Abstract
Streptococcus suis is an emerging zoonotic pathogen causing severe infections in pigs and humans. Thirty-three serotypes of S. suis have been identified using serum agglutination. The capsular polysaccharides synthesis (cps) locus is usually conserved among different strains of the same serotype. The cps loci of 15 serotypes have been sequenced, while the loci of the other serotypes remain unknown. In the present study, two to six serotype-specific genes of each of eight serotypes, i.e., serotypes 3, 4, 5, 8, 10, 19, 23, and 25, were identified using cross-hybridization with 93 nucleic acid probes specific to genes in the cps locus, and serotype-specific PCR assays for rapid and sensitive detection of the eight serotypes were then developed. The PCR typing results of the 148 serologically typeable isolates were completely consistent with agglutination results. Furthermore, some autoagglutinating, acapsular, and multiagglutinating strains which could not be differentiated by traditional serum agglutination assays were positive in the PCR assays. Use of the PCR assays with clinical tonsillar specimens showed that the assays are sensitive and able to identify samples with autoagglutinating isolates. To our knowledge, this is the first study to identify the serotype-specific genes of the eight Streptococcus suis serotypes and develop rapid and sensitive PCR assays for the eight serotypes which can be identified only by serum agglutination.
INTRODUCTION
Streptococcus suis is a Gram-positive bacterium and an important pathogen of pigs (11, 35). Infection of S. suis is endemic in nearly all countries with an extensive pig industry, and it is also a zoonotic disease (33). Humans can be infected with S. suis when they handle infected pig carcasses or meat, especially if they have exposed cuts and abrasions on their hands (33). Human infection can be severe, with meningitis, septicemia, endocarditis, and deafness being possible outcomes of infection. S. suis can localize in the upper respiratory tracts of pigs, especially on the palatine tonsils and in the nasal cavities (6). Carrier pigs, harboring the organism in their palatine tonsils, play an important role in disseminating S. suis (6, 14). Various authors have determined the prevalence of carrier pigs of S. suis based on tonsillar samples from slaughter pigs (4, 7). Attempts to control the disease are still hampered by the lack of effective vaccines, sensitive diagnostics, and sufficient knowledge about the epidemiology of the disease.
So far, based on the presence of specific capsular polysaccharides, 33 serotypes of S. suis have been identified (15, 28). Among them, serotype 2 is the major one circulating in pigs (11, 35). Other serotypes are also frequently isolated from the diseased pigs, especially serotypes 3, 4, and 8. For example, among the 407 S. suis strains isolated from diseased pigs in China analyzed by Wei et al. (32), serotype 2 (43.2%) was the most prevalent, followed by serotype 3 (14.7%) and serotypes 4, 8, 5, 7, and 1/2 (3.2 to 6.4%). Serotypes 6, 9, 10, 11, 12, 13, 14, 15, 16, 17, 19, 20, 21, 22, 23, 25, and 28 were also found in China. The strains isolated from diseased pigs in Canada mainly belong to serotypes 2 and 3, followed by serotypes 1/2, 4, 7, and 8 (21). In Denmark, serotypes 2, 3, 4, 7, and 8 were found to be the most prevalent in pigs (1). The prevalent strains in Japan belong to serotypes 2, 3, 7, and 1/2 (16). Among S. suis isolated from pigs with polyserositis in Korea, serotype 3 (29%) and serotype 4 (21%) are the most prevalent (18).
S. suis isolates are generally serotyped by serum agglutination method, based on the presence of specific capsular polysaccharides (13). This typing method is laborious and time-consuming and can be performed only on isolated colonies. Among the 33 serotypes of S. suis, the cps loci of serotypes 1, 2, 3, 4, 5, 7, 8, 9, 10, 14, 16, 19, 23, 25, and 1/2 have been sequenced, while the loci of the other serotypes remain unknown (30). Based on serotype-specific genes in the cps locus, rapid and sensitive PCR assays specific for serotype 1 (and 14), 2 (and 1/2), 7, 9, and 16 have been described (26, 27, 31). Until now, no rapid and sensitive PCR typing method for identifying other S. suis serotypes has been available. According to the above epidemiological information, it is important to develop rapid and sensitive PCR typing methods for other prevalent serotypes.
In the present study, the serotype-specific genes of serotypes 3, 4, 5, 8, 10, 19, 23, and 25 in the cps locus were screened by nucleic acid cross-hybridization, and PCR assays to type these eight serotypes were developed based on their serotype-specific genes. The PCR assays were evaluated using reference strains and clinical isolates and samples.
MATERIALS AND METHODS
Bacterial strains.
S. suis reference strains (serotypes 1 to 31, 33, and 1/2) were obtained from M. Gottschalk (Faculty of Veterinary Medicine, University of Montreal, Montreal, Canada) (12). Streptococcus agalactiae 1886 (group B), Streptococcus dysgalactiae ATCC 9926 (group C), Streptococcus equi subsp. zooepidemicus ATCC 35246 (group C), and Streptococcus mutans UA159 (group C) were kept by Nanjing Agricultural University. One hundred ninety-five Chinese S. suis isolates were isolated from pigs between 2005 and 2010. The details of the isolates are shown in Table S1 in the supplemental material. S. suis strains were grown in Todd-Hewitt broth (Oxoid, United Kingdom) or on plates with Columbia agar blood base (Oxoid) containing 6% (vol/vol) sheep blood overnight.
Screening of serotype-specific genes.
Serotype-specific genes were identified using nucleic acid cross-hybridization, as described previously (26, 27, 31). Briefly, genomic DNAs of the eight serotypes reference strains were extracted using a Wizard genomic-DNA purification kit (Promega, Madison, WI). Except the conserved genes (cpsA to cpsD) and transposase genes (30), DNA fragments of the genes in each serotype cps locus were amplified by PCR (see Table S2 in the supplemental material) according to the cps locus sequences of S. suis serotypes 3, 4, 5, 8, 10, 19, 23, and 25 (GenBank accession no. JF273646 to JF273648, JF273650, JF273652, and JF273654 to JF273656). The amplified fragments were labeled as DNA probes using a DIG High Prime I DNA labeling and detection starter kit (Roche Applied Science, Germany) and hybridized to genomic DNAs of reference strains of all 33 serotypes, according to the instructions. As a positive control, a probe specific for S. suis 16S rRNA genes was used in the nucleic acid cross-hybridization. The DNA on the blots was hybridized at 65°C with appropriate DNA probes. After hybridization, the membranes were washed twice with a solution comprising 40 mM sodium phosphate (pH 7.2), 1 mM EDTA, and 5% sodium dodecyl sulfate (SDS) for 30 min at 65°C and twice with a solution comprising 40 mM sodium phosphate (pH 7.2) and 1 mM EDTA–1% SDS for 30 min at 65°C (25).
Development of the PCR typing methods.
The primers located in the serotype-specific genes (Table 1) were designed to develop the PCR assays for serotypes 3, 4, 5, 8, 10, 19, 23, and 25. The reference strains of the 33 S. suis serotypes and other Streptococcus strains (1886, ATCC 9926, ATCC 35246, and UA159) were detected by the PCR assays. The PCR mixture (25 μl) contained 1.25 U Taq DNA polymerase (TaKaRa, Japan), 2.5 μl 10× PCR buffer, 2 μl deoxynucleoside triphosphate (dNTP) mixture (0.25 mM for each dNTP), 0.5 μl upstream primer (10 pM), 0.5 μl downstream primer (10 pM), and 0.5 μl broth culture (or genomic DNA). DNA amplification was carried out in a Perkin Elmer 9600 thermal cycler. The sequences of the primers and the PCR programs of each serotype are listed in Table 2.
Table 1.
Serotype-specific genes and function of the translated proteins in S. suis serotypes 3, 4, 5, 8, 10, 19, 23, and 25
| Serotype | Serotype-specific gene | Protein function |
|---|---|---|
| 3 | cps3H | Galactose phosphate transferase |
| cps3I | Acetyltransferase | |
| cps3J | Glycosyl transferase | |
| cps3K | Flippase | |
| cps3La | Wzy polysaccharide polymerase | |
| 4 | cps4La | Glycosyl transferase |
| cps4M | Flippase | |
| 5 | cps5Ia | Glycosyl transferase |
| cps5J | Acetyltransferase | |
| cps5N | Acetyltransferase | |
| cps5P | Acetyltransferase | |
| 8 | cps8H | Glycosyl transferase |
| cps8I | Phosphotransferase | |
| cps8J | Glycosyl transferase | |
| cps8Ka | Wzy polysaccharide polymerase | |
| cps8L | UDP-N-acetylglucosamine 2-epimerase | |
| 10 | cps10Ma | Wzy polysaccharide polymerase |
| cps10N | Epimerase | |
| cps10Q | Glycerophosphotransferase | |
| cps10R | Cytidylyltransferase | |
| 19 | cps19I | Acetyltransferase |
| cps19L | Wzy polysaccharide polymerase | |
| cps19M | Acetyltransferase | |
| cps19Na | Flippase | |
| cps19O | Glycosyl transferase | |
| 23 | cps23H | Glycosyl transferase |
| cps23Ja | Glycosyl transferase | |
| cps23K | Polysaccharide pyruvyl transferase | |
| 25 | cps25F | Glycosyl transferase |
| cps25Ga | Glycosyl transferase | |
| cps25H | Phosphotransferase | |
| cps25I | Glycosyl transferase | |
| cps25K | Acetyltransferase | |
| cps25L | Acetyltransferase |
Gene selected to develop the PCR method.
Table 2.
Sequences of the primers and the PCR programs used to identify S. suis serotypes 3, 4, 5, 8, 10, 19, 23, and 25
| Serotype | Primer | Sequence (5′→3′) | PCR program | Amplicon length (bp) |
|---|---|---|---|---|
| 3 | cps3L-P1 | AGTTGGTATTTGTTTAGG | 5 min at 94°C and 30 amplification cycles (94°C at 30 s, 30 s at 50°C, and 30 s at 72°C) | 340 |
| cps3L-P2 | GGAAGATTTAGCAGTTTT | |||
| 4 | cps4L-P1 | TACTTCAGTTAGGTTCCTCGCA | 5 min at 94°C and 30 cycles of 30 s at 94°C, 30 s at 50°C, and 30 s at 72°C | 690 |
| cps4L-P2 | CTCTCCACTTATTCACTTTCCC | |||
| 5 | cps5I-P1 | TTTTCGTTGTATTTTCCAAA | 5 min at 94°C and 30 cycles of 30 s at 94°C, 30 s at 50°C, and 30 s at 72°C | 262 |
| cps5I-P2 | TCCAAACATTATCCCCTATT | |||
| 8 | cps8K-P1 | ATCTGTTGTAATGGTCGC | 5 min at 94°C and 30 cycles of 30 s at 94°C, 30 s at 50°C, and 1 min at 72°C | 1,200 |
| cps8K-P2 | CTATTCCTCTTGGTGTGA | |||
| 10 | cps10M-P1 | TCACTACCACGGAATGCT | 5 min at 94°C and 30 cycles of 30 s at 94°C, 30 s at 50°C, and 1 min at 72°C | 898 |
| cps10M-P2 | CAAAGGTCACCCCAGAAT | |||
| 19 | cps19N-P1 | AGGCTACAAAGCGATAGAACT | 5 min at 94°C and 30 cycles of 30 s at 94°C, 30 s at 50°C, and 30 s at 72°C | 391 |
| cps19N-P2 | ACCAAAGAATCAACCAATAAA | |||
| 23 | cps23J-P1 | GCTATCAGGAATACACAAAG | 5 min at 94°C and 30 cycles of 30 s at 94°C, 30 s at 50°C, and 1 min at 72°C | 758 |
| cps23J-P2 | AGTGGAATAACAACAAAAAT | |||
| 25 | cps25G-P1 | GATTTTTGTGTGACTGTGGG | 5 min at 94°C and 30 cycles of 30 s at 94°C, 30 s at 50°C, and 30 s at 72°C | 478 |
| cps25G-P2 | TTCGGTAACTTCTCGTGT |
Evaluation of PCR typing methods.
To evaluate the sensitivity of the PCR assays, 10-fold serially diluted broth culture of S. suis reference strains, from 1 × 107 CFU/ml to 1 CFU/ml, were detected by the PCR typing methods. The methods were also used to detect the broth culture of 195 S. suis isolates from pigs in China. The isolates were also serotyped by antiserum agglutination (13). The serotyping antiserum produced by rabbits was prepared according to reported methods (13). The capsule was detected with a capsular staining kit (Genmed Scientifics).
Detection of S. suis in clinical samples.
Tonsils from 100 pigs were collected from herds suspected of having S. suis infection and stored at −20°C until the detection assay. A 200-μl portion of each macerated tonsillar specimen was incubated overnight at 37°C in 5 ml Todd-Hewitt broth with 0.25% streptococcus selective supplement (Oxoid, United Kingdom). The broth culture was assayed in the eight-serotype-specific PCR. To evaluate the results obtained with the PCR assays, these tonsillar specimens were subjected to an extended bacteriological examination (34). Tonsillar specimens were plated, and colonies were lifted onto sterilized GeneScreen Plus membranes (New England Nuclear) and hybridized with the serotype-specific probes, the genes of which were identical to those used to develop the serotype-specific PCR method. Hybridizing colonies were subcultured, characterized, and serotyped by standard procedures (8, 13).
Sequencing the cps locus of three clinical isolates.
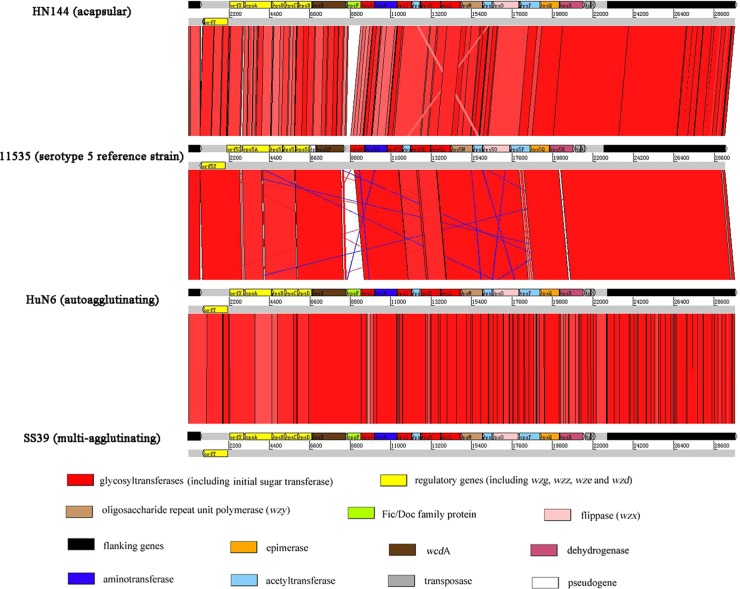
To determine why the PCR was positive for some serologically untypeable isolates, the entire cps locus was sequenced and analyzed for the three clinical isolates (1 multiagglutinating, 1 acapsular, and 1 autoagglutinating), which were positive in serotype 5 PCR detection. Amplification, sequencing, and annotation of the cps locus were performed as described previously (30). The cps loci of the three isolates were compared with that of the serotype 5 reference strain 11535. Visual representation of the alignments using nucleotide similarities (TBLASTX) of the cps locus were performed with the Artemis comparison tool (ACT) (5).
Nucleotide sequence accession numbers.
The sequences of the cps loci of isolates HuN6, HN144, and SS39 have been deposited in GenBank under accession numbers JX105395 to JX105397.
RESULTS
Screening of serotype-specific genes.
The 16S rRNA gene probe hybridized with almost equal intensities with all serotypes. The results of cross-hybridization experiments are shown in Table S3 in the supplemental material. Most of the probes specific for genes in the cps locus can react with the genomic DNAs of 2 to 22 serotypes. Two (serotype 4) to six (serotype 25) serotype-specific genes were identified in each serotype through the probe hybridization profiles. The serotype-specific genes mainly encode glycosyltransferase, acetyltransferase, polysaccharide polymerase, or flippase (Table 1).
Development of the PCR typing methods.
The detection results for chromosomal DNA of the 33 S. suis reference strains using the PCR typing methods showed that each of the reference strains of the eight serotypes was specifically detected by the PCR assays. The reference strains for the other 25 serotypes and other Streptococcus strains were negative in all the eight serotype-specific PCR assays. At least 5 CFU (0.5 μl, 1 × 104 CFU/ml) of all reference strains of the eight serotypes in broth culture could be detected in one reaction (25 μl).
Evaluation of the PCR typing methods.
The detection results of the 195 clinical isolates by the PCR assays and agglutination are listed in Table S1 in the supplemental material. One hundred forty-eight isolates could be serotyped by serum agglutination, while the other 47 isolates were autoagglutinating, acapsular, or multiagglutinating. Except the acapsular isolates, multiagglutinating and autoagglutinating isolates were encapsulated. PCR typing of the 148 serologically typeable isolates as serotypes 3, 4, 5, 8, 10, 19, 23, and 25 was completely consistent with identifications made by traditional serum agglutination assays, while isolates of other serotypes were negative in the eight-serotype PCR typing. Of the 47 serologically untypeable isolates, three (one autoagglutinating, one acapsular, and one multiagglutinating) were positive in the serotype 5 PCR assay, whereas the others were negative in all eight serotype-specific PCR assays (see Table S1 in the supplemental material).
cps loci of three serologically untypeable isolates.
The entire cps loci of the aforementioned three isolates (HuN6, HN144, and SS39) were amplified and sequenced, and sequences of 29,716 bp, 29,718 bp, and 29,718 bp were obtained, respectively. Differences between these sequences and those of the serotype 5 reference strains are shown in Fig. 1. The serotype-specific gene (cps5I) detected in serotype 5 typing PCR were confirmed to exist in the three serologically untypeable isolates. The only obvious difference in the cps loci was wcdA and its flanking genes. Compared to the cps locus of the serotype 5 reference strain (30), wcdA in the three isolates is longer at the 5′ end, resulting in translation of a lengthened CapD-like protein (capsule depolymerase). The sequence upstream of wcdA in the three isolates lost a pseudogene. In the cps loci of the three isolates, the gene downstream of wcdA (fic) could produce the Fic/Doc (filamentation induced by cAMP/death on curing) family protein originally described as being required for cyclic AMP (cAMP)-induced filamentation (17, 19). The Fic/Doc family gene was lost in the cps locus of the serotype 5 reference strain.
Fig 1.
Comparisons of the cps loci of isolates HuN6, HN144, and SS39 and the serotype 5 reference strain. The only obvious difference in the cps loci was wcdA and its flanking genes. The red and blue bands represent forward and reverse matches, respectively.
Detection in clinical samples.
We subsequently analyzed tonsillar specimens by PCR assays and by bacteriological examination to compare the two typing methods. All tonsillar specimens bacteriologically positive for a specific serotype were also positive in the particular serotype-specific PCR (Table 3). Moreover, tonsils negative for a specific serotype by PCR were also negative for this particular serotype or phenotype by the bacteriological method. Two samples positive in the PCR tested bacteriologically negative (one of five for serotype 3; one of three for serotype 5). The isolates from these two samples were positive for autoagglutination by the serological methods. To corroborate this and to exclude false-positive PCR results, we repeatedly tested the samples both by PCR and by bacteriological examination. The same results were obtained.
Table 3.
Comparison of PCR assays and bacteriological examination (BE) for 100 tonsillar specimens from herds with suspected S. suis infection
| Serotype | No. of tonsillar specimens |
||||
|---|---|---|---|---|---|
| PCR+, BE+ | PCR+, BE− | PCR−, BE− | PCR−, BE+ | Total PCR+ | |
| 3 | 4 | 1 | 95 | 0 | 5 |
| 4 | 3 | 0 | 97 | 0 | 3 |
| 5 | 2 | 1 | 97 | 0 | 3 |
| 8 | 2 | 0 | 98 | 0 | 2 |
| 10 | 0 | 0 | 100 | 0 | 0 |
| 19 | 1 | 0 | 99 | 0 | 1 |
| 23 | 0 | 0 | 100 | 0 | 0 |
| 25 | 1 | 0 | 99 | 0 | 1 |
DISCUSSION
Capsular polysaccharides are an extremely diverse range of molecules that may differ not only by monosaccharide units but also in how these units are joined together (24). The biosynthesis of capsular polysaccharides requires a complex pathway, and generally, the genes involved in this process are clustered in cps locus (24, 26, 27). In S. suis, cps locus covers multiple genes encoding regulatory proteins, glycosyltransferase, polysaccharide polymerase, flippase, and some transferases (30). According to the presence of specific capsular antigenic determinants, traditional antiserum agglutination is performed to serotype some pathogens (2, 22, 29), as well as S. suis (13). The agglutination method is laborious and time-consuming and can be performed only on isolated colonies. Although several serotypes of S. suis circulate in pigs (16, 18, 21, 32), except for the PCR assays specific to serotypes 1 (and 14), 2 (and 1/2), 7, 9, and 16 (26, 27, 31), no rapid PCR methods can be used to type the other serotypes. The PCR assays for serotypes 1 (and 14), 2 (and 1/2), 7, 9, and 16 were developed based on serotype-specific genes in the cps locus (26, 27, 31), which is usually conserved in different strains of the same serotype (3). The sequenced cps locus in the eight serotypes (serotypes 3, 4, 5, 8, 10, 19, 23, and 25) can be used to develop S. suis serotype-specific PCR assays (30).
In this study, 93 genes in the eight serotypes cps loci were amplified and labeled as probes for the cross-hybridization experiments with chromosomal DNAs from 33 serotypes. Two to six genes in the cps locus of each serotype were identified as serotype-specific genes, one of which was selected to develop a rapid PCR assay for that serotype. The PCR assays can be used in the detection and typing of reference strains and clinical isolates and samples for the eight S. suis serotypes. The PCR typing results for the reference strains and the 148 serologically typeable isolates were completely consistent with those of traditional serum agglutination assays. The results indicate that the genes identified (Table 1) in cross-hybridization experiments and PCR assays are serotype specific. The functions of proteins translated from the serotype-specific genes were mainly glycosyltransferase, acetyltransferase, polysaccharide polymerase, and flippase, which are important to the unique structure of capsular polysaccharide (10).
The sensitivity of the PCR assays was evaluated and found to be at least 5 CFU in reference strain detection. The clinical tonsillar specimen detection showed that the PCR assays are sensitive and able to detect the samples with autoagglutinating isolates which cannot be identified by the regular bacteriological examination.
Among the 195 clinical isolates assayed, three (HuN6, HN144, and SS39) which did not agglutinate with antiserum to serotype 5 were positive in the serotype 5 PCR typing. The cps loci of the three serologically untypeable isolates were sequenced. The serotype-specific gene (cps5I) detected in serotype 5 typing PCR was found in these isolates, indicating that the serotype 5 PCR typing results were not false positives. Mutation in the cps locus, even at a single point, can change the capsular phenotype (20). The comparison results showed that the only obvious difference in the cps loci was wcdA and its flanking genes. wcdA and fic may be important to the capsular phenotype. Compared to serotype 5 reference strains, the cps loci of the three serologically untypeable isolates contained a longer wcdA gene, associated with the translation of a CapD-like protein lengthened at the C terminus. The length difference of the CapD-like protein may change the protein activation and influence the capsule anchor to the cell wall envelope. It was also possible that not only the genes in the cps locus but also some other known or unknown genes outside the locus could regulate the capsular phenotype (9). Isolates which can agglutinate with the antiserum to serotype 1 and 2 are recognized as S. suis serotype 1/2 (23). Therefore, the multiagglutinating isolates could potentially be new serotypes. Once cross-absorption experiments with the other serotype antiserum and isolates have been carried out, the multiagglutinating isolate (SS39) will be considered a new serotype. Although the cps loci of HN144 and HuN6 (autoagglutinating) were similar to that of SS39 (acapsular), the capsular phenotypes were different. According to the present study, it is uncertain whether HN144 and HuN6 are potentially new serotypes.
In conclusion, the PCR assays were effective in identifying S. suis serotypes 3, 4, 5, 8, 10, 19, 23, and 25 by testing whole bacterial cells and infected tonsillar specimens. PCR can potentially be used for typing the eight serotypes with both high specificity and high sensitivity, avoiding the problems associated with serologic assays. The PCR assays can be used for clinical diagnostics, epidemiology studies, and disease control involving these eight S. suis serotypes. Multiplex PCR serotyping assays will be developed and described in a future work.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Basic Research Program of China (2012CB518804), Special Fund for the Public Welfare Industry of Chinese Ministry of Agriculture (201303041), Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), and the Fund for Key Technique Introduction of the Important Animal Disease Prevention from MOA (2006-G57).
We thank Jiming Chen for his comments on data analysis.
Footnotes
Published ahead of print 8 August 2012
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Aarestrup FM, Jorsal SE, Jensen NE. 1998. Serological characterization and antimicrobial susceptibility of Streptococcus suis isolates from diagnostic samples in Denmark during 1995 and 1996. Vet. Microbiol. 60:59–66 [DOI] [PubMed] [Google Scholar]
- 2. Bentley SD, et al. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2:e31 doi:10.1371/journal.pgen.0020031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boulnois GJ. 1989. Genetics of capsular polysaccharide production in bacteria. Symp. Soc. Exp. Biol. 43:417–422 [PubMed] [Google Scholar]
- 4. Breton J, Mitchell WR, Rosendal S. 1986. Streptococcus suis in slaughter pigs and abattoir workers. Can. J. Vet. Res. 50:338–341 [PMC free article] [PubMed] [Google Scholar]
- 5. Carver TJ, et al. 2005. ACT: the Artemis Comparison Tool. Bioinformatics 21:3422–3423 [DOI] [PubMed] [Google Scholar]
- 6. Clifton-Hadley FA, Alexander TJ. 1980. The carrier site and carrier rate of Streptococcus suis type II in pigs. Vet. Rec. 107:40–41 [DOI] [PubMed] [Google Scholar]
- 7. Clifton-Hadley FA, Alexander TJ, Enright MR, Guise J. 1984. Monitoring herds for Streptococcus suis type 2 by sampling tonsils of slaughter pigs. Vet. Rec. 115:562–564 [DOI] [PubMed] [Google Scholar]
- 8. Devriese LA, Ceyssens K, Hommez J, Kilpper-Balz R, Schleifer KH. 1991. Characteristics of different Streptococcus suis ecovars and description of a simplified identification method. Vet. Microbiol. 26:141–150 [DOI] [PubMed] [Google Scholar]
- 9. Fittipaldi N, et al. 2007. Potential use of an unencapsulated and aromatic amino acid-auxotrophic Streptococcus suis mutant as a live attenuated vaccine in swine. Vaccine 25:3524–3535 [DOI] [PubMed] [Google Scholar]
- 10. Forsee WT, Cartee RT, Yother J. 2000. Biosynthesis of type 3 capsular polysaccharide in Streptococcus pneumoniae. Enzymatic chain release by an abortive translocation process. J. Biol. Chem. 275:25972–25978 [DOI] [PubMed] [Google Scholar]
- 11. Gottschalk M, Xu J, Calzas C, Segura M. 2010. Streptococcus suis: a new emerging or an old neglected zoonotic pathogen? Future Microbiol. 5:371–391 [DOI] [PubMed] [Google Scholar]
- 12. Harel J, Higgins R, Gottschalk M, Bigras-Poulin M. 1994. Genomic relatedness among reference strains of different Streptococcus suis serotypes. Can. J. Vet. Res. 58:259–262 [PMC free article] [PubMed] [Google Scholar]
- 13. Higgins R, Gottschalk M. 1990. An update on Streptococcus suis identification. J. Vet. Diagn. Invest. 2:249–252 [DOI] [PubMed] [Google Scholar]
- 14. Higgins R, Gottschalk M. (ed). 1999. Streptococcal diseases. Iowa State University Press, Ames, IA [Google Scholar]
- 15. Hill JE, et al. 2005. Biochemical analysis, cpn60 and 16S rDNA sequence data indicate that Streptococcus suis serotypes 32 and 34, isolated from pigs, are Streptococcus orisratti. Vet. Microbiol. 107:63–69 [DOI] [PubMed] [Google Scholar]
- 16. Kataoka Y, Sugimoto C, Nakazawa M, Morozumi T, Kashiwazaki M. 1993. The epidemiological studies of Streptococcus suis infections in Japan from 1987 to 1991. J. Vet. Med. Sci. 55:623–626 [DOI] [PubMed] [Google Scholar]
- 17. Kawamukai M, et al. 1988. Cloning of the fic-1 gene involved in cell filamentation induced by cyclic AMP and construction of a Δfic Escherichia coli strain. J. Bacteriol. 170:3864–3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim D, et al. 2010. Distribution of capsular serotypes and virulence markers of Streptococcus suis isolated from pigs with polyserositis in Korea. Can. J. Vet. Res. 74:314–316 [PMC free article] [PubMed] [Google Scholar]
- 19. Komano T, Utsumi R, Kawamukai M. 1991. Functional analysis of the fic gene involved in regulation of cell division. Res. Microbiol. 142:269–277 [DOI] [PubMed] [Google Scholar]
- 20. Melchiorre S, et al. 2012. Point mutations in wchA are responsible for the non-typability of two invasive Streptococcus pneumoniae isolates. Microbiology 158:338–344 [DOI] [PubMed] [Google Scholar]
- 21. Messier S, Lacouture S, Gottschalk M. 2008. Distribution of Streptococcus suis capsular types from 2001 to 2007. Can. Vet. J. 49:461–462 [PMC free article] [PubMed] [Google Scholar]
- 22. Onokodi JK, Wauters G. 1981. Capsular typing of klebsiellae by coagglutination and latex agglutination. J. Clin. Microbiol. 13:609–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Perch B, Kjems E, Slot P, Pedersen KB. 1981. Biochemical and serological properties of R, S, and RS streptococci. Acta Pathol. Microbiol. Scand. B 89:167–171 [DOI] [PubMed] [Google Scholar]
- 24. Roberts IS. 1996. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu. Rev. Microbiol. 50:285–315 [DOI] [PubMed] [Google Scholar]
- 25. Sambrook J, Russell DW. (ed). 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 26. Smith HE, van Bruijnsvoort L, Buijs H, Wisselink HJ, Smits MA. 1999. Rapid PCR test for Streptococcus suis serotype 7. FEMS Microbiol. Lett. 178:265–270 [DOI] [PubMed] [Google Scholar]
- 27. Smith HE, et al. 1999. The cps genes of Streptococcus suis serotypes 1, 2, and 9: development of rapid serotype-specific PCR assays. J. Clin. Microbiol. 37:3146–3152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Staats JJ, Feder I, Okwumabua O, Chengappa MM. 1997. Streptococcus suis: past and present. Vet. Res. Commun. 21:381–407 [DOI] [PubMed] [Google Scholar]
- 29. Verdier I, et al. 2007. Identification of the capsular polysaccharides in Staphylococcus aureus clinical isolates by PCR and agglutination tests. J. Clin. Microbiol. 45:725–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang K, Fan W, Cai L, Huang B, Lu C. 2011. Genetic analysis of the capsular polysaccharide synthesis locus in 15 Streptococcus suis serotypes. FEMS Microbiol. Lett. 324:117–124 [DOI] [PubMed] [Google Scholar]
- 31. Wang K, Fan W, Wisselink H, Lu C. 2011. The cps locus of Streptococcus suis serotype 16: development of a serotype-specific PCR assay. Vet. Microbiol. 153:403–406 [DOI] [PubMed] [Google Scholar]
- 32. Wei Z, et al. 2009. Characterization of Streptococcus suis isolates from the diseased pigs in China between 2003 and 2007. Vet. Microbiol. 137:196–201 [DOI] [PubMed] [Google Scholar]
- 33. Wertheim HF, Nghia HD, Taylor W, Schultsz C. 2009. Streptococcus suis: an emerging human pathogen. Clin. Infect. Dis. 48:617–625 [DOI] [PubMed] [Google Scholar]
- 34. Wisselink HJ, et al. 1999. Detection of virulent strains of Streptococcus suis type 2 and highly virulent strains of Streptococcus suis type 1 in tonsillar specimens of pigs by PCR. Vet. Microbiol. 67:143–157 [DOI] [PubMed] [Google Scholar]
- 35. World Health Organization 2005. Outbreak associated with Streptococcus suis in pigs, China. Wkly. Epidemiol. Rec. 80:269–270 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.