Abstract
Temporal and seasonal trends in Campylobacter genotypes causing human gastroenteritis were investigated in a 6-year study of 3,300 recent isolates from Oxfordshire, United Kingdom. Genotypes (sequence types [ST]) were defined using multilocus sequence typing and assigned to a clonal complex (a cluster of related strains that share four or more identical alleles with a previously defined central genotype). A previously undescribed clonal complex (ST-464) was identified which, together with ST-42, ST-45, and ST-52 complexes, showed increasing incidence. Concurrently, the incidence of ST-574, ST-607, and ST-658 complexes declined. The relative frequencies of three clonal complexes (ST-45, ST-283, and ST-42) peaked during summer and those of two (ST-353 and ST-403) peaked during winter. Nine clonal complexes (ST-22, ST-45, ST-48, ST-61, ST-257, ST-283, ST-403, ST-658, and ST-677) were significantly associated with ciprofloxacin sensitivity (P < 0.05). Seven clonal complexes (ST-49, ST-206, ST-354, ST-446, ST-460, ST-464, and ST-607) were associated with ciprofloxacin resistance (P < 0.05). Clonal complexes exhibited changing incidence and differences in seasonality and antibiotic resistance phenotype. These data also demonstrated that detailed surveillance at a single site captures information which reflects that observed nationally.
INTRODUCTION
Campylobacter jejuni and Campylobacter coli are the most frequent causes of acute bacterial gastroenteritis in humans, representing an unrelenting worldwide public health problem. C. jejuni accounts for over 90% of cases, with the majority of the remainder caused by C. coli (14). The annual incidence of diagnosed human infections in 2008 was 92 per 100,000 individuals (16) in England and Wales, compared to 13 per 100,000 in the United States (4). However, estimates of underreporting indicate that the actual incidences are 7- and 38-fold higher, respectively (4, 47). Incidence varies with age, being highest among those under five years of age (16), and overall, males are more frequently affected, with a male-to-female sex ratio of 1.2 to 1 (28). Many wild and farmed avian and mammalian species carry campylobacters as commensal members of the gastrointestinal microbiota, with human infection resulting from either direct contact with contaminated feces or indirect transmission via contaminated food.
Multilocus sequence typing (MLST) studies have determined C. jejuni and C. coli population structures to be essentially nonclonal but comprising a large number of genetically related isolates, or clonal complexes (8, 10). Outbreak investigation (39), case control (35), and “natural” experiments (46, 50) have implicated contaminated poultry meat as a major cause of clinical infection. These findings have been strongly supported by source attribution models that have employed MLST data to estimate the contribution made by different potential infection sources to the burden of human disease (41, 42). While emphasizing the importance of contaminated poultry in this regard, these analyses also support a role for bovine, ovine, and other sources (7, 41, 42).
Reports from various temperate countries have shown that the incidence of human campylobacteriosis is at its highest consistently during the same week each year, although this time point shows variation among countries (26, 28, 38). This pronounced seasonality may be coincident with increased Campylobacter prevalence in reservoirs for infection and seasonal changes in human behavior that affect exposure (38). Variation in the prevalence of Campylobacter in various reservoirs has been found in some studies to correspond to the seasonal pattern of human campylobacteriosis (44), but this finding has not been universal (18). Seasonal variation in the incidence of human infection caused by certain clonal complexes has been described, notably increased incidences of sequence type 45 (ST-45) and ST-283 complex during the early summer (29, 43).
As human Campylobacter infection is generally self-limiting, antimicrobial therapy is not routinely recommended in the United Kingdom, but where symptoms are severe or prolonged, ciprofloxacin and clarithromycin are the treatments of choice (1). In Oxfordshire, United Kingdom, the proportion of clinical ciprofloxacin-resistant Campylobacter isolates increased from 3% in 1991 to 37.5% in 2008 (5). Resistance levels of 30 to 50% have been reported in Cambodia (2006) (30), 23.0% in the United States (2009) (49), and 71.4% in rural India (2002) (20), and more recently the level was 97.9% in China (17). An association between poultry consumption and acquisition of ciprofloxacin-resistant Campylobacter has been described, and some studies have detected possible associations between clonal complex and antibiotic resistance (15, 24). In Australia, where these drugs have not been approved for use in food-producing animals, levels of fluoroquinolone resistance remain low (48).
Here, MLST was used (8, 10) to study genotypes causing Campylobacter infection in a defined human population (approximately 640,000 individuals in 2009, corresponding to approximately 1% of the total United Kingdom population) living in the catchment area of the Clinical Microbiology Laboratory, John Radcliffe Hospital, Oxfordshire, United Kingdom. These data permitted the investigation of clonal complex seasonality and ciprofloxacin resistance. Additionally, the data (available at http://pubmlst.org/perl/bigsdb/bigsdb.pl?db=pubmlst_campylobacter_isolates) provided a baseline to inform on the impact of proposed interventions intended to reduce Campylobacter infection rates in England and Wales and provided a comparator for other human and veterinary Campylobacter epidemiological studies.
MATERIALS AND METHODS
Isolates.
The Campylobacter isolates included in this study were cultured from human stool samples submitted to the Clinical Microbiology Laboratory, Oxford University Hospitals NHS Trust, Oxford, United Kingdom, between 15 September 2003 and 14 September 2009. The specimens were from both hospital and community subjects. The size of the population served is approximately 640,000, representing around 1% of the United Kingdom population. Fecal samples were emulsified in cysteine selenite broth, plated onto Campylobacter blood-free selective agar (E & O Laboratories Limited, Bonnybridge, United Kingdom), and incubated at 42°C for 48 h under microaerophilic conditions. Bacterial colonies identified as oxidase-positive, catalase-producing, curved Gram-negative rods, with typical colonial morphology when subcultured to blood agar, were reported as Campylobacter species.
For long-term storage, isolates were emulsified in Luria broth containing 5% glycerol and stored at −80°C. The susceptibilities of the Campylobacter isolates to ciprofloxacin (1 μg) and erythromycin (5 μg) were determined using a modified Stokes disk diffusion method (Becton, Dickinson, Oxford, United Kingdom) (2). For isolates with reduced zones, the MIC was determined by Etest (bioMérieux Clinical Diagnostics). The method was subject to internal and external quality assurance (National External Quality Assessment Service [NEQAS]) with satisfactory performance. The isolation date was recorded for each isolate, together with corresponding anonymized details of patient age, sex, and recent travel outside the United Kingdom.
Multilocus sequence typing.
Bacterial DNA samples were produced from cultured Campylobacter cells suspended in molecular biology-grade water (125 ml; Sigma-Aldrich Company Ltd., Dorset, United Kingdom). The suspension was vortex mixed briefly and heated at 100°C for 10 min, debris was removed by centrifugation at 13,000 × g for 10 min, and the supernatant, containing chromosomal DNA, was removed and stored at −20°C. When further DNA template was required, isolates were recultured on Columbia blood agar plates (Oxoid Ltd., Basingstoke, United Kingdom) and incubated at 42°C for 48 h under microaerophilic conditions.
Multilocus sequence typing was performed as described previously (8, 10, 32). Briefly, PCR amplification of the seven housekeeping loci was performed using either oligonucleotide primers designed to amplify the relevant loci from both C. jejuni and C. coli isolates (32) or those described previously (8, 10). Amplification products were purified by precipitation with 20% polyethylene glycol-2.5 M NaCl (11), and nucleotide sequencing was performed at least once on each DNA strand using the PCR primers and BigDye ready reaction mix (version 3; Applied Biosystems, Foster City, CA). Previously described and newly identified alleles, sequence types (ST), and clonal complexes were assigned by querying the MLST database located at http://pubmlst.org/campylobacter/ (21), which held details of 88,859 sequences, 5,705 MLST profiles, and 13,069 isolates as of 9 January 2012. Potential central genotypes of emergent or previously undescribed clonal complexes were identified by their frequency within the data set and the pubMLST database. Candidate central genotype allelic profiles were then tested for the presence of single-, double-, and triple-locus variants in the pubMLST database, and clonal complex members sharing four or more identical alleles with the putative central genotype were identified with the BURST algorithm, available at the website detailed above.
Statistical analyses.
Frequency distributions of continuous or categorical variables within populations were compared by either the t test, Pearson's chi-square statistic, or, where sample sizes were small, Fisher's exact test. These and logistic regression analyses of secular and seasonal trends of clonal complexes were carried out using StataIC 10 (StataCorpLP, Texas). Genetic differentiation between populations was assessed using Fst pairwise comparisons of concatenated sequences for the seven MLST alleles with ARLEQUIN software version 3 (12). The diversity of STs within and between clonal complexes among populations was assessed using a modified version of Simpson's index of diversity (19), based on the probability of two unrelated strains being of the same type.
RESULTS
Isolates.
A total of 4,291 Campylobacter isolates, each representing a unique infection, were cultured between September 2003 and September 2009. Isolates were genotyped by MLST, and the ST was determined for 3,300 (76.9%) (Table 1). The STs were used to identify isolates to the species level (41): 91.6% were C. jejuni and 8.4% C. coli. The remaining 991 isolates were partially typed or untyped due to loss of viability during storage. Comparison of patient demographic data (sex, age, and month of isolation) and isolate antibiotic resistance data (ciprofloxacin and erythromycin) for the total isolates (4,291) versus isolates with complete allelic profiles (3,300) confirmed that the genotyped cases were representative of the collection. Subsequent analyses include only the 3,300 cases for which the ST of the Campylobacter isolates was determined.
Table 1.
Summary of isolates genotyped over the 6-year study period
| Yr of study | No. (%) of isolates |
||
|---|---|---|---|
| Unique | Assigned to sequence type | Assigned to clonal complex | |
| September 2003–September 2004 | 584 | 575 (98.5) | 530 |
| September 2004–September 2005 | 637 | 531 (83.4) | 495 |
| September 2005–September 2006 | 680 | 537 (79.0) | 496 |
| September 2006–September 2007 | 841 | 613 (72.9) | 560 |
| September 2007–September 2008 | 778 | 468 (60.2) | 408 |
| September 2008–September 2009 | 771 | 576 (74.7) | 503 |
Age, sex, and seasonal distribution of cases.
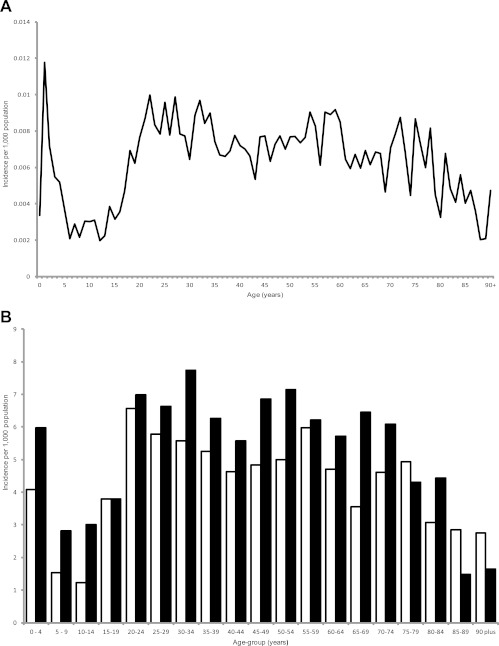
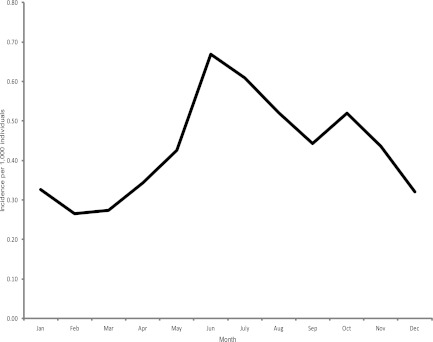
The age distribution of the 3,300 cases stratified by year of age was examined. The highest rates of illness occurred in children under two and young adults (Fig. 1A). The sex distribution of cases within the John Radcliffe Hospital Microbiology Laboratory catchment area was examined by 5-year age group. The proportion of illness in males was greater in all groups except those aged 75 to 79 years and individuals over 85 years of age, where numbers of cases in females were higher, and those aged 15 to 19, where no sex bias was observed (Fig. 1B). The seasonal distribution of cases showed a marked summer peak (Fig. 2).
Fig 1.
Age distribution of Campylobacter cases in the John Radcliffe Hospital catchment area, Oxfordshire, United Kingdom, between mid-September 2003 and mid-September 2009. (A) Incidence of infection per 1,000 individuals; (B) incidence among females (□) and males (■) per 1,000 age-related individuals.
Fig 2.
Seasonality of human Campylobacter genotyped isolates from the John Radcliffe Hospital catchment area, Oxfordshire, United Kingdom, between 2003 and 2009.
MLST and clonal complexes.
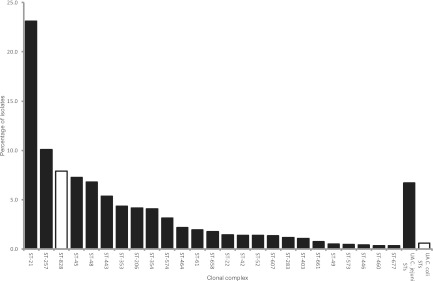
The 3,300 isolates comprised 368 C. jejuni and 70 C. coli STs, each of which was assigned to a clonal complex using http://pubmlst.org/campylobacter. A total of 157 (26.8%) STs could not be assigned to a previously defined clonal complex; 137 were C. jejuni, and 20 were C. coli. These unassigned STs were investigated for the presence of undescribed central genotypes, leading to the identification of the C. jejuni ST-464 complex, containing eight STs and 71 isolates. In total, 35 C. jejuni clonal complexes and 1 C. coli clonal complex (ST-828 complex) were present within the data set, with 149 (25.4%) STs remaining unassigned. Six clonal complexes (ST-21, ST-257, ST-828, ST-45, ST-48, and ST-443 complexes) represented 59.8% of all isolates, ranging from 22.8% to 5.3%, respectively (Fig. 3). In total, C. coli belonging to the ST-828 complex or unassigned to an ST comprised 8.4% of isolates.
Fig 3.
Relative proportion of clonal complexes represented more than 10 times in the data set in decreasing order of abundance. UA indicates sequence types that are not assigned to a clonal complex. Isolates belonging to C. coli, indicated by white bars, either were assigned to the ST-828 complex or belonged to sequence types unassigned to a clonal complex.
Temporal trends in clonal complexes.
The temporal distribution of the 20 most abundant clonal complexes was analyzed using logistic regression analysis, which identified a year-on-year increase in the number of isolates assigned to ST-42, ST-45, ST-464, and ST-52 clonal complexes, whereas isolates belonging to ST-574, ST-607, and ST-658 complexes decreased in incidence over the study period. The overall trends were summarized as odds ratios representing the average increase or decrease in probability of an isolate belonging to that clonal complex per year of the study (Table 2). Although these seven clonal complexes do show an overall linear trend across the 6-year period, none are monotonic and there are substantial year-to-year variations, which in some cases (ST-42, ST-45, and ST-607 complexes) equal or exceed the overall change from year 1 to year 6.
Table 2.
Overall trends and odds ratiosa
| Clonal complex | % of isolates in complex |
Odds ratiob | P value | CIc | |||||
|---|---|---|---|---|---|---|---|---|---|
| Yr 1 | Yr 2 | Yr 3 | Yr 4 | Yr 5 | Yr 6 | ||||
| ST-42 | 1.6 | 0.6 | 0.6 | 1.3 | 1.7 | 2.4 | 1.227 | 0.02 | 1.033–1.459 |
| ST-45 | 6.8 | 6.6 | 8.5 | 4.9 | 7.9 | 8.7 | 1.087 | 0.03 | 1.008–1.172 |
| ST-52 | 0.5 | 0.8 | 1.7 | 1.8 | 1.3 | 2.1 | 1.218 | 0.025 | 1.025–1.447 |
| ST-464 | 0.9 | 0.4 | 1.7 | 1.3 | 3.2 | 5.6 | 1.612 | <0.001 | 1.367–1.877 |
| ST-574 | 5.2 | 3.2 | 2.4 | 3.3 | 1.7 | 2.4 | 0.846 | 0.004 | 0.754–0.948 |
| ST-607 | 1.4 | 2.7 | 1.7 | 0.7 | 0.4 | 1.0 | 0.818 | 0.025 | 0.686–0.976 |
| ST-658 | 2.6 | 2.3 | 1.7 | 1.8 | 0.9 | 1.2 | 0.824 | 0.012 | 0.708–0.959 |
Overall trends and odds ratios representing the average increase or decrease in probability of an isolate belonging to that clonal complex per year of the study.
Changing odds per year.
95% confidence interval.
Seasonality of clonal complexes.
The data were examined for seasonal variation in the clonal complexes that occurred during the summer peak in incidence (June to September), compared to those that occurred during the rest of the year, using Pearson's chi-square test. The composition of clonal complexes that occurred between June and September was significantly different from that which occurred during the rest of the year (P value with the chi-square test [χ2 P] < 0.001).
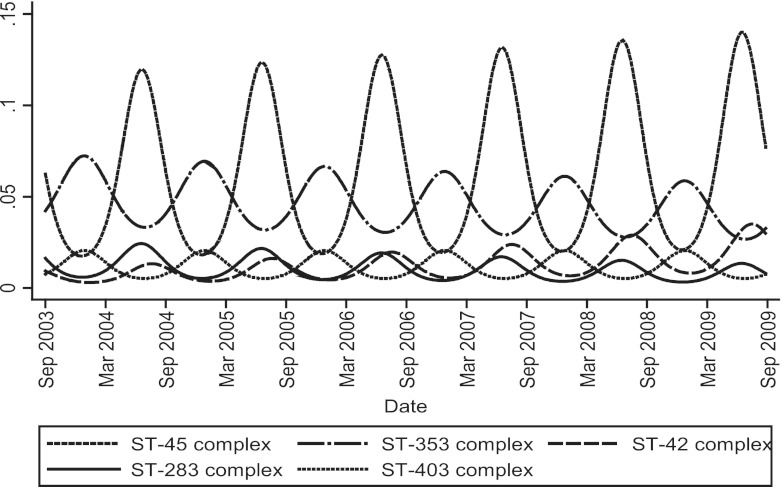
A logistic regression model was then used to identify significant seasonal trends for individual clonal complexes. Isolates belonging to ST-283, ST-42, and ST-45 complexes peaked as a proportion of the total during June and July (P = 0.013, P = 0.018, and P < 0.001, respectively) (Fig. 4). ST-353 and ST-403 complexes (P < 0.005 and P = 0.021, respectively) were proportionally most prevalent during November. Variation that deviated from overall harmonic seasonal patterns was detected for the ST-257 complex (χ2 P = 0.008), which was lower than expected in July and November, and for the ST-354 complex (χ2 P < 0.001), which peaked in November (data not shown).
Fig 4.
Seasonal and longer-term trends observed using a harmonic regression model of the data for clonal complexes whose frequency as a proportion of the total incidence were significantly higher during summer (ST-45, ST-283, and ST-42 complexes) or winter months (ST-353 and ST-403 complexes).
Antibiotic resistance and clonal complex.
Following primary culture, 3,262 isolates (98.9%) were examined by disk diffusion for sensitivity to ciprofloxacin (1 μg) and 3,265 (98.9%) isolates for sensitivity to erythromycin (5 μg). The proportion (percentage) of all ciprofloxacin-sensitive isolates (2,243; 69.8%) and all resistant isolates (972; 30.2%) associated with each of the 25 clonal complexes represented 10 times or more in the data set was determined. Isolates unassigned to a clonal complex were considered as a separate group for each species. Equivalent calculations were made for erythromycin-sensitive (3,167; 98.2%) and -resistant (57; 1.8%) isolates.
Nine clonal complexes (ST-22, ST-45, ST-48, ST-61, ST-257, ST-283, ST-403, ST-658, and ST-677) were significantly associated with ciprofloxacin sensitivity (P < 0.05) (Table 3), and seven clonal complexes (ST-49, ST-206, ST-354, ST-446, and ST-460, along with the newly described ST-464 and ST-607 complexes) were significantly associated with ciprofloxacin resistance, as were unassigned C. jejuni STs. One clonal complex (ST-257) was significantly associated with erythromycin-sensitive isolates, whereas the only significant association between either of the antibiotics and C. coli isolates was that between both ST-828 complex and unassigned C. coli STs with erythromycin resistance.
Table 3.
Clonal complexes represented by 10 or more isolates, demonstrating a nonrandom association with antibiotic sensitivity or resistance as determined by Pearson's χ2 or Fisher's exact testc
| Clonal complex | Ciprofloxacin |
Erythromycin |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sensitive |
Resistant |
P value | Sensitive |
Resistant |
P value | |||||
| No. of isolates | % | No. of isolates | % | No. of isolates | % | No. of isolates | % | |||
| ST-21 complex | 531 | 71.5 | 212 | 28.5 | 0.250 | 731 | 98.4 | 12 | 1.6 | 0.718 |
| ST-22 complex | 39 | 84.8 | 7 | 15.2 | 0.026 | 46 | 100 | 0 | 0 | 1a |
| ST-42 complex | 35 | 77.8 | 10 | 22.2 | 0.274 | 45 | 100 | 0 | 0 | 1a |
| ST-45 complex | 213 | 90.6 | 22 | 9.4 | <0.001 | 229 | 97.5 | 6 | 2.6 | 0.366 |
| ST-48 complex | 182 | 82.7 | 38 | 17.3 | <0.001 | 215 | 97.7 | 5 | 2.3 | 0.556 |
| ST-49 complex | 8 | 47.1 | 9 | 52.9 | 0.041 | 17 | 100 | 0 | 0 | 1a |
| ST-52 complex | 29 | 64.4 | 16 | 35.6 | 0.434 | 45 | 100 | 0 | 0 | 1a |
| ST-61 complex | 57 | 93.4 | 4 | 6.6 | <0.001a | 62 | 100 | 0 | 0 | 0.628a |
| ST-206 complex | 80 | 59.7 | 54 | 40.3 | 0.010 | 133 | 99.3 | 1 | 0.8 | 0.731a |
| ST-257 complex | 275 | 84.9 | 49 | 15.1 | <0.001 | 323 | 99.7 | 1 | 0.3 | 0.041a |
| ST-283 complex | 34 | 91.9 | 3 | 8.1 | 0.002a | 37 | 100 | 0 | 0 | 1a |
| ST-353 complex | 94 | 67.6 | 45 | 32.4 | 0.574 | 138 | 99.3 | 1 | 0.7 | 0.516a |
| ST-354 complex | 28 | 21.4 | 103 | 78.6 | <0.001 | 131 | 100 | 0 | 0 | 0.172a |
| ST-403 complex | 31 | 91.2 | 3 | 8.8 | 0.004a | 34 | 100 | 0 | 0 | 1a |
| ST-443 complex | 122 | 70.1 | 52 | 29.9 | 0.918 | 170 | 97.1 | 5 | 2.9 | 0.261 |
| ST-446 complex | 3 | 21.4 | 11 | 78.6 | <0.001a | 13 | 92.9 | 1 | 7.1 | 0.221a |
| ST-460 complex | 4 | 36.4 | 7 | 63.6 | 0.023a | 11 | 100 | 0 | 0 | 1a |
| ST-464 complex | 14 | 20.6 | 54 | 79.4 | <0.001 | 67 | 98.5 | 1 | 1.5 | 1a |
| ST-573 complex | 13 | 86.7 | 2 | 13.3 | 0.257a | 15 | 100 | 0 | 0 | 1a |
| ST-574 complex | 63 | 61.8 | 39 | 38.2 | 0.074 | 102 | 100 | 0 | 0 | 0.262a |
| ST-607 complex | 18 | 42.9 | 24 | 57.1 | <0.001 | 42 | 97.7 | 1 | 2.3 | 0.538a |
| ST-658 complex | 48 | 82.8 | 10 | 17.2 | 0.030 | 58 | 100 | 0 | 0 | 0.626a |
| ST-661 complex | 19 | 82.7 | 4 | 17.4 | 0.254a | 23 | 100 | 0 | 0 | 1a |
| ST-677 complex | 11 | 100 | 0 | 0.0 | 0.041a | 11 | 100 | 0 | 0 | 1a |
| ST-828 complexb | 170 | 65.9 | 88 | 34.1 | 0.158 | 245 | 95.0 | 13 | 5.0 | <0.001 |
| UA C. jejuni STs | 112 | 53.5 | 97 | 46.4 | <0.001 | 208 | 96.7 | 7 | 3.3 | 0.087 |
| UA C. coli STsb | 10 | 52.6 | 9 | 47.4 | 0.103 | 16 | 84.2 | 3 | 15.8 | 0.004a |
| Total | 2,243 | 972 | 3,167 | 57 | ||||||
Values calculated using Fisher's exact test (values without this superscript were calculated using Pearson's chi-square test).
C. coli isolates.
Underlined values have statistically significant P values. Data were calculated with Pearson's chi-square test unless noted otherwise. UA, sequence types unassigned to a clonal complex.
Patients reporting recent travel outside the United Kingdom (n = 381) were five times more likely to have a ciprofloxacin-resistant Campylobacter than those who had not (χ2 P < 0.001). No difference in the proportion of erythromycin-resistant isolates was detected between travelers and nontravelers.
Patient demographics and genotype.
The MLST and clonal complex data for each of the 3,300 isolates were examined to detect associations among infecting genotype and patient age, gender, and recent travel outside the United Kingdom. Seven 10-year age groups and an eighth, comprising individuals aged 70 or over, were used to segregate the STs. Simpson's index of diversity was calculated for each group (see Fig. S1 in the supplemental material). This revealed no difference in the genotypic diversity of isolates infecting each group (diversity index [DI], 0.973 to 0.980). Similarly, differences in nucleotide content as indexed by FST values calculated from concatenated nucleotide sequences of the MLST loci indicated that the maximum level of sequence differentiation between the STs of each 10-year age group was <1%.
Clonal complexes affecting males and females were similar (χ2 P = 0.338), as were the levels of nucleotide sequence diversity for isolates infecting both genders (male DI, 0.975 to 0.979; female DI, 0.973 to 0.978). Recent travel outside the United Kingdom was reported by 11.5% of individuals, isolates from whom belonged to clonal complexes that were significantly different (P < 0.001) from those from subjects who had not recently been abroad.
DISCUSSION
Sequence-based characterization, and especially MLST, of Campylobacter isolates has become the principal tool for understanding the molecular epidemiology of this important pathogen (10). Such studies have established that although genetically diverse, Campylobacter populations are highly structured and that the clonal complex is an informative unit of analysis (9). A key insight has been that particular clonal complexes are associated with the colonization of certain host species, which has permitted the genetic attribution of cases of human disease to likely infection sources and in so doing has principally implicated retail chicken meat products in a variety of settings (34, 42, 52). Longitudinal surveillance of the genotypes of human clinical isolates therefore provides a means of monitoring changes in infection sources or the appearance of new disease-associated Campylobacter genotypes. The present 6-year study examined temporal and seasonal trends in the nature and properties of the Campylobacter clonal complexes responsible for human disease in a defined, centrally located United Kingdom region (Oxfordshire) which represents approximately 1% of United Kingdom residents.
Patterns of age-related illness and gender bias within the present study were largely as reported for England and Wales (16). In common with similar studies of human infection (31, 33, 41, 43), the Campylobacter genotypes responsible for human disease were highly diverse, with a predominance of genotypes similar to those seen previously. However, investigation of isolates having an ST unassigned to a clonal complex identified 62 isolates of ST-464. This ST was first submitted to the pubMLST Campylobacter isolate database from a human isolate cultured in Japan in 2001. A further 29 ST-464 isolates and 54 isolates representing 47 single-, double-, and triple-locus variants of this ST have since been submitted to the database from Canada, China, Europe, Japan, and Thailand (January 2012). Furthermore, ST-464 was isolated from chicken breast meat in the United States in 2002 (51) and ST-464 together with single-, double-, and triple-locus variants from either chicken or human stools in Northern China between 2003 and 2008 (53). On the basis of the above data, ST-464 was assigned as the central genotype of a novel clonal complex. Furthermore, these findings illustrate that the global diversity of Campylobacter isolates responsible for human disease remains incompletely understood.
There was evidence for temporal changes in the clonal complexes causing human disease over the 6-year study period: four clonal complexes (ST-42, ST-45, ST-52, and ST-464) increased in relative incidence, whereas three complexes (ST-574, ST-607, and ST-658) decreased (Table 2). The ST-42 complex has been reported to be mainly isolated from ruminants, ST-45 complex from chicken, wild bird, and environmental sources (6, 13, 41, 44), and ST-464 complex predominantly from chickens (21, 51, 53), while the ST-52 complex has a less well-defined host range. Both ST-574 and ST-607 complex isolates are largely chicken associated (21, 41). Therefore, changes in the relative incidence of different clonal complexes did not provide any evidence for changing importance of different reservoirs of human infection. The annual periodicity observed was typical of temperate regions, with a marked summer peak in human disease incidence between June and September (Fig. 3) (26, 38). The most prevalent clonal complexes overall were ST-21, ST-257, ST-828, ST-45, ST-48, and ST-443 (Fig. 3), which were also among the most frequently isolated, and of comparable incidence with those in a similar-sized study of human campylobacteriosis in Scotland from 2005 until 2006 (41). These data therefore established that detailed surveillance at a single sentinel surveillance site captures information that reflects that observed nationally.
Evidence of seasonality contributing significantly to the summer peak in disease incidence was observed for three clonal complexes; ST-45 (June), ST-283 (July), and ST-42 (July). This pattern has been reported previously for ST-45 (22, 29, 44) and ST-283 complexes (29) but not for the ST-42 complex. The ST-45 and ST-283 complexes have both been identified as chicken associated (7, 21, 41), and the ST-45 complex has additionally been identified from a wide range of wild animal sources (27), but the ST-42 complex has been predominantly associated with ruminant hosts (6, 13, 21). While MLST data for isolates belonging to ST-45 and ST-283 complexes confirms a recent common ancestor (29), no similarity in genotype or host association was observed between these two complexes and isolates belonging to the ST-42 complex. The present study identified two clonal complexes (ST-353 and ST-403) that peak in relative incidence during the winter (November). The former is predominantly isolated from chickens, as well as cases of human infection, but the latter has a much wider host range which largely excludes poultry (21, 41). The basis for their shared seasonal pattern is therefore unclear and does not appear to be associated with a shared host. Seasonal differences are likely to reflect the Campylobacter genotypes that are present in the food chain or, possibly, exposure of the human population to different infection sources.
Rapidly increasing fluoroquinolone resistance in Campylobacter strains causing human disease is a matter of public health importance as has been noted previously in the Oxfordshire data set (5). The mean values for ciprofloxacin (30.1%) and erythromycin (1.7%) resistance phenotypes over the 6-year period were similar to those recently reported (5, 25), although even higher levels of fluoroquinolone resistance have been observed (17, 20, 30). Similarly, higher rates of ciprofloxacin-resistant isolates among patients reporting recent travel outside the United Kingdom confirm a previous study (37). The poultry production industry is often seen as the major source of antimicrobial resistance in Campylobacter (36).There was a statistically significant association between ciprofloxacin resistance and seven C. jejuni clonal complexes; ST-49, ST-206, ST-354, ST-446, ST-460, ST-464, and ST-607 (Table 3). Previous reports have identified an association between ST-21, ST-206, ST-353, and ST-354 complexes and ciprofloxacin resistance (15, 24, 25, 51), although each of these investigations included fewer than 200 geographically diverse isolates. Furthermore, variation occurred between studies in the methodology and breakpoints used to define ciprofloxacin-resistant strains. Eight of the clonal complexes associated with ciprofloxacin resistance have not been associated with a single reservoir of infection, two (ST-354 and ST-464) are chicken associated (41, 51, 53), and one (ST-206) is predominantly isolated from ruminants (41). Of nine clonal complexes associated with ciprofloxacin sensitivity, two (ST-257 and ST-283) are chicken associated (40) and one (ST-45) has been isolated from a variety of hosts, including poultry (41, 44).
These results suggest a complex acquisition pathway and raise the question whether the food industry, or one subset of it such as poultry production, is a dominating influence in the distribution of ciprofloxacin resistance among campylobacters. Levels of resistance to erythromycin remained static and low (<2%) throughout the study, and the only associations observed were between erythromycin resistance and C. coli isolates and between ST-257 complex and erythromycin sensitivity, the former in agreement with reports from others (51). However, unlike fluoroquinolones, the fitness cost to low-level erythromycin-resistant isolates results in a lack of stability in the absence of selective pressure (3, 23), which may in part explain the difference in levels of resistance to these antibiotics in clinical isolates.
The continuing high burden of human campylobacteriosis remains an unresolved public health problem of high importance. Genetic attribution studies have identified the principal source of infection as retail meat, especially chicken, in a number of countries, including the United Kingdom. Molecular data have led to interventions in the poultry industry in New Zealand which have proved to be effective in reducing human disease (40); however, New Zealand poultry production is limited to three major suppliers (33) and the extent to which this will be possible in countries with more complex industries remains unclear. In any case, ongoing surveillance of the genotypes causing human disease is essential if the impact of such interventions is to be measured and understood, particularly as the predominant genotypes in human disease show pronounced changes over time and new genotypes are frequently observed, reflecting the size and diversity of the Campylobacter populations that infect chickens. The present study demonstrates that such surveillance is feasible. In a country such as the United Kingdom where nationally distributed foods are the major source of infection, this can be achieved by intensive surveillance at relatively few sites.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the United Kingdom Department for Environment, Food and Rural Affairs (DEFRA) (grant numbers OZ0611 and OZ0615) and made use of the Campylobacter MLST website (http://pubmlst.org/campylobacter/) (21) developed by Keith Jolley, sited at the University of Oxford, and funded by The Wellcome Trust (grant number 087622/Z/08/Z). K.E.D. was supported by the NIHR Biomedical Centre, Oxford, United Kingdom. M.C.J.M. is a Wellcome Trust Senior Research Fellow.
We extend our thanks to the staff of the Clinical Microbiology Laboratory, John Radcliffe Hospital, Oxford, United Kingdom, for their assistance in collecting Campylobacter isolates.
Footnotes
Published ahead of print 18 July 2012
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. British National Formulary March 2012. British National Formulary, online edition. http://www.medicinescomplete.com/mc/bnf/current/102035.htm
- 2. Brown DF, Kothari D. 1978. Antimicrobial-susceptibility testing of rapidly growing pathogenic bacteria. II. A field trial of four disc-diffusion methods. J. Antimicrob. Chemother. 4:27–38 [DOI] [PubMed] [Google Scholar]
- 3. Caldwell DB, Wang Y, Lin J. 2008. Development, stability, and molecular mechanisms of macrolide resistance in Campylobacter jejuni. Antimicrob. Agents Chemother. 52:3947–3954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. CDC 2010. Campylobacter: general information. http://www.cdc.gov/nczved/divisions/dfbmd/diseases/campylobacter/
- 5. Cody AJ, Clarke L, Bowler IC, Dingle KE. 2010. Ciprofloxacin-resistant campylobacteriosis in the UK. Lancet 376:1987. [DOI] [PubMed] [Google Scholar]
- 6. Colles FM, Jones K, Harding RM, Maiden MC. 2003. Genetic diversity of Campylobacter jejuni isolates from farm animals and the farm environment. Appl. Environ. Microbiol. 69:7409–7413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de Haan CP, Kivisto RI, Hakkinen M, Corander J, Hanninen ML. 2010. Multilocus sequence types of Finnish bovine Campylobacter jejuni isolates and their attribution to human infections. BMC Microbiol. 10:200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dingle KE, Colles FM, Falush D, Maiden MC. 2005. Sequence typing and comparison of population biology of Campylobacter coli and Campylobacter jejuni. J. Clin. Microbiol. 43:340–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dingle KE, et al. 2002. Molecular characterization of Campylobacter jejuni clones: a basis for epidemiologic investigation. Emerg. Infect. Dis. 8:949–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dingle KE, et al. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Embley TM. 1991. The linear PCR reaction: a simple and robust method for sequencing amplified rRNA genes. Lett. Appl. Microbiol. 13:171–174 [DOI] [PubMed] [Google Scholar]
- 12. Excoffier L, Laval G, Schneider S. 2007. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. Online 1:47–50 [PMC free article] [PubMed] [Google Scholar]
- 13. French NP, et al. 2009. Molecular epidemiology of Campylobacter jejuni isolates from wild-bird fecal material in children's playgrounds. Appl. Environ. Microbiol. 75:779–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gillespie IA, et al. 2006. Investigating vomiting and/or bloody diarrhoea in Campylobacter jejuni infection. J. Med. Microbiol. 55:741–746 [DOI] [PubMed] [Google Scholar]
- 15. Habib I, Miller WG, Uyttendaele M, Houf K, De Zutter L. 2009. Clonal population structure and antimicrobial resistance of Campylobacter jejuni in chicken meat from Belgium. Appl. Environ. Microbiol. 75:4264–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Health Protection Agency 2009. Campylobacter infections by age and sex, England and Wales, 2000–2010 http://www.hpa.org.uk/Topics/InfectiousDiseases/InfectionsAZ/Campylobacter/EpidemiologicalData/campyDataEWsex19892009/
- 17. Hou FQ, Sun XT, Wang GQ. 2012. Clinical manifestations of Campylobacter jejuni infection in adolescents and adults, and change in antibiotic resistance of the pathogen over the past 16 years. Scand. J. Infect. Dis. 44:439–443 [DOI] [PubMed] [Google Scholar]
- 18. Humphrey TJ, Henley A, Lanning DG. 1993. The colonization of broiler chickens with Campylobacter jejuni: some epidemiological investigations. Epidemiol. Infect. 110:601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hunter PR, Gaston MA. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jain D, Sinha S, Prasad KN, Pandey CM. 2005. Campylobacter species and drug resistance in a north Indian rural community. Trans. R. Soc. Trop. Med. Hyg. 99:207–214 [DOI] [PubMed] [Google Scholar]
- 21. Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. Bioinformatics 11:595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jorgensen F, et al. 2011. Influence of season and geography on Campylobacter jejuni and C. coli subtypes in housed broiler flocks reared in Great Britain. Appl. Environ. Microbiol. 77:3741–3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim JS, Carver DK, Kathariou S. 2006. Natural transformation-mediated transfer of erythromycin resistance in Campylobacter coli strains from turkeys and swine. Appl. Environ. Microbiol. 72:1316–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kinana AD, et al. 2006. Genetic diversity and quinolone resistance in Campylobacter jejuni isolates from poultry in Senegal. Appl. Environ. Microbiol. 72:3309–3313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kittl S, Kuhnert P, Hachler H, Korczak BM. 2011. Comparison of genotypes and antibiotic resistance of Campylobacter jejuni isolated from humans and slaughtered chickens in Switzerland. J. Appl. Microbiol. 110:513–520 [DOI] [PubMed] [Google Scholar]
- 26. Kovats RS, et al. 2005. Climate variability and Campylobacter infection: an international study. Int. J. Biometeorol. 49:207–214 [DOI] [PubMed] [Google Scholar]
- 27. Kwan PS, et al. 2008. Molecular epidemiology of Campylobacter jejuni populations in dairy cattle, wildlife, and the environment in a farmland area. Appl. Environ. Microbiol. 74:5130–5138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Louis VR, et al. 2005. Temperature-driven Campylobacter seasonality in England and Wales. Appl. Environ. Microbiol. 71:85–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McCarthy ND, et al. 2012. Molecular epidemiology of human Campylobacter jejuni shows association between seasonal and international patterns of disease. Epidemiol. Infect. 28:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Meng CY, et al. 2011. Etiology of diarrhea in young children and patterns of antibiotic resistance in Cambodia. Pediatr. Infect. Dis. J. 30:331–335 [DOI] [PubMed] [Google Scholar]
- 31. Mickan L, et al. 2007. Multilocus sequence typing of Campylobacter jejuni isolates from New South Wales, Australia. J. Appl. Microbiol. 102:144–152 [DOI] [PubMed] [Google Scholar]
- 32. Miller WG, et al. 2005. Extended multilocus sequence typing system for Campylobacter coli, C. lari, C. upsaliensis, and C. helveticus. J. Clin. Microbiol. 43:2315–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mullner P, et al. 2010. Molecular epidemiology of Campylobacter jejuni in a geographically isolated country with a uniquely structured poultry industry. Appl. Environ. Microbiol. 76:2145–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mullner P, et al. 2009. Assigning the source of human campylobacteriosis in New Zealand: a comparative genetic and epidemiological approach. Infect. Genet. Evol. 9:1311–1319 [DOI] [PubMed] [Google Scholar]
- 35. Neimann J, Engberg J, Molbak K, Wegener HC. 2003. A case-control study of risk factors for sporadic Campylobacter infections in Denmark. Epidemiol. Infect. 130:353–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nelson JM, Chiller TM, Powers JH, Angulo FJ. 2007. Fluoroquinolone-resistant Campylobacter species and the withdrawal of fluoroquinolones from use in poultry: a public health success story. Clin. Infect. Dis. 44:977–980 [DOI] [PubMed] [Google Scholar]
- 37. Niederer L, et al. 2012. Genotypes and antibiotic resistances of Campylobacter jejuni and Campylobacter coli isolates from domestic and travel-associated human cases. Appl. Environ. Microbiol. 78:288–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nylen G, et al. 2002. The seasonal distribution of Campylobacter infection in nine European countries and New Zealand. Epidemiol. Infect. 128:383–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pebody RG, Ryan MJ, Wall PG. 1997. Outbreaks of Campylobacter infection: rare events for a common pathogen. Commun. Dis. Rep. CDR Rev. 7:R33–R37 [PubMed] [Google Scholar]
- 40. Sears A, et al. 2011. Marked campylobacteriosis decline after interventions aimed at poultry, New Zealand. Emerg. Infect. Dis. 17:1007–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sheppard SK, et al. 2009. Campylobacter genotypes from food animals, environmental sources and clinical disease in Scotland 2005/6. Int. J. Food Microbiol. 134:96–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sheppard SK, et al. 2009. Campylobacter genotyping to determine the source of human infection. Clin. Infect. Dis. 48:1072–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sopwith W, et al. 2006. Campylobacter jejuni multilocus sequence types in humans, northwest England, 2003–2004. Emerg. Infect. Dis. 12:1500–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sopwith W, et al. 2008. Identification of potential environmentally adapted Campylobacter jejuni strain, United Kingdom. Emerg. Infect. Dis. 14:1769–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Reference deleted.
- 46. Stern NJ, et al. 2003. Campylobacter spp. in Icelandic poultry operations and human disease. Epidemiol. Infect. 130:23–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tam CC, et al. 2012. Longitudinal study of infectious intestinal disease in the UK (IID2 study): incidence in the community and presenting to general practice. Gut 61:69–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Unicomb LE, et al. 2006. Low-level fluoroquinolone resistance among Campylobacter jejuni isolates in Australia. Clin. Infect. Dis. 42:1368–1374 [DOI] [PubMed] [Google Scholar]
- 49. US Food and Drug Administration 2009. U.S. Food and Drug Administration Executive Report 2009. www.fda.gov/downloads/AnimalVeterinary/SafetyHealth/AntimicrobialResistance/NationalAntimicrobialResistanceMonitoringSystem/ucm269042.pdf
- 50. Vellinga A, Van Loock F. 2002. The dioxin crisis as experiment to determine poultry-related Campylobacter enteritis. Emerg. Infect. Dis. 8:19–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang X, et al. 2011. Antimicrobial resistance and molecular subtyping of Campylobacter jejuni and Campylobacter coli from retail meats. J. Food. Prot. 74:616–621 [DOI] [PubMed] [Google Scholar]
- 52. Wilson DJ, et al. 2008. Tracing the source of campylobacteriosis. PLoS Genet. 4:e1000203 doi:10.1371/journal.pgen.1000203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang M, et al. 2010. Molecular typing and antimicrobial susceptibility profiles of Campylobacter jejuni isolates from north China. J. Med. Microbiol. 59:1171–1177 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.