance was not detected. Low-level mupirocin resistance was detected in 8 (9.4%) patients (Fig. 1). Of patients with MSSA only, one patient had a strain positive for both USA300 and PVL (1/19; 5.3%) and one patient had a strain positive for USA400. All of the MSSA isolates were sensitive to mupirocin.
Clinical cultures obtained in the 90 days following the index nasal screen result were positive for MRSA in 30/130 (23%) patients. The positive cultures were consistent with an NHSN-defined MRSA infection in 25 of 30 patients, including 19 of 85 (22.4%) patients with MRSA recovered on culture of the index nasal screen and 6 of 45 (13.3%) patients without MRSA recovered on culture (P = 0.12; chi-square test). The MRSA infection rate was also not significantly different between patients whose index nasal strain was a USA300 MRSA (2/13; 18.2%) and those with a non-USA300 MRSA (17/72; 23%; P = 0.72, Fisher's exact test).
The detection of nasal MRSA by the Xpert MRSA test was classified by timing of detection relative to admission. Sixty-three patients had no previous nasal MRSA testing in our health care system, 47 converted between hospitalizations, and 20 converted during hospitalization (more than 72 h after admission). None of the 13 patients with MRSA that were identified as USA300 converted during hospitalization; all converted between hospitalizations or were newly identified as MRSA positive in our system. Low-level resistance to mupirocin was higher among documented in-hospital converters (6/20; 30%) than among out-of-hospital converters (2/110; 1.8%; P < 0.001).
Follow-up nasal cultures.
Analysis of nasal cultures from follow-up swabs were available for 70/130 (53.8%) patients. Among these, 57/130 (43.8%) had follow-up swabs during the initial hospitalization, and 13/130 (10%) had repeat hospitalizations. The number of follow-up swabs ranged from 1 to 14 swabs per patient during a single admission. Agreement between the initial swab and follow-up swabs for persistence of MRSA occurred in 69/70 (98.6%) of patients. Repeat hospitalization was also not associated with any discordance in mecA nasal carriage status. Among the 13 patients rehospitalized within the 6-month study period, all were still positive for MRSA at readmission.
DISCUSSION
Nasal screening for MRSA carriage in all hospitalized patients is a resource-intensive process that remains controversial (7, 10, 21). Implementation of this program in the VA system has been associated with significant reductions in infection rates (9). Since much of the infrastructure for molecular testing is in place, evaluations of additional molecular characteristics that further improve infection prevention efforts or clinical management decisions have the potential to be reasonably cost-effective. However, our findings do not support expanding the MRSA surveillance program to include detection of mupA or genes for USA300 or PVL, all of which were found either at low frequency or were not significantly associated with MRSA infection risk in our population of newly identified nasal carriers.
Since most VA hospitals currently utilize MRSA PCR assays for screening, few data on cocolonization with MSSA or the prevalence of mecA dropouts are available as part of the standard MRSA Prevention Program. The latter phenomenon would result in a positive MRSA result by the Xpert MRSA screen but a negative result for phenotypic resistance to methicillin or carriage of the mecA gene. The mechanism for the false-positive detection appears to be related to loss of part of the mecA gene (2). A previous study found that 7.7% of 248 S. aureus isolates from an academic hospital were mecA dropouts, similar to what we identified in our population (1). Other studies have reported rates as high as 25% (2, 6). The prevalence of these MSSA isolates with mecA remnants has not been reported in the acute care veteran population but has a potentially magnified effect, given the large-scale screening program involving over 1 million screening tests per year (9). Whether grouping of these patients with patients who are true MRSA carriers results in adverse consequences, such as transmission and subsequent infection, cannot be easily determined without a dedicated study using both culture- and PCR-based methods. The newer generation of molecular tests may help to alleviate this misclassification (2).
Although we did not identify the mupA gene or high-level resistance to mupirocin in our cohort, we did find a relatively high rate of low-level mupirocin resistance. This was primarily identified among patients who converted in the hospital and was only present in MRSA strains. The higher frequency of low-level mupirocin resistance in MRSA strains compared to MSSA strains has been noted in previous clinical studies (12, 19). However, previous studies of low-level mupirocin resistance have not stratified the strains by acquisition location or identified newly positive patients (13, 14). Our data provide some insight into the epidemiology of these strains, as none of the isolates were USA300 and none of the patients were inpatients known to have received mupirocin ointment in the previous year. Studies have shown that the spontaneous mutation rate to mupirocin that results in low-level resistance is lower than the rates that result in resistance to ciprofloxacin and rifampin (18). A recent study found a coassociation between chlorhexidine resistance and low-level mupirocin resistance in hospitalized patients who had been decolonized with mupirocin and failed eradication (13). The clinical implications of low-level mupirocin resistance are still not clear, but increasing rates may be observed in the setting of increasing mupirocin use and warrant continued periodic assessment (14, 22).
Analysis of isolates recovered from follow-up swabs of known MRSA-positive patients indicated that repeated testing for MRSA nasal colonization over a 6-month period was not informative. MRSA colonization status was stable during hospitalization as well as between hospitalizations. The one patient who had a change from a positive Xpert MRSA to a negative Xpert MRSA nasal screen during the same hospitalization had an invasive MRSA skin and soft tissue infection at the same time as his positive nasal screen result and was on vancomycin therapy at the time of the follow-up screen. Thus, continued nasal surveillance of nasal MRSA-positive patients during the same hospitalization or subsequent hospitalizations could be eliminated, resulting in significant potential cost savings. Given the range of 1 to 14 repeat tests in the 57 patients in our cohort, and an estimated cost of $45 dollars per test, this could save between $2,565 (for 57 patients with one repeat) to $35,910 (57 patients × 14 tests each) at a single facility over 6 months. These numbers are based on our study cohort only and could be much higher based on facility census data. Avoiding repeat testing on readmission of known positive patients would result in additional cost savings. The actual cost savings resulting from not only the number of lab tests performed but also the time and resources needed for performing the screening would need to be evaluated in a dedicated cost-effectiveness study.
Our study was limited by being performed in a single center on a sample of admitted patients. This was minimized by inclusion of a relatively large sample size for such a comprehensive molecular and microbiological assessment of nasal S. aureus carriage. We did not perform broth enrichment, which could explain our high rate of cultures without MRSA isolation. However, broth enrichment is not feasible as part of a standard screening program, and we correctly classified these patients as likely positive based on the higher sensitivity of PCR compared to culturing, rather than as false negatives. Overall, the data serve to support additional, multicenter investigations of phenomena with potentially significant infection prevention implications, such as mecA dropout and low-level mupirocin resistance.
The strengths of the study include the comprehensive and longitudinal assessment of nasal carriage status. Our findings support the VA MRSA Prevention Program while providing insights for potential cost reduction strategies. A universal screening program with an iterative algorithm that requires repeat screening only at certain intervals or in certain at-risk populations could be just as effective for MRSA infection reduction and yet be less costly. Further large-scale studies are needed to evaluate the potential cost-effectiveness of this strategy.
ACKNOWLEDGMENTS
This work was supported in part by the National Center for Occupational Health and Infection Control, Office of Public Health and Environmental Hazards, Veterans Health Administration, Gainesville, FL.
We thank the VA Boston Microbiology staff and Infection Prevention staff for their contributions to the MRSA Prevention program and to this study.
Footnotes
Published ahead of print 25 July 2012
REFERENCES
- 1. Arbefeville SS, et al. 2011. Prevalence and genetic relatedness of methicillin-susceptible Staphylococcus aureus isolates detected by the Xpert MRSA nasal assay. J. Clin. Microbiol. 49:2996–2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Blanc DS, et al. 2011. High proportion of wrongly identified methicillin-resistant Staphylococcus aureus carriers by use of a rapid commercial PCR assay due to presence of staphylococcal cassette chromosome element lacking the mecA gene. J. Clin. Microbiol. 49:722–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. British Society for Antimicrobial Chemotherapy 2012. Methods for antimicrobial susceptibility testing. British Society for Antimicrobial Chemotherapy, Birmingham, United Kingdom [Google Scholar]
- 4. Clinical Laboratory Standards Institute 2012. Performance standards for antimicrobial disk susceptibility tests; approved standard, 11th ed, MO2-A11 Clinical Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 5. Davis KA, Stewart JJ, Crouch HK, Florez CE, Hospenthal DR. 2004. Methicillin-resistant Staphylococcus aureus (MRSA) nares colonization at hospital admission and its effect on subsequent MRSA infection. Clin. Infect. Dis. 39:776–782 [DOI] [PubMed] [Google Scholar]
- 6. Desjardins M, Guibord C, Lalonde B, Toye B, Ramotar K. 2006. Evaluation of the IDI-MRSA assay for detection of methicillin-resistant staphylococcus aureus from nasal and rectal specimens pooled in a selective broth. J. Clin. Microbiol. 44:1219–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Harbarth S, et al. 2008. Universal screening for methicillin-resistant Staphylococcus aureus at hospital admission and nosocomial infection in surgical patients. JAMA 299:1149–1157 [DOI] [PubMed] [Google Scholar]
- 8. Huang SS, et al. 2011. Methicillin-resistant Staphylococcus aureus infection and hospitalization in high-risk patients in the year following detection. PLoS One 6:e24340 doi:10.1371/journal.pone.0024340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jain R, et al. 2011. Veterans Affairs initiative to prevent methicillin-resistant Staphylococcus aureus infections. N. Engl. J. Med. 364:1419–1430 [DOI] [PubMed] [Google Scholar]
- 10. Kluytmans J, Harbarth S. 2011. Control of MRSA in intensive care units. BMJ 343:d5885 (Comment.) [DOI] [PubMed] [Google Scholar]
- 11. Kreisel KM, et al. 2011. USA300 methicillin-resistant Staphylococcus aureus bacteremia and the risk of severe sepsis: is USA300 methicillin-resistant Staphylococcus aureus associated with more severe infections? Diagn. Microbiol. Infect. Dis. 70:285–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. LaPlante KL, Caffrey AR, Gupta K. 2010. Prevention of surgical-site infections. N. Engl. J. Med. 362:1540–1541 (Author reply, 362:1542–1543.) [PubMed] [Google Scholar]
- 13. Lee AS, et al. 2011. Impact of combined low-level mupirocin and genotypic chlorhexidine resistance on persistent methicillin-resistant Staphylococcus aureus carriage after decolonization therapy: a case-control study. Clin. Infect. Dis. 52:1422–1430 [DOI] [PubMed] [Google Scholar]
- 14. Lee AS, et al. 2011. Trends in mupirocin resistance in meticillin-resistant Staphylococcus aureus and mupirocin consumption at a tertiary care hospital. J. Hosp. Infect. 77:360–362 [DOI] [PubMed] [Google Scholar]
- 15. Lim MS, Marshall CL, Spelman D. 2006. Carriage of multiple subtypes of methicillin-resistant Staphylococcus aureus by intensive care unit patients. Infect. Control Hosp. Epidemiol. 27:1063–1067 [DOI] [PubMed] [Google Scholar]
- 16. Maslow JN, et al. 1995. Variation and persistence of methicillin-resistant Staphylococcus aureus strains among individual patients over extended periods of time. Eur. J. Clin. Microbiol. Infect. Dis. 14:282–290 [DOI] [PubMed] [Google Scholar]
- 17. Scanvic A, et al. 2001. Duration of colonization by methicillin-resistant Staphylococcus aureus after hospital discharge and risk factors for prolonged carriage. Clin. Infect. Dis. 32:1393–1398 [DOI] [PubMed] [Google Scholar]
- 18. Schmitz FJ, et al. 2000. Development of resistance to ciprofloxacin, rifampin, and mupirocin in methicillin-susceptible and -resistant Staphylococcus aureus isolates. Antimicrob. Agents Chemother. 44:3229–3231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schmitz FJ, et al. 1998. The prevalence of low- and high-level mupirocin resistance in staphylococci from 19 European hospitals. J. Antimicrob. Chemother. 42:489–495 [DOI] [PubMed] [Google Scholar]
- 20. Shurland SM, et al. 2012. USA300 methicillin-resistant S. aureus (USA300 MRSA) colonization and the risk of MRSA infection in residents of extended-care facilities. Epidemiol. Infect. 140:390–399 [DOI] [PubMed] [Google Scholar]
- 21. Tacconelli E, et al. 2009. Rapid screening tests for methicillin-resistant Staphylococcus aureus at hospital admission: systematic review and meta-analysis. Lancet Infect. Dis. 9:546–554 [DOI] [PubMed] [Google Scholar]
- 22. Thomas CM, Hothersall J, Willis CL, Simpson TJ. 2010. Resistance to and synthesis of the antibiotic mupirocin. Nat. Rev. Microbiol. 8:281–289 [DOI] [PubMed] [Google Scholar]
- 23. Tracy LA, et al. 2011. Staphylococcus aureus infections in US veterans, Maryland, USA, 1999–2008. Emerg. Infect. Dis. 17:441–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang K, McClure JA, Elsayed S, Louie T, Conly JM. 2008. Novel multiplex PCR assay for simultaneous identification of community-associated methicillin-resistant Staphylococcus aureus strains USA300 and USA400 and detection of mecA and Panton-Valentine leukocidin genes, with discrimination of Staphylococcus aureus from coagulase-negative staphylococci. J. Clin. Microbiol. 46:1118–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang K, et al. 2004. New quadriplex PCR assay for detection of methicillin and mupirocin resistance and simultaneous discrimination of Staphylococcus aureus from coagulase-negative staphylococci. J. Clin. Microbiol. 42:4947–4955 [DOI] [PMC free article] [PubMed] [Google Scholar]