Abstract
Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) has been found to be an accurate, rapid, and inexpensive method for the identification of bacteria and yeasts. Previous evaluations have compared the accuracy, time to identification, and costs of the MALDI-TOF MS method against standard identification systems or commercial panels. In this prospective study, we compared a protocol incorporating MALDI-TOF MS (MALDI protocol) with the current standard identification protocols (standard protocol) to determine the performance in actual practice using a specimen-based, bench-by-bench approach. The potential impact on time to identification (TTI) and costs had MALDI-TOF MS been the first-line identification method was quantitated. The MALDI protocol includes supplementary tests, notably for Streptococcus pneumoniae and Shigella, and indications for repeat MALDI-TOF MS attempts, often not measured in previous studies. A total of 952 isolates (824 bacterial isolates and 128 yeast isolates) recovered from 2,214 specimens were assessed using the MALDI protocol. Compared with standard protocols, the MALDI protocol provided identifications 1.45 days earlier on average (P < 0.001). In our laboratory, we anticipate that the incorporation of the MALDI protocol can reduce reagent and labor costs of identification by $102,424 or 56.9% within 12 months. The model included the fixed annual costs of the MALDI-TOF MS, such as the cost of protein standards and instrument maintenance, and the annual prevalence of organisms encountered in our laboratory. This comprehensive cost analysis model can be generalized to other moderate- to high-volume laboratories.
INTRODUCTION
Matrix-assisted laser desorption ionization–time of flight mass spectroscopy (MALDI-TOF MS) is increasingly utilized by clinical microbiology laboratories for the identification of bacteria and yeasts (7, 18, 19, 21, 22). This proteomic method has been found to be accurate, rapid, and inexpensive to perform (8, 11, 16). To date, published studies have principally focused on comparisons of MALDI-TOF MS to phenotypic and genotypic methods of identification for specific categories of organisms (1, 15, 19, 21). Therefore, the current literature may not adequately reflect the actual performance on clinical specimens and the full impact on workflow on a daily basis in a busy, complex clinical laboratory. Accuracy studies commonly cite the limitations of MALDI-TOF MS to differentiate genetically similar organisms, such as Streptococcus pneumoniae from other members of the Streptococcus mitis group, and Shigella species from Escherichia coli. Because clinical microbiology laboratories that utilize MALDI-TOF MS would implement or retain supplemental testing such as an optochin disk test and indole, adequate assessment has to include supplemental tests.
Previous comparisons of time to identification (TTI) and costs between MALDI-TOF MS methods and biochemical identification systems essentially reflected the difference of procedural times and costs per run (8, 16). In actual practice, the difference in TTI may be greater, because MALDI-TOF MS requires only a small amount of organisms (inoculant volume) and can be performed directly from a single colony from typical primary culture plates. In contrast, most identification methods, such as automated identification systems, require a larger inoculant volume, and subculture is needed. Subcultures are also required in many instances when conventional identification tests can be performed only from nonselective medium types. MALDI-TOF MS methods can reduce or eliminate the need for tests performed to screen for pathogens, such as coagulase for staphylococci or triple sugar iron (TSI) agar for enteric bacterial pathogens; cost assessments should account for these changes. Most studies have not considered the auxiliary costs of MALDI-TOF MS instrument servicing and controls and other practical variables in the implementation. For example, cost savings will depend on the prevalence of each species encountered in a laboratory and whether the MALDI-TOF MS has been verified to independently identify that species. Thorough evaluations to assess all potential factors that might contribute to costs and savings related to implementation of a MALDI-TOF MS identification system are warranted, particularly considering the capital expense of the MALDI-TOF MS instrument.
Using a “specimen” rather than “organism” approach, we prospectively evaluated the applied effects of a MALDI-TOF MS identification protocol on actual practice. The TTI of a MALDI-TOF MS identification protocol (MALDI protocol) was compared in parallel with our standard identification protocols (standard protocol) for clinically significant bacteria and yeasts. The potential annual cost savings was estimated using a comprehensive model that takes into account the costs of reagents and labor, measured performance, instrument maintenance expenses, and organism prevalence.
MATERIALS AND METHODS
Standard identification protocols (standard protocol).
The clinical microbiology laboratory at the Johns Hopkins Hospital is a College of American Pathologists (CAP)-accredited laboratory that meets or exceeds all state and federal requirements. This full service laboratory provides testing for three hospitals and 21 outpatient clinics and processes about 175,000 specimens annually for bacterial and fungal cultures. Bacterial and yeast cultures are routinely read once daily on the day shift, 7 days per week.
Specimens submitted for bacterial culture are assigned to one of eight workstations (“benches”) after inoculation: blood cultures; sterile body fluid cultures; urine cultures; stool cultures; aerobic wound cultures; anaerobic wound and tissue cultures; respiratory tract cultures; and cystic fibrosis respiratory tract cultures. Bacteria were most frequently identified by the following: the Phoenix microbial identification system (PHX) (BD Diagnostics, Sparks, MD); latex agglutination kits (LA) (Pro-Lab Diagnostics, Richmond Hill, ON, Canada); manual biochemical tests (multiple manufacturers); and cell wall fatty acid analysis using gas-liquid chromatography (GLC) (MIDI, Inc., Newark, DE). A yeast bench analyzed cultures from primary fungal media or subcultures of isolates first recovered on the bacteriology benches. Most yeasts are identified by germ tube, biochemical tests, CHROMagar Candida (BD Diagnostics) examination and/or sugar fermentation tests plus microscopic examination. For bacteria and yeasts that are not identifiable by other methods, 16S rRNA gene and internal transcribed spacer (ITS)/D1D2 sequencing (Applied Biosystems, Foster City CA), respectively, were performed once weekly in-house. The standard approaches to identification of bacteria and yeasts are summarized in Table S1 in the supplemental material.
MALDI-TOF MS identification protocol (MALDI protocol).
MALDI-TOF MS was performed using the Bruker Microflex instrument, Biotyper software v. 3.0, and database v. 3.1.2 (Bruker Daltonik GmbH, Bremen, Germany) according to the manufacturer's instructions. Details of these procedures have been published elsewhere (8). As the ideal identification strategy incorporating MALDI-TOF MS has not been established, the MALDI protocol was developed based on the manufacturer's suggestions and findings from other recent studies (14, 16, 17). This protocol consisted of repeat MALDI-TOF MS attempts and the incorporation of supplementary tests. An attempt is defined as a single procedure where an isolate is processed by either the direct transfer procedure (DTP) or by formic acid extraction procedure (FEP) “spotted” in duplicate onto a reusable 96-well polished steel target plate and then read by the MALDI-TOF MS instrument. The formic acid procedure consisted of the following steps. A large single colony was suspended in 300 μl of water in a microcentrifuge tube, and 900 μl of ethanol was added. After a 2-min centrifugation step, the ethanol solution was removed, and the pellet was air dried. The pellet was resuspended in 50 μl of 70% formic acid, and 50 μl of 100% acetonitrile was added. The tube was centrifuged for 2 min. A 1-μl aliquot of the supernatant was added to the steel target and allowed to dry before application of 1 μl of matrix. The highest scoring organism identification and the corresponding spectral scores in both duplicates (paired identifications and paired scores) determined whether the results were acceptable. The algorithms for accepting results or repeating attempts are as follows.
Bacterial isolates were processed initially (first attempt) by the DTP. The species-level identification result was accepted if the paired identifications were of the same species, and the paired scores were ≥2.0 and ≥1.7. Genus-level identification was not incorporated in this study. A repeat attempt (second attempt) was performed for isolates with unacceptable initial results. While this may be performed the same day, the second attempt occurred the next day from subcultures to minimize disruption of workflow during the study. The criteria for result acceptability for the second attempt were the same as for the first attempt. If the results were still unacceptable, then the isolates were processed by FEP, and MALDI-TOF MS was repeated for the third and final attempt. As recommended by the manufacturer, if a Gram-positive bacillus was not identified in the first attempt by DTP, FEP was performed on the second (and final) attempt. These final attempts, processed by FEP, have a lower acceptable paired-score cutoff (≥1.7 for each score). Yeast isolates were processed by FEP. A species-level identification result was accepted if the paired identifications were of the same species, and the paired scores were ≥1.7 (for each score). A second (and final) attempt was performed if the result from the first attempt was unacceptable.
The MALDI protocol included supplementary tests to address the misidentification of Streptococcus pneumoniae and Shigella species. All isolates identified as S. pneumoniae and viridans group streptococci, except those from the Streptococcus anginosus complex, required either the optochin disk test (Remel, Lenexa, KS) or the BBL Pneumoslide LA test (BD Diagnostics). Non-lactose-fermenting isolates identified as E. coli from stool specimens required a positive indole test (BD Diagnostics) to rule out Shigella.
Time to identification analysis.
The time to identification (TTI) is the number of days between when an isolate was first visualized on the primary media to when the identification is finalized by each protocol. The necessity of multiple attempts and supplementary tests in the MALDI protocol were reflected in the calculations. TTI analyses were included only for the isolates that had their MALDI protocol finalized on weekdays. For the TTI analysis, each isolate was classified into one of 20 organism groups with each group having similar standard protocols.
Cost analysis for each species.
The cost analysis was based on the prevalence of species encountered in the laboratory and the cost to identify an isolate of a certain species (cost per isolate). This cost per isolate has two values. The first, reagent cost per isolate, was the summed costs of all tests required to identify that isolate multiplied by the probability of that test being applied. The equation is expressed as follows:
The “reagent cost of test n” was derived from laboratory accounting data. The “average number of test n applied per isolate” varied from species to species. This “average” value was based on the reagent consumption data from the study and accounted for tests that occasionally required repeat attempts or were variably performed. An individual example for one bacterial species of the reagent cost per isolate calculation is provided in Table S2a in the supplemental material. If the reagent consumption data were not available for a certain species, as could occur if that species had not been encountered during the study, then the “average” values were derived from the expected reagent usage as described in the laboratory standard operating procedure (SOP). The second cost per isolate value, labor time per isolate, was similarly calculated except the “reagent cost of test n” was replaced with “hands-on labor time of test n”. Table S2b shows an example.
The cost per isolate values for both the standard and MALDI protocols were calculated for each species. The annual cost of each species was calculated for both protocols by applying the cost per isolate values to the prevalence of that species encountered in our laboratory during the preceding 12 months. Labor time values were converted to labor costs by multiplying the labor time by the 2010 national mean hourly medical technologist (MT) wage of $26.16 (13).
Estimated annual savings.
The difference of annual costs between the two protocols represents the savings that can be accrued if the MALDI protocol had been the primary identification method for the laboratory. In addition to the difference of costs to identify each species, other costs for the MALDI protocol were determined. This included the fixed annual costs of bacterial test standard (BTS) controls (performed in duplicate daily for each of the 9 target plates), daily target plate cleaning costs, and instrument maintenance fees (amortized over 5 years). The costs of alternative tests required for species that were poorly identified by the MALDI protocol were calculated. The calculation was the cost of standard protocol methods multiplied by the likelihood of the MALDI protocol failing to identify an isolate of a certain species (i.e., unacceptable results). The likelihood value was based on the accuracy of the MALDI protocol measured in this study (data not shown).
Finally, the cost savings also depends on which species the MALDI-TOF MS has been verified to identify. For this study, only the species or species groups (for example, viridans group streptococci) that were encountered more than 50 times (“common” species) in the preceding 12 months were considered eligible for cost savings that can be accrued annually. This threshold of 50 represents the number of isolates required for verification of the MALDI-TOF MS as an identification method (9). Conversely, species encountered less than 50 times (“uncommon” species) would not demonstrate any cost savings, until the MALDI-TOF MS has been verified for that species, because their identification costs are the sum of both protocols. Isolates would first be processed by the MALDI protocol, where the results would not be accepted and then processed by the standard protocol. Overall, the estimated annual cost difference between the two protocols represents the potential cost savings that can be achieved within 12 months of MALDI-TOF MS implementation at our institution.
Data analysis.
Evaluation of the TTI was performed using the Wilcoxon matched-pairs signed-rank test. Data were managed using Microsoft Office Access 2007 (Microsoft Corp., Redmond, WA) or Microsoft Office Excel 2007 (Microsoft Corp.), and analyzed with Stata 9.2 (Stata Corporation, College Station, TX).
RESULTS
Study design.
Over a 12-week period, clinical specimens were assessed “bench by bench” on a rotating basis, one bench per week. All laboratory benches were assigned as the study bench for 1 week, except for the blood, anaerobic wound/tissue, and yeast benches, which were assigned for 2 weeks. Only specimens that were first processed on weekdays were included in the study. The study evaluated isolates that required full identification, as per the laboratory SOP. These isolates were processed by the standard and MALDI protocols at the same time from primary culture media. The MALDI protocol was performed only on weekdays during the day shift with the exception of public holidays.
Medical technologists (MTs) were trained on the MALDI protocols prior to and during the study. The MT on the study bench performed both the standard protocol and spotted the target for the MALDI protocol. Isolates were batched onto a single target. Once or twice a day, targets were analyzed on the MALDI-TOF MS instrument by a different MT from the one who performed the standard protocol. The MT on the study bench was blinded to the results from the MALDI-TOF MS. The observer recorded the following information daily from the study bench: (i) the identification results; (ii) the reagents consumed for the identification of each isolate; and (iii) the TTI for both protocols. MALDI-TOF MS results were available only to the observer, who in turn instructed the study bench MT if additional attempts were necessary in accordance with the protocol. Data were directly recorded into an Access 2007 (Microsoft Corp., Redmond WA) database at the bench.
Study isolates.
During the study period, 2,214 patient specimens were processed by 17 MTs. Of the 991 positive specimens, 357 (36.0%) contained multiple isolates. In total, 952 isolates (824 bacterial isolates and 128 yeast isolates) were fully identified by both the standard and MALDI protocols and enrolled in the study. Some isolates were not enrolled for the following reasons: they were first recovered on a day when the MALDI protocol was not performed (for example, weekends or public holidays); they were only partially identified as they were considered normal flora; or the MT did not adhere to the MALDI protocol. The overall accuracy of the MALDI protocol was determined to be 98.3% (data not shown).
Time to identification comparison.
TTI calculations were available for 911 isolates completely processed during the Monday through Friday observation period each week. On the first day of workup, 87.2% of the isolates were identified by the MALDI protocol, and by the second day, 97.8% were identified (see Table S3 in the supplemental material). In comparison, 9.4% were identified by the standard protocol on the first day, and 61.5% by the second day. Acceptable MALDI protocol results did not occur on the first day of workup due to mixed isolates in primary media or insufficient growth. TTI was longer in those isolates that grew slowly and required FEP, as higher inoculant volumes for FEP are needed. The longest TTI was 5 days for an Actinomyces sp. isolate.
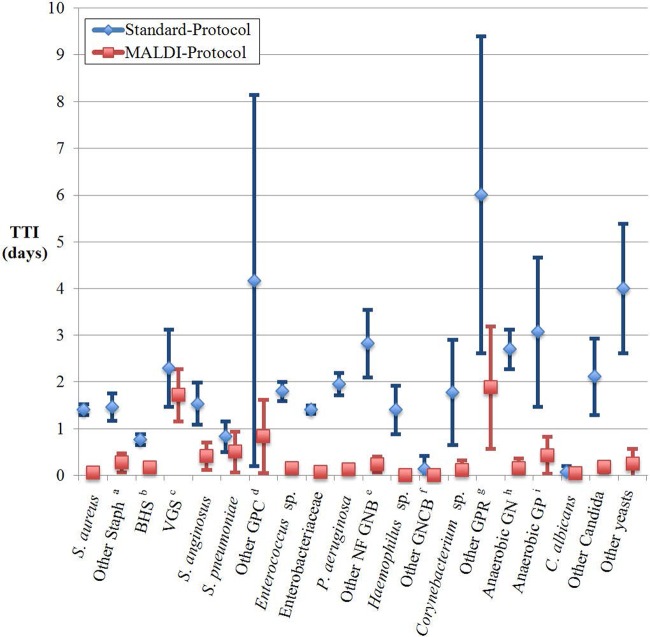
The MALDI protocol yielded results more quickly than the standard protocol for most isolates when assessed by organism groups (P < 0.05) (Fig. 1) (see Table S3 in the supplemental material). The differences in TTI were calculated based upon time in days not hours (Table 1). Overall, the MALDI protocol provided identifications 1.45 days (P < 0.05) earlier on average and was at least 1 day more rapid in identifying the majority of organism groups. Isolates that required gene sequencing as part of the standard protocol had the largest differences in TTI because sequencing was batched once a week. By excluding the 10 isolates that required gene sequencing, the mean difference in TTI between the two protocols was 1.38 days (P < 0.05) for all organisms. The distribution of isolates in relation to the difference in TTI is displayed in Table 1. For example, 66.1% of Staphylococcus aureus isolates were identified 1 day earlier by the MALDI protocol.
Fig 1.
Time to identification (TTI) and 95% confidence interval by MALDI and standard protocols. The blue and red symbols represent the mean TTI by the standard and MALDI protocols, and the error bars represent the 95% confidence intervals (95% CI). The organisms identified were as follows: Other Stapha, staphylococci other than S. aureus; BHSb, beta-hemolytic Streptococcus; VGSc, viridans group Streptococcus; GPCd, Gram-positive cocci; members of the Enterobacteriaceae family; NF GNBe, glucose-nonfermenting Gram-negative bacilli; GNCBf, fastidious Gram-negative coccobacilli; GPRg, Gram-positive rod; Anaerobic GNh, anaerobic Gram-negative bacteria; Anaerobic GPi, anaerobic Gram-positive bacteria.
Table 1.
Difference in time to identification (TTI) between the standard protocol and MALDI protocol
| Organism or group | No. of isolates | Mean no. of days isolate identified earlier | Proportion (%) identified earlier by the MALDI protocol after the following no. of days of workup: |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| <0a | 0b | 1 | 2 | 3 | 4 | 5 | 6 | >6 | |||
| Staphylococcus aureus | 109 | 1.35 | 1.8 | 66.1 | 28.4 | 2.8 | 0.9 | ||||
| Other staphc | 26 | 1.19 | 7.7 | 65.4 | 26.9 | ||||||
| BHSd | 72 | 0.60 | 1.4 | 38.9 | 58.3 | 1.4 | |||||
| VGSe | 7 | 0.57 | 42.9 | 57.1 | |||||||
| Streptococcus anginosus | 17 | 1.12 | 41.2 | 29.4 | 5.9 | 23.5 | |||||
| Streptococcus pneumoniae | 6 | 0.33 | 66.7 | 33.3 | |||||||
| Other GPCf | 6 | 3.33 | 33.3 | 16.7 | 16.7 | 16.7 | 16.7 | ||||
| Enterococcus sp. | 78 | 1.64 | 1.3 | 51.3 | 34.6 | 9.0 | 2.6 | 1.3 | |||
| Enterobacteriaceae | 284 | 1.34 | 2.8 | 69.4 | 23.2 | 2.1 | 1.1 | 1.1 | 0.4 | ||
| Pseudomonas aeruginosa | 77 | 1.82 | 41.6 | 49.4 | 2.6 | 1.3 | 2.6 | 2.6 | |||
| Other NF GNBg | 39 | 2.59 | 2.6 | 30.8 | 35.9 | 2.6 | 15.4 | 5.1 | 5.1 | 2.6 | |
| Haemophilus sp. | 10 | 1.40 | 80.0 | 20.0 | |||||||
| Other GNCBh | 7 | 0.14 | 85.7 | 14.3 | |||||||
| Corynebacterium sp. | 9 | 1.67 | 22.2 | 33.3 | 22.2 | 22.2 | |||||
| Other GPRi | 8 | 4.13 | 12.5 | 37.5 | 12.5 | 37.5 | |||||
| Anaerobic GNj | 26 | 2.54 | 3.8 | 65.4 | 7.7 | 19.2 | 3.8 | ||||
| Anaerobic GPk | 14 | 2.64 | 21.4 | 14.3 | 28.6 | 7.1 | 7.1 | 14.3 | 7.1 | ||
| Candida albicans | 52 | 0.04 | 3.8 | 92.3 | 1.9 | 1.9 | |||||
| Other Candida sp. | 56 | 1.93 | 8.9 | 67.9 | 7.1 | 3.6 | 3.6 | 3.6 | 5.4 | ||
| Other yeasts | 8 | 3.75 | 25.0 | 37.5 | 12.5 | 12.5 | 12.5 | ||||
| All organismsl | 911 | 1.45 | 0.4 | 13.5 | 52.7 | 23.6 | 3.7 | 2.7 | 1.1 | 1.1 | 1.1 |
Proportion identified earlier by standard protocol (range, 1 to 2 days).
Proportion identified where the TTI by MALDI and standard protocols were equal.
Other staph, staphylococci other than S. aureus.
BHS, beta-hemolytic Streptococcus.
VGS, viridans group Streptococcus.
GPC, Gram-positive cocci.
NF GNB, non-glucose-fermenting Gram-negative bacilli.
GNCB, fastidious Gram-negative coccobacilli.
GPR, Gram-positive rod.
Anaerobic GN, anaerobic Gram-negative bacteria.
Anaerobic GP, anaerobic Gram-positive bacteria.
The total values for all organisms are shown in boldface type.
Estimated cost savings with the MALDI protocol in 1 year.
In the 12 months preceding the study, our laboratory identified 47,845 bacteria and yeasts representing 279 species. The 55 “common” species (i.e., those encountered at least 50 times in the preceding year) represented 46,417 (97.0%) of isolates. Fifty-one of these species were encountered at least once during the study. Their cost per isolate values were derived from the study reagent consumption data. The reagent consumption data for each species were compared with the laboratory SOP and found to be similar for most species. The four species not encountered during the 12-week observation period, Staphylococcus saprophyticus, Providencia stuartii, Salmonella enteritidis, and Campylobacter jejuni/coli, had their cost per isolate values estimated from the laboratory SOP. The cost per isolate values by both the standard and MALDI protocols were determined for all 55 species. The values do not include the costs of media from the primary cultures or subcultures or susceptibility testing; these are required by both protocols, so their costs cancel out.
The MALDI protocol reagent costs were $0.35 and $0.79 per test for nonextracted and formic acid (FA)-extracted isolates in duplicate, respectively (see Table S4 in the supplemental material). The fixed annual cost of the protocol was estimated at $31,273 (Table S5). The cost per isolate of species poorly identified by the MALDI protocol, such as Prevotella species, Peptostreptococcus species, and Cryptococcus neoformans, included the costs of alternate tests. The costs to identify the 224 “uncommon” species (1,428 isolates per year) by the MALDI protocol were based on the average reagent consumption data from all isolates of “uncommon” species encountered during the study.
Multiplying the cost per isolate of each of the 55 “common” species by their prevalence, the cumulative cost of identification by the standard protocol is $189,969 (Tables 2 and 3). The cumulative costs of the MALDI protocol of the 55 common species, the 224 “uncommon” species, and fixed costs are $87,556. The total savings that can be achieved by our laboratory within the first 12 months is $102,413 or 53.9% less than the standard protocol. The five most commonly identified species in this laboratory, Pseudomonas aeruginosa, E. coli, S. aureus, Streptococcus agalactiae, and Enterococcus faecalis, represented 65.3% of the potential savings. As the MALDI protocol is verified for additional species, further cost savings can be achieved. For example, if a threshold of 30 isolates were required for verification instead of 50, an additional 8 “uncommon” species or species groups can be identified by the MALDI protocol alone. Because both protocols would not be required in parallel, these 8 species represent 319 isolates and would increase the cost savings to $106,061.
Table 2.
Reagent costs and labor time between the standard protocol and MALDI protocol
| Species or species group | No. of isolates per year | Standard protocol |
MALDI protocol |
||
|---|---|---|---|---|---|
| Reagent cost ($) | Labor time (h:min) | Reagent cost ($) | Labor time (h:min) | ||
| Staphylococcus aureus | 7,059 | 15,747.53 | 76:38 | 2,523.63 | 93:21 |
| Staphylococcus lugdunensis | 65 | 180.99 | 1:44 | 48.11 | 2:58 |
| Staphylococcus saprophyticus | 63 | 243.81 | 1:03 | 21.87 | 0:47 |
| Streptococcus anginosus group | 418 | 3,824.28 | 53:05 | 240.17 | 11:19 |
| Streptococcus pyogenes | 1,419 | 3,683.99 | 55:48 | 492.55 | 17:44 |
| Streptococcus agalactiae | 3,516 | 10,866.87 | 199:06 | 1,292.22 | 46:32 |
| Streptococcus dysgalactiae | 376 | 1,278.06 | 24:01 | 142.38 | 5:07 |
| Streptococcus pneumoniae | 338 | 1,095.08 | 27:13 | 786.29 | 32:23 |
| Viridans streptococcus group | 699 | 7,099.89 | 90:48 | 613.70 | 40:13 |
| Enterococcus faecium | 909 | 3,718.73 | 21:00 | 347.07 | 12:29 |
| Enterococcus faecalis | 3,346 | 10,630.48 | 32:21 | 1,254.34 | 45:10 |
| Enterococcus raffinosus | 72 | 791.30 | 10:12 | 202.32 | 6:22 |
| Enterococcus species | 76 | 839.60 | 10:13 | 163.90 | 3:29 |
| Micrococcus luteus | 54 | 102.02 | 0:42 | 39.97 | 2:28 |
| Citrobacter freundii group | 263 | 524.64 | 3:24 | 91.29 | 3:17 |
| Citrobacter koseri | 260 | 510.64 | 3:20 | 90.25 | 3:15 |
| Citrobacter sp. | 58 | 110.75 | 2:05 | 20.13 | 0:43 |
| Enterobacter aerogenes | 381 | 715.46 | 4:23 | 132.25 | 4:45 |
| Enterobacter cloacae group | 883 | 1,921.35 | 10:55 | 306.50 | 11:02 |
| Enterobacter sp. | 51 | 428.95 | 5:42 | 35.41 | 1:16 |
| Escherichia coli | 8,816 | 17,445.65 | 109:53 | 3,158.82 | 113:45 |
| Klebsiella oxytoca | 322 | 676.80 | 3:34 | 111.77 | 4:01 |
| Klebsiella pneumoniae | 2,884 | 5,598.89 | 32:33 | 1,090.44 | 39:16 |
| Morganella morganii | 201 | 611.22 | 3:49 | 69.77 | 2:30 |
| Proteus mirabilis | 1,315 | 5,050.28 | 48:01 | 456.45 | 16:26 |
| Proteus vulgaris/penneri | 57 | 122.20 | 0:39 | 23.74 | 0:51 |
| Providencia stuartii | 82 | 151.52 | 0:54 | 28.46 | 1:01 |
| Salmonella enteritidis | 118 | 218.05 | 1:18 | 40.96 | 1:28 |
| Acinetobacter baumannii group | 284 | 1,185.44 | 3:39 | 107.54 | 3:52 |
| Achromobacter sp. | 190 | 2,460.26 | 2:57 | 190.10 | 10:29 |
| Acinetobacter sp. | 65 | 968.87 | 1:37 | 22.56 | 0:48 |
| Burkholderia cepacia | 85 | 8,187.99 | 1:39 | 7,195.85 | 1:03 |
| Pseudomonas aeruginosa | 4,974 | 22,792.85 | 70:15 | 2,039.79 | 82:39 |
| Pseudomonas fluorescens/putida | 61 | 801.10 | 0:58 | 21.17 | 0:45 |
| Serratia marcescens | 374 | 783.35 | 4:19 | 129.82 | 4:40 |
| Stenotrophomonas maltophilia | 732 | 3,187.41 | 12:50 | 293.17 | 10:33 |
| Campylobacter jejuni/coli | 57 | 267.34 | 4:42 | 19.79 | 0:42 |
| Haemophilus influenzae | 404 | 739.41 | 12:30 | 140.23 | 5:03 |
| Haemophilus species, but not H. influenzae | 95 | 110.27 | 1:03 | 32.98 | 1:11 |
| Moraxella catarrhalis | 120 | 317.10 | 2:40 | 41.65 | 1:30 |
| Actinomyces sp. | 59 | 5,623.30 | 0:09 | 77.10 | 5:02 |
| Corynebacterium striatum | 201 | 1,568.12 | 0:39 | 83.72 | 3:00 |
| Propionibacterium acnes | 184 | 446.99 | 4:09 | 63.87 | 2:18 |
| Bacteroides fragilis | 465 | 4,068.84 | 36:04 | 161.41 | 5:48 |
| Clostridium species, but not C. perfringens | 78 | 3,439.08 | 9:27 | 47.52 | 2:42 |
| Peptostreptococcus sp. | 154 | 608.17 | 14:48 | 401.44 | 15:09 |
| Prevotella sp. | 254 | 1,699.49 | 17:15 | 585.00 | 12:31 |
| Candida albicans | 1,675 | 1,045.88 | 22:47 | 1,401.85 | 118:52 |
| Candida glabrata | 933 | 890.81 | 64:52 | 787.86 | 66:48 |
| Candida krusei/inconspicua | 147 | 365.74 | 10:52 | 115.57 | 9:48 |
| Candida lusitaniae | 59 | 27.14 | 3:26 | 46.39 | 3:56 |
| Candida parapsilosis | 247 | 471.03 | 20:08 | 224.07 | 19:00 |
| Candida tropicalis | 294 | 2,029.46 | 25:31 | 295.35 | 25:02 |
| Cryptococcus neoformans | 74 | 107.97 | 6:34 | 130.16 | 9:19 |
| Saccharomyces cerevisiae | 51 | 262.87 | 6:43 | 40.10 | 3:24 |
| “Uncommon” speciesa | 1,428 | –b | – | 1,093.03 | 69:14 |
| All speciesc | 47,845 | 158,645.33 | 1,197:24 | 29,613.84 | 1,019:28 |
Species where less than 50 isolates are encountered per year.
These values are not required to estimate annual savings, as they are cancelled out in the cost differences.
The total values for all species are shown in boldface type.
Table 3.
Estimated annual costs for the standard protocol and MALDI protocol
| Item | Standard protocol cost ($) | MALDI protocol cost ($) |
|---|---|---|
| Annual reagent costs | 158,645.33 | 29,613.84 |
| Annual labor costs (based on reagent time) | 31,323.98 | 26,669.34 |
| Fixed annual costs of MALDI protocola | 31,272.53 | |
| Totalb | 189,969.31 | 87,555.70 |
See Table S5 in the supplemental material.
The total costs for all items are shown in boldface type.
The labor time savings of 178 h ($4,655) reflects the overall effect of the MALDI protocol (Table 3). Some species required more labor with the MALDI protocol, whereas others required less. For example, based on the 2011 annual numbers, the 1,675 isolates of Candida albicans identified would have required an estimated 96 h more labor time with the MALDI protocol, whereas the 5,311 beta-hemolytic streptococci would have required 210 h less.
DISCUSSION
Our study prospectively compared the MALDI protocol with an entire repertoire of biochemical, immunological, and genotypic tests to elucidate its performance, TTI, and costs of identification. Of these three measures, performance as related to accuracy is the most described (2, 3, 5, 16). Such studies usually cite known limitations with S. pneumoniae and Shigella and may not explicitly state the workup algorithm and the frequency of repeated attempts when using MALDI-TOF MS. Our study accounted for duplicate smears, repeat runs, and supplementary tests that maximize accuracy, hence reflecting what a laboratory may implement in actual practice.
The MALDI protocol has a rapid TTI due to the MALDI-TOF MS process and the low inoculant volume requirements. This study is the first to show the TTI reduction considering both of these advantages, whereas other studies have compared the MALDI-TOF MS process only with biochemical identification systems (8, 16). Instead of a 6- to 24-h difference, we anticipate that the MALDI protocol can provide finalized identifications at least 1 day earlier for most organisms. An even greater difference of TTI is observed in organisms that are biochemically inert, fastidious, and/or slow-growing, such as the mucoid P. aeruginosa strains, HACEK (Haemophilus parainfluenzae, Haemophilus aphrophilus, Haemophilus paraphrophilus, Actinobacillus actinomycetemcomitans, Cardiobacterium hominis, Eikenella corrodens, and Kingella species) group, and anaerobic bacteria. Non-C. albicans yeasts can also be more rapidly identified even with the additional extraction step. On the other hand, beta-hemolytic streptococci, viridans group streptococci, S. pneumoniae, Micrococcus luteus, Moraxella catarrhalis, and C. albicans had a mean difference of less than 1 day. There are two explanations why these organisms were not identified more quickly by the MALDI protocol: (i) the availability of rapid tests for these species (for example, germ tube to identify C. albicans) or (ii) the delay caused by performance of supplementary tests required for the MALDI protocol workup (for example, to differentiate S. pneumoniae from S. mitis). The comparison of the MALDI protocol with rapid presumptive-identification methods, such as those available for S. aureus and E. coli, was not assessed in this study, although those methods are likely faster than the MALDI protocol, as they can be performed directly at the bench.
Furthermore, the difference in TTI between the two protocols is likely underestimated in isolates that required repeat MALDI-TOF MS attempts. These repeat attempts were typically performed the next day to minimize interruption to the routine workflow during the study, as specimens that already have been processed are not typically reevaluated the same day, and sufficient organism is available the next day for extraction if required. In actual practice, repeat attempts can be performed the same day. Isolates that fail to be identified a second time should be considered for alternative identification tests to minimize the TTI. A recent study utilizing the same instrument found that MALDI-TOF MS could provide an identification for 93% of isolates within 24 h, which is slightly higher than the value we measured (87.2%) (12). As the MTs become more experienced with the MALDI-TOF MS procedure and repeat attempts are processed the same day, we expect that the TTI for the MALDI protocol will be further reduced than measured during our study. Despite the improvement in TTI, we do not anticipate that the time to susceptibility results for most organisms will change.
The cost analysis considers our laboratory's characteristics, such as SOP and organism prevalence; performance of the identification system; verification of the system; and the reagent and labor costs for the individual test components. This analysis is designed to best approximate the direct cost savings from incorporating a MALDI-TOF MS protocol as first-line identification in a moderate- to large-volume laboratory, potentially replacing many routine phenotypic and genotypic methods. This analysis incorporates other direct costs of the MALDI-TOF MS identification system, including the costs of auxiliary tests and reagents. Thorough cost evaluations are essential to justify the high upfront costs of the instrument. We believe that this cost analysis model is applicable to other moderate- or large-volume laboratories if similar data can be obtained or estimated.
While this study was a thorough examination, the projected cost savings from this study may be underestimated because of the difficulty of assessing some measures. The cost savings did not account for indirect cost savings from decreased wastes, quality control of replaced tests, and the training and labor associated with these retired procedures. A recent retrospective cost assessment in a laboratory that processes 83% of our volume found a cost savings of $177,090 annually with MALDI protocol implementation, an overall 89.3% savings (12). In addition to reduced reagent costs, the authors noted savings from reduced waste, subculture media, and DNA sequencing, but no loss of staff. We anticipate that additional savings will accrue with time, as more organisms are verified for MALDI-TOF MS identification. MT experience will decrease the need for repeat MALTI-TOF MS attempts, and the database updates will reduce the need for supplementary tests for organisms that the current version is unable to identify or discriminate. Conversely, the cost savings calculated in other studies, which estimated potential savings of 80 to 95% (8, 16), may be overestimated. Those studies did not account for the instrument service and other auxiliary test costs, which we determined to cost more than the reagents required for the MALDI protocol.
The overall effect of MALDI-TOF MS implementation on labor costs is difficult to determine, although we anticipate that there will be very little change. Subjectively, most of the labor time spent on the bench is the assessment of primary cultures and the performance of subcultures. Subcultures are still needed for routine susceptibility testing for 82.4% of organisms; those organisms that do not, such as most beta-hemolytic streptococci, anaerobic bacteria, enteric pathogens, and yeasts may require less labor with MALDI-TOF MS implementation. A single MALDI protocol for most species would provide a more streamlined workflow than current practice, with several protocols for different species. The reduction or elimination of certain routine tests could further reduce the labor spent on quality control and technologist training associated with these tests. However, any savings realized by the aforementioned reductions may potentially be neutralized by the time needed to manage and maintain the MALDI-TOF MS instrument.
Our study defines additional parameters with the use of the MALDI-TOF MS, but the ideal algorithm for a MALDI protocol still requires further elucidation. Repeat MALDI-TOF MS attempts were required for 10.2% of isolates. Most of these actually had the correct identification on the first attempt (data not shown), but the spectral scores were not acceptable (i.e., both scores were <2). As previously mentioned, this rate is anticipated to decrease over time with improved MT expertise at smearing the target plate and further expansion of the spectral database. Alternatively, changing the interpretation strategy on isolates with spectral scores of ≥1.7 but <2 could also reduce the need for repeat attempts. Some proposed strategies include assessing the spectral scores in conjunction with the number of entries in the database (4), comparing the difference of scores between the first and second best matches (10), and formic acid extraction for all isolates (14). We observed that certain species, such as Streptococcus anginosus complex, can be correctly identified if the paired scores are ≥1.7 but <2, and the paired identifications are the same. A species-dependent threshold can be determined during a site's verification of the MALDI-TOF MS. This concept has been advocated by others (6, 14, 20), and individual species-dependent thresholds might be part of a solution.
Several limitations to our study exist. Some of the isolates processed may have been repeat isolates from the same patient, and the actual number of unique isolates assessed is less than 952. However, this study was designed to compare the different protocols and assess cost and TTI reduction with MALDI-TOF MS applied to current laboratory practices. The performance of MALDI-TOF MS for less-commonly identified organisms may not be reflective of actual performance in terms of accuracy, cost, and TTI, warranting greater numbers for sufficient evaluation. Many of these organisms have a higher workup cost, as currently they may be identified only by GLC and sequencing, thus disproportionately contributing to the reagent costs of the laboratory. Despite this, the study demonstrated that most of the financial benefit of the MALDI protocol occurs with commonly identified organisms. Similarly, another limitation to the study is that certain organisms, such as Shigella, were not observed during the observation period, and the reagent costs for these “absent” organisms were estimates.
The MALDI-TOF MS system is robust with different MT users and a broad range of specimens, reflecting the potential as the primary identification system of bacteria and yeasts in a diverse, high-volume clinical laboratory. The MALDI protocol demonstrated reduction of costs and TTI, while maintaining similar accuracy as the standard protocol. We provided a model to better approximate cost savings that can be applied to other laboratories. Studies to correlate the reduction of TTI with clinical outcomes and further cost analysis studies in the microbiology laboratory are warranted.
Supplementary Material
ACKNOWLEDGMENTS
We thank the following medical technologists for their interest and participation in the study: Filipos Abraham, Ana Boyer, Latetia Bryan, Michael Camp, Rachel Green, Denise Jones, Maria Karais, Warda Memon, Jo-Anne Miller, Jamie Prestridge, Mark Romagnoli, Sherin Shahegh, Abi Thompson, Christi Vargas, Teresa Wakefield, and Chelsea Weik. We acknowledge Qilu Yu and David Dowdy's statistical insight. We are also grateful to Bruker Daltonic, Inc., Billerica, MA, for loaning the equipment and instrument and providing the reagents necessary for the study and Markita Weaver for providing training and technical support.
Footnotes
Published ahead of print 1 August 2012
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Alatoom AA, Cunningham SA, Ihde SM, Mandrekar J, Patel R. 2011. Comparison of direct colony method versus extraction method for identification of gram-positive cocci by use of Bruker Biotyper matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 49:2868–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bader O, et al. 2011. Improved clinical laboratory identification of human pathogenic yeasts by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Microbiol. Infect. 17:1359–1365 [DOI] [PubMed] [Google Scholar]
- 3. Benagli C, Rossi V, Dolina M, Tonolla M, Petrini O. 2011. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for the identification of clinically relevant bacteria. PLoS One 6:e16424 doi:10.1371/journal.pone.0016424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bessede E, et al. 2011. Matrix-assisted laser-desorption/ionization biotyper: experience in the routine of a university hospital. Clin. Microbiol. Infect. 17:533–538 [DOI] [PubMed] [Google Scholar]
- 5. Bizzini A, Durussel C, Bille J, Greub G, Prod'hom G. 2010. Performance of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of bacterial strains routinely isolated in a clinical microbiology laboratory. J. Clin. Microbiol. 48:1549–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bizzini A, et al. 2011. Matrix-assisted laser desorption ionization-time of flight mass spectrometry as an alternative to 16S rRNA gene sequencing for identification of difficult-to-identify bacterial strains. J. Clin. Microbiol. 49:693–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carbonnelle E, et al. 2011. MALDI-TOF mass spectrometry tools for bacterial identification in clinical microbiology laboratory. Clin. Biochem. 44:104–109 [DOI] [PubMed] [Google Scholar]
- 8. Cherkaoui A, et al. 2010. Comparison of two matrix-assisted laser desorption ionization–time of flight mass spectrometry methods with conventional phenotypic identification for routine identification of bacteria to the species level. J. Clin. Microbiol. 48:1169–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Clark RB, Lewinski MA, Loeffelholz MJ, Tibbetts RJ. 2009. Cumitech 31A, Verification and validation of procedures in the clinical microbiology laboratory. ASM Press, Washington, DC [Google Scholar]
- 10. Degand N, et al. 2008. Matrix-assisted laser desorption ionization–time of flight mass spectrometry for identification of nonfermenting gram-negative bacilli isolated from cystic fibrosis patients. J. Clin. Microbiol. 46:3361–3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dhiman N, Hall L, Wohlfiel SL, Buckwalter SP, Wengenack NL. 2011. Performance and cost analysis of matrix-assisted laser desorption ionization–time of flight mass spectrometry for routine identification of yeast. J. Clin. Microbiol. 49:1614–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gaillot O, et al. 2011. Cost-effectiveness of switch to matrix-assisted laser desorption ionization–time of flight mass spectrometry for routine bacterial identification. J. Clin. Microbiol. 49:4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Garcia E, et al. 2011. American Society for Clinical Pathology's 2010 wage survey of U.S. clinical laboratories. Lab. Med. 42:141–146 [DOI] [PubMed] [Google Scholar]
- 14. Justesen US, et al. 2011. Species identification of clinical isolates of anaerobic bacteria: a comparison of two matrix-assisted laser desorption ionization–time of flight mass spectrometry systems. J. Clin. Microbiol. 49:4314–4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martiny D, et al. 2012. Comparison of the Microflex LT and Vitek(R) MS systems for the routine identification of bacteria by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 50:1313–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Neville SA, et al. 2011. Utility of matrix-assisted laser desorption ionization-time of flight mass spectrometry following introduction for routine laboratory bacterial identification. J. Clin. Microbiol. 49:2980–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Saffert RT, et al. 2011. Comparison of Bruker Biotyper matrix-assisted laser desorption ionization–time of flight mass spectrometer to BD Phoenix automated microbiology system for identification of gram-negative bacilli. J. Clin. Microbiol. 49:887–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Seng P, et al. 2009. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis. 49:543–551 [DOI] [PubMed] [Google Scholar]
- 19. Stevenson LG, Drake SK, Shea YR, Zelazny AM, Murray PR. 2010. Evaluation of matrix-assisted laser desorption ionization–time of flight mass spectrometry for identification of clinically important yeast species. J. Clin. Microbiol. 48:3482–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Szabados F, et al. 2012. Evaluation of species-specific score cutoff values of routinely isolated clinically relevant bacteria using a direct smear preparation for matrix-assisted laser desorption/ionization time-of-flight mass spectrometry-based bacterial identification. Eur. J. Clin. Microbiol. Infect. Dis. 31:1109–1119 [DOI] [PubMed] [Google Scholar]
- 21. Veloo AC, Knoester M, Degener JE, Kuijper EJ. 2011. Comparison of two matrix-assisted laser desorption ionization-time of flight mass spectrometry methods for the identification of clinically relevant anaerobic bacteria. Clin. Microbiol. Infect. 17:1501–1506 [DOI] [PubMed] [Google Scholar]
- 22. Wieser A, Schneider L, Jung J, Schubert S. 2012. MALDI-TOF MS in microbiological diagnostics-identification of microorganisms and beyond. Appl. Microbiol. Biotechnol. 93:965–974 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.