Abstract
Standardized one-step real-time RT-PCR assay detected enterovirus RNA in cardiac biopsy samples from 4 of 20 patients suffering from idiopathic dilated cardiomyopathy (IDCM). The median viral load was 287 copies per microgram of total extracted nucleic acids, with positive- to negative-strand RNA ratios ranging from 2 to 20. These results demonstrate enterovirus persistence in the heart of IDCM patients, characterized by low viral loads and low positive- to negative-RNA ratios.
TEXT
Enteroviruses (EV), especially group B coxsackieviruses (CVB), are considered a common cause of acute myocarditis in children and young adults, a disease which is a precursor to 10 to 20% of chronic myocarditis cases as well as dilated cardiomyopathy (prevalence = 7 cases/100,000), characterized by an increase in both myocardial mass and volume, compromising cardiac contractility and ultimately resulting in poor left ventricular function (8, 9, 12, 14). Dilated cardiomyopathy is considered to be the second leading cause of heart transplantation worldwide after ischemic heart disease. The concept that persistent endomyocardial EV infection could be the etiologic cause of a subset of idiopathic dilated cardiomyopathy (IDCM) cases is supported by the detection in up to 35% of explanted heart tissues from end-stage IDCM patients of EV RNA and the viral capsid protein VP1 in the absence of viruses that can be cultivated by classical techniques (2, 3, 13). However, the viral molecular mechanisms involved in the progression of acute myocarditis to chronic myocarditis and subsequently to IDCM are currently poorly understood. In 2008, Chapman et al. (7) reported for the first time the isolation from human heart tissue of a CVB2 strain with genomic 5′-terminal deletions (TD). These mutations induced significant slowing of viral replication and a lowering of virus titer in cell culture models where an absence of classical cytopathic effects was associated with an abnormal positive- to negative-strand viral RNA ratio close to 1 rather than the high positive- to negative-strand ratios normally seen in wild-type virus infected cells (11). These findings demonstrated the existence of unexpected EV-TD genomic populations in clinical samples, thereby a mechanism by which EV can persist in heart long after the acute infection cycle.
In this context, the aim of the present study was to validate a sensitive and standardized one-step real-time RT-PCR assay capable of detecting and quantifying EV RNA (both positive and negative strands) of wild-type as well as TD EV strains in cardiac tissue samples obtained from IDCM patients. Reverse transcription and PCR were carried out using a Superscript III Platinum one-step quantitative reverse transcription-PCR (qRT-PCR) kit (Invitrogen, Life Technologies, Saint-Aubin, France) containing 200 nM forward primer (5′-CCCTGAATGCGGCTAATCC-3′, positions 456 to 474), 200 nM reverse primer (5′-ATTGTCACCATAAGCAGCCA-3′, positions 582 to 601), and 100 nM probe (FAM-5′-AACCGACTACTTTGGGTGTCCGTGTTTC-3′-TAMRA, positions 539 to 566) (17). Reverse transcription was performed at 55°C for 45 min, the RT was heat inactivated at 95°C for 2 min, and then the cDNA was amplified in 45 cycles as follows: denaturation at 94°C for 15 s, annealing at 63°C for 1 min, and an extension step at 68°C for 30 s. To validate detection and quantitation of EV in cardiac biopsy specimens, serial dilutions of positive- and negative-strand transcripts of wild-type and TD CVB3 clones kindly provided by N. M. Chapman (University of Nebraska Medical Center, Omaha, NE) were tested. Both wild-type and TD positive- and negative-strand RNA molecules were detected in vitro by the one-step RT-PCR assay. The sensitivity and the reproducibility of the RT-PCR assay were then determined using serial 10-fold dilutions of the transcripts ranging from 3 × 106 to 30 copies diluted in DNA and RNA extracts of EV-negative cardiac tissues with similar results in terms of standard curves, regardless of the transcripts used for the experiments. The threshold of viral RNA detection was found repeatedly to be 30 copies per well for both wild-type and TD EV RNA transcripts for both positive- and negative-strand RNAs. This sensitivity of detection, is crucial because TD mutants replicate slowly and to low titers (7, 10, 11). This will be important for future work and may explain past inability to detect RNA despite viral capsid protein VP1 detection (1).
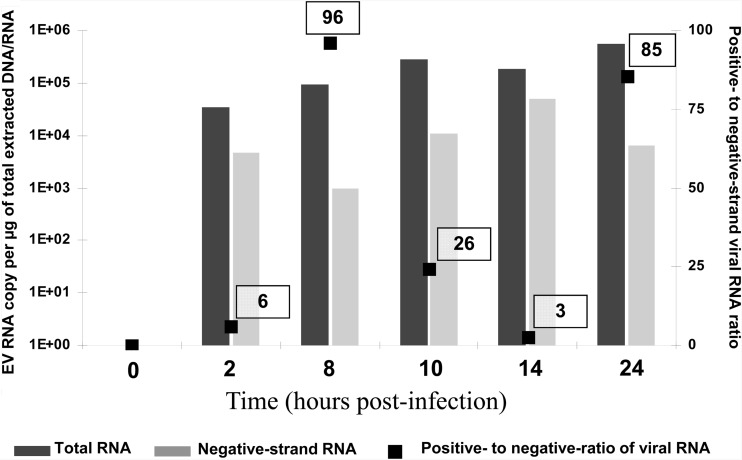
Being able to quantitate both positive- and negative-strand viral RNA titers in clinical samples would permit the measurement of the positive- to negative-strand viral RNA ratio to facilitate an assessment of EV persistence mechanisms potentially associated with the development of IDCM. To validate the specific negative-strand viral RNA isolation, a time course of CVB3 replication model was performed in MA-104 cells (rhesus monkey kidney cell line), which were infected at a multiplicity of infection of 1 50% tissue culture infective dose (TCID50) with the CVB3 prototype strain Nancy and collected at 0, 2, 8, 10, 14, and 24 h postinfection. Negative-strand RNA was isolated from total RNA molecules by annealing a biotinylated negative-strand specific primer (E3REV; 5′-GGAACCGACTACTTTGGGTGTCCGTG-3′) and binding to streptavidin-labeled magnetic beads (Invitrogen, Life Technologies, Saint-Aubin, France) (7, 11). Purified negative-strand and total viral RNA molecules were quantified with a one-step real-time RT-PCR assay using serial dilutions of the transcripts for the generation of the standard curves. The positive- to negative-strand viral RNA ratio was then determined using the following calculation: (total EV RNA − negative-strand EV RNA)/negative-strand EV RNA. Positive- to negative-strand viral RNA ratios assessed at 8 h and 24 h postinfection were 96 and 85, respectively, consistent with the high positive-to-negative ratios normally seen in wild-type-virus-infected-cell models (Fig. 1).
Fig 1.
Levels of total viral RNA and negative-strand viral RNA and positive- to negative-strand viral RNA ratios during the first 24 h of CVB3 replication in MA-104 cells.
Ninety-nine paraffin-embedded cardiac biopsy specimens collected by the department of pathology of the European Hospital Georges Pompidou (Paris, France) were investigated. These samples were explanted heart tissue samples collected between 2000 and 2009 from a population of 20 adult patients demonstrating histological findings compatible with IDCM (4). As control samples, heart tissue was selected from 10 adult patients who had died accidentally or by suicide (CHU, Reims France). EV RNA was detected in 6 (9%) of the 99 samples analyzed, corresponding to 4 (20%) of the 20 IDCM patients, whereas no viral RNA was detected in the heart tissue samples taken from the 10 control subjects. These results confirm those published by our group and others that have reported EV RNA detection in up to 35% of cardiac explants in subjects with IDCM (2, 5, 9, 13). The median viral load assessed was 287 copies/μg of total extracted nucleic acids, with a range of 10 to 3,368 copies/μg (Table 1). These results are in agreement with those obtained from murine heart tissues chronically infected with CVB3 (3, 15). In contrast, this median viral load was 500 times lower than that obtained in our model of MA-104 cells (1.83 × 105 copies) infected with the wild-type CVB3 Nancy strain, which produces an acute and lytic viral infection (Fig. 1).
Table 1.
EV RNA loads and viral RNA ratios assessed in the four EV-positive IDCM patients
| Patient | Cardiac anatomic location | EV RNA load (copies/μg of total extracted RNA) |
Ratio of positive- to negative-strand viral RNA | |
|---|---|---|---|---|
| Total | Negative-strand EV | |||
| 1 | Septum | 127 | 40 | 2.2 |
| Septum | 357 | 120 | 2 | |
| 2 | Septum | 217 | 61 | 2.5 |
| Right ventricle | 183 | 45 | 3.1 | |
| 3 | Right ventricle | 2,108 | 347 | 5 |
| 4 | Left ventricle | 3,368 | 160 | 20 |
Negative-strand RNA molecules were detected in the four EV-positive IDCM patients. The ratios observed in explanted heart tissues samples ranged from 2 to 20, indicating several levels of viral genome replication activities, though all were consistent with a chronic viral infection (Table 1). These ratios were closer to those observed (4/1) in a persistent CVB infection of muscle in mice afflicted with chronic inflammatory myopathy (16). Moreover, they are also consistent with CVB TD strain infections, which were previously reported to have low levels of viral replication, with positive- to negative-strand viral RNA ratios of 2 to 3 in chronically infected mice and humans (7, 11). In contrast, the experimental curve obtained in the MA-104 cell infection model revealed an average positive- to negative-strand viral RNA ratio of 47/1, which is in accordance with ratios previously observed (40/1 to 75/1) in similar cellular models of active EV infections (11, 16).
In summary, a standardized one-step real-time RT-PCR assay was validated for wild-type and TD EV strains detection in human heart tissues. This reliable method detected EV RNA in up to 20% of IDCM patients, in agreement with previously published studies. Moreover, this quantitative molecular tool provided information fundamental to the diagnosis and understanding of EV cardiac infections by demonstrating low viral loads and low positive- to negative-strand RNA ratios in the human heart samples analyzed, which were clearly consistent with chronic or persistent viral infection (2, 6, 13). Further experiments using molecular cloning and sequencing of the 5′ nontranslated region (NTR) of the EV strains detected in IDCM patients are under way and will help identify and characterize the potential presence of EV strains presenting with terminal deletions in the 5′ NTR.
ACKNOWLEDGMENTS
Fanny Renois is supported by an official grant from the French Army department (Bourse DGA: Délégation Générale de l'Armement, Ministère de la Défense, Topic: Microbiology, infectious diseases, 2009–2012).
We thank N. M. Chapman and S. Tracy for graciously reading the manuscript prior to submission for publication.
None of the authors has a commercial or other association that might pose a conflict of interest (e.g., pharmaceutical stock ownership or consultancy). This work was supported in part by a grant for Clinical and Virological research (SFR-CAP santé /EA-4684) from the Medical University and School of Medicine of Reims, France. No extra funding was used. There is no commercial relationship or any potential conflict of interest of any nature.
Footnotes
Published ahead of print 25 July 2012
REFERENCES
- 1. Andréoletti L, et al. 2007. Active coxsackieviral B infection is associated with disruption of dystrophin in endomyocardial tissue of patients who died suddenly of acute myocardial infarction. J. Am. Coll. Cardiol. 50:2207–2214 [DOI] [PubMed] [Google Scholar]
- 2. Andréoletti L, et al. 2000. Enteroviruses can persist with or without active viral replication in cardiac tissue of patients with end-stage ischemic or dilated cardiomyopathy. J. Infect. Dis. 182:1222–1227 [DOI] [PubMed] [Google Scholar]
- 3. Andréoletti L, et al. 1996. Detection of enteroviral RNA by polymerase chain reaction in endomyocardial tissue of patients with chronic cardiac diseases. J. Med. Virol. 48:53–59 [DOI] [PubMed] [Google Scholar]
- 4. Aretz HT, et al. 1987. Myocarditis: a histopathologic definition and classification. Am. J. Cardiovasc. Pathol. 1:3–14 [PubMed] [Google Scholar]
- 5. Baboonian C, Treasure T. 1997. Meta-analysis of the association of enteroviruses with human heart disease. Heart 78:539–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bock CT, Klingel K, Kandolf R. 2010. Human parvovirus B19-associated myocarditis. N. Engl. J. Med. 362:1248–1249 [DOI] [PubMed] [Google Scholar]
- 7. Chapman NM, Kim KS, Drescher KM, Oka K, Tracy S. 2008. 5′ terminal deletions in the genome of a coxsackievirus B2 strain occurred naturally in human heart. Virology 375:480–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cooper LT, et al. 2007. The role of endomyocardial biopsy in the management of cardiovascular disease: a scientific statement from the American Heart Association, the American College of Cardiology, and the European Society of Cardiology. Endorsed by the Heart Failure Society of America and the Heart Failure Association of the European Society of Cardiology. J. Am. Coll. Cardiol. 50:1914–1931 [DOI] [PubMed] [Google Scholar]
- 9. Dennert R, Crijns HJ, Heymans S. 2008. Acute viral myocarditis. Eur. Heart J. 29:2073–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim KS, Chapman NM, Tracy S. 2008. Replication of coxsackievirus B3 in primary cell cultures generates novel viral genome deletions. J. Virol. 82:2033–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim KS, et al. 2005. 5′-terminal deletions occur in coxsackievirus B3 during replication in murine hearts and cardiac myocyte cultures and correlate with encapsidation of negative-strand viral RNA. J. Virol. 79:7024–7041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim KS, Hufnagel G, Chapman NM, Tracy S. 2001. The group B coxsackieviruses and myocarditis. Rev. Med. Virol. 11:355–368 [DOI] [PubMed] [Google Scholar]
- 13. Li Y, et al. 2000. Enteroviral capsid protein VP1 is present in myocardial tissues from some patients with myocarditis or dilated cardiomyopathy. Circulation 101:231–234 [DOI] [PubMed] [Google Scholar]
- 14. Rakar S, et al. 1997. Epidemiology of dilated cardiomyopathy. A prospective post-mortem study of 5252 necropsies. The Heart Muscle Disease Study Group. Eur. Heart J. 18:117–123 [DOI] [PubMed] [Google Scholar]
- 15. Reetoo KN, et al. 2000. Quantitative analysis of viral RNA kinetics in coxsackievirus B3-induced murine myocarditis: biphasic pattern of clearance following acute infection, with persistence of residual viral RNA throughout and beyond the inflammatory phase of disease. J. Gen. Virol. 81:2755–2762 [DOI] [PubMed] [Google Scholar]
- 16. Tam PE, Messner RP. 1999. Molecular mechanisms of coxsackievirus persistence in chronic inflammatory myopathy: viral RNA persists through formation of a double-stranded complex without associated genomic mutations or evolution. J. Virol. 73:10113–10121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Verstrepen WA, Kuhn S, Kockx MM, Van De Vyvere ME, Mertens AH. 2001. Rapid detection of enterovirus RNA in cerebrospinal fluid specimens with a novel single-tube real-time reverse transcription-PCR assay. J. Clin. Microbiol. 39:4093–4096 [DOI] [PMC free article] [PubMed] [Google Scholar]