Abstract
No clinical isolates have been reported for the recently described thermoactinomycete Kroppenstedtia eburnea. Between 2006 and 2011, we obtained 14 clinical isolates from patients in 9 U.S. states. Here we report growth characteristics, 16S rRNA gene sequencing, matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry-based identification, and antimicrobial susceptibility profiles of this recently described organism.
TEXT
Members of the family Thermoactinomycetaceae are aerobic, thermophilic or mesophilic, Gram-positive, filamentous bacteria. They have primarily been isolated from environmental sources, including soil, buildings, humidifiers, and fresh and salt water, as well as from foods such as cereal grains and dairy products (2, 3, 5, 10, 12, 14, 15). Inhalation of several species has been linked to hypersensitivity pneumonitis, but reports of isolation of thermoactinomycetes from clinical specimens are rare (4, 14). Kroppenstedtia eburnea was recently described as a new genus and species within this family (12). The type strain was recovered in a contract manufacturing organization, and no clinical isolates have previously been reported. A review of partial 16S rRNA gene (16S) sequencing results in our laboratory over a 5-year period revealed 14 clinical isolates of K. eburnea. Here we report their growth characteristics, morphological properties, partial 16S sequences, proteomic profiles obtained by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS), and antimicrobial susceptibility.
K. eburnea isolates were collected from blood (n = 11), cerebrospinal fluid (CSF; n = 1), peritoneal fluid (n = 1), and skin (n = 1, Table 1) in 9 U.S. states between February 2006 and March 2011 and were referred to ARUP Laboratories for identification. Limited clinical information could be obtained for 3 patients (for case data, see Table 1).
Table 1.
Demographic information, source, and year of isolation for 14 patients from whom Kroppenstedtia eburnea was isolateda
| Patient | Case history | Sex | Age (yr) | State | Isolate | Yr | Source |
|---|---|---|---|---|---|---|---|
| 1 | M | <1 | SC | A | 2006 | Blood | |
| 2 | F | 61 | CA | B | 2006 | Skin, abdominal wall | |
| 3 | M | 82 | CO | C | 2007 | Blood | |
| 4 | F | 31 | MA | D | 2008 | Blood | |
| 5 | M | 80 | IL | E | 2008 | Blood | |
| 6 | M | 78 | MA | F | 2009 | Blood | |
| 7 | F | 56 | IL | G | 2009 | Blood | |
| 8 | F | 65 | ME | H | 2009 | Blood | |
| 9 | 1 | F | 59 | IN | I | 2010 | CSF |
| 10 | F | 64 | OH | J | 2010 | Blood | |
| 11 | F | 71 | IN | K | 2010 | Peritoneal fluid | |
| 12 | F | 68 | OH | L | 2010 | Blood | |
| 13 | 2 | F | 65 | NY | M | 2011 | Blood |
| 14 | 3 | F | 70 | IL | N | 2011 | Blood |
Case histories 1 through 3 are provided in the main text. F, female; M, male.
Case 1.
A 59-year-old female from Indiana treated with chemotherapy for metastatic colon carcinoma was admitted to an emergency department (ED) with altered mental status. CSF culture revealed growth of branching Gram-positive rods on sheep blood agar (SBA) by day 3. CSF chemistry was unremarkable, and no leukocytes were noted. Blood cultures remained negative. The white blood cell (WBC) count was 3.6 × 109 cells/liter (64% neutrophils; no bands), the hemoglobin (Hb) concentration was 11.8 g/dl, and the hematocrit (HCT) was 36.7%. Antibacterial therapy was not initiated, and the patient's neurologic symptoms largely resolved.
Case 2.
A 65-year-old female from New York was evaluated in an ED for altered mental status after a fall 5 days prior. A physical examination, vital signs, and magnetic resonance imaging of the head revealed no abnormalities, blood was drawn for culture, and the patient was discharged. The blood culture showed growth of filamentous, branching Gram-positive rods on day 4 that grew as flat, dry, colonies within 24 h in subculture on SBA and chocolate agar. The patient was contacted and reportedly felt well. Treatment was not initiated, and repeat blood cultures remained negative.
Case 3.
A 70-year-old female from Illinois with rheumatoid arthritis on chemotherapy for breast cancer was admitted to an ED with hypotension and shortness of breath. The patient had leukocytosis but was afebrile. A blood culture demonstrated growth of Gram-positive rods. The patient received no antibacterial therapy, and subsequent blood cultures remained negative.
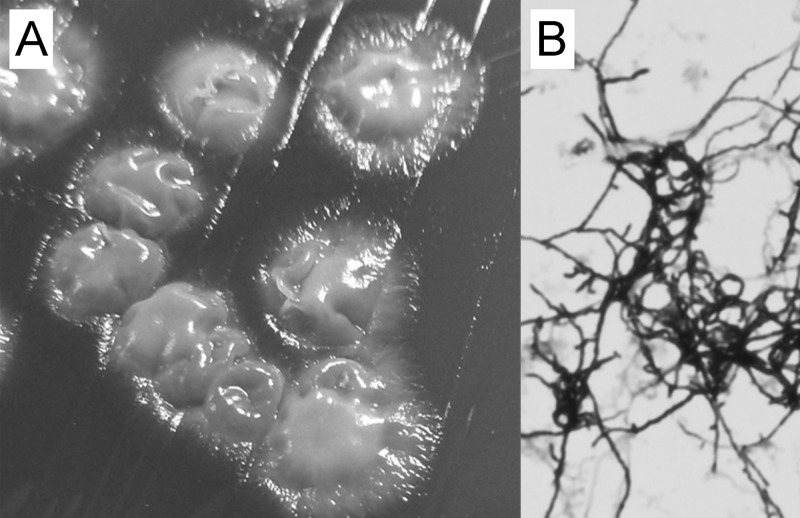
Twelve of the 14 isolates could be retrieved from a frozen repository and were subcultured at least twice on Columbia SBA. When incubated at 37°C in ambient air, isolates grew more rapidly on buffered charcoal yeast extract (BCYE) agar (visible after 24 h) than on SBA and chocolate agar (visible after 48 h) but failed to grow on Middlebrook 7H11 and Sabouraud dextrose agars. Individual, off-white, adherent colonies with irregular margins and a dull surface were visible after 48 (BCYE) and 72 (SBA) h (Fig. 1A, older colonies). As expected, K. eburnea isolates grew more rapidly at 50°C than at 42°C or 30°C (12). Microscopic examination revealed Gram-positive, elongated, septate hyphae with moderate branching (Fig. 1B) that were negative by acid-fast and auramine O staining.
Fig 1.
Colony morphology on Columbia sheep blood agar (A) and microscopic Gram-stain morphology (original magnification, ×1,000) after subculturing in Mueller-Hinton broth (B).
Partial 16S sequencing was performed as previously described, and sequences were interpreted according to CLSI guidelines (6, 9). Sequences (JN986812 to JN986825) were aligned with those of thermoactinomycete type strains, and a phylogenetic tree (Fig. 2A) was constructed using the neighbor-joining method in Molecular Evolutionary Genetics Analysis version 4.1 (11). Sequences showed 100% similarity to the K. eburnea type strain (FN665656) when considering corresponding bases a match to IUPAC ambiguity symbols (i.e., “G” or “A” for “R”).
Fig 2.
(A) Phylogenetic tree based on partial 16S sequences of clinical and type strains of K. eburnea and related taxa. A clinical D. activa isolate was used for comparison; Aneurinibacillus thermoaerophilus and Bacillus fortis were used as outgroups. Percent bootstrap values are based on 1,000 iterations. (B and C) MALDI-TOF spectra and fingerprints (B) and principal component analysis-based clustering (C); clinical isolates of D. activa and Bacillus cereus were used for comparison. Intensity values are presented in arbitrary units; m/z, mass/charge ratio.
Single bacterial colonies subcultured on SBA were analyzed on a microflex LRF MALDI-TOF mass spectrometer (Bruker Daltonics) in linear positive-ion mode using FlexControl 3.0 software (Bruker Daltonics) as reported elsewhere (7). A clinical isolate and the type strain of the closely related Desmospora activa species (Fig. 2A; AM940019) were included for comparison (14). Isolates could be readily identified as K. eburnea and differentiated from D. activa after addition of reference spectra for both organisms to the Biotyper database (version 3.0; Bruker Daltonics) (median score, 2.4; range, 2.0 to 2.7). Using principal component analysis, mass spectra of clinical isolates from K. eburnea (n = 12) and D. activa (n = 1) clustered with the respective type strains (Biotyper version 3.0; Fig. 2B and C).
The antimicrobial susceptibility of 11 clinical isolates was tested against a panel of 14 antimicrobial drugs. Individual colonies grown on SBA were incubated in Middlebrook 7H9 broth containing 3-mm-diameter glass beads at 37°C for 48 h and mixed every 8 to 12 h by vortexing. Suspensions were adjusted to a turbidity of 1.0 McFarland, 70 μl was inoculated into 11 ml of cation-adjusted Mueller-Hinton broth, and 100 μl of the resulting dilutions was inoculated into each well of microtiter plates with lyophilized antimicrobials (Sensititre Rapid Growing Mycobacteria Plate [RAPMYCO]; Trek Diagnostic Systems). Plates were incubated at 37°C for 72 h and read according to CLSI guidelines for aerobic actinomycetes (13). MICs were within ±1 dilution for all isolates and were generally low, with the notable exception of clarithromycin (≥32 μg/ml; Table 2).
Table 2.
Antimicrobial susceptibility profiles for 12 clinical isolates of Kroppenstedtia eburneaa
| Isolate | MIC (μg/ml) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T/S | CIP | MXF | AMI | DOX | TGC | CLA | LZD | IMI | FEP | AUG | AXO | MIN | TOB | |
| B | ≤0.25/4.8 | ≤0.12 | ≤0.25 | ≤1 | ≤0.12 | ≤0.015 | ≥32 | ≤1 | ≤2 | 4 | ≤2/1 | ≤4 | ≤1 | ≤1 |
| C | ≤0.25/4.8 | ≤0.12 | ≤0.25 | ≤1 | ≤0.12 | ≤0.015 | ≥32 | ≤1 | ≤2 | 4 | 4/2 | ≤4 | ≤1 | ≤1 |
| D | ≤0.25/4.8 | ≤0.12 | ≤0.25 | ≤1 | ≤0.12 | ≤0.015 | ≥32 | ≤1 | ≤2 | 4 | ≤2/1 | ≤4 | ≤1 | ≤1 |
| E | ≤0.25/4.8 | ≤0.12 | ≤0.25 | ≤1 | ≤0.12 | ≤0.015 | ≥32 | ≤1 | ≤2 | 8 | 4/2 | ≤4 | ≤1 | ≤1 |
| F | ≤0.25/4.8 | ≤0.12 | ≤0.25 | ≤1 | ≤0.12 | ≤0.015 | ≥32 | ≤1 | ≤2 | 8 | 8/4 | 8 | ≤1 | ≤1 |
| G | ≤0.25/4.8 | ≤0.12 | ≤0.25 | ≤1 | ≤0.12 | ≤0.015 | ≥32 | ≤1 | ≤2 | 8 | ≤2/1 | ≤4 | ≤1 | ≤1 |
| H | ≤0.25/4.8 | ≤0.12 | ≤0.25 | ≤1 | ≤0.12 | ≤0.015 | ≥32 | ≤1 | ≤2 | 4 | ≤2/1 | ≤4 | ≤1 | ≤1 |
| I | ≤0.25/4.8 | ≤0.12 | ≤0.25 | ≤1 | ≤0.12 | ≤0.015 | ≥32 | ≤1 | ≤2 | 8 | 4/2 | ≤4 | ≤1 | ≤1 |
| J | ≤0.25/4.8 | ≤0.12 | ≤0.25 | ≤1 | ≤0.12 | ≤0.015 | ≥32 | ≤1 | ≤2 | 8 | 4/2 | ≤4 | ≤1 | ≤1 |
| L | ≤0.25/4.8 | ≤0.12 | ≤0.25 | ≤1 | ≤0.12 | ≤0.015 | ≥32 | ≤1 | ≤2 | 8 | 4/2 | ≤4 | ≤1 | ≤1 |
| M | ≤0.25/4.8 | ≤0.12 | ≤0.25 | ≤1 | ≤0.12 | 0.06 | ≥32 | ≤1 | ≤2 | 8 | 4/2 | ≤4 | ≤1 | ≤1 |
| N | ≤0.25/4.8 | ≤0.12 | ≤0.25 | ≤1 | ≤0.12 | ≤0.015 | ≥32 | ≤1 | ≤2 | 8 | ≤2/1 | ≤4 | ≤1 | ≤1 |
Abbreviations: T/S, trimethoprim-sulfamethoxazole; CLA, clarithromycin; AXO, ceftriaxone; CIP, ciprofloxacin; LZD, linezolid; MIN, minocycline; MXF, moxifloxacin; IMI, imipenem; TOB, tobramycin; AMI, amikacin; DOX, doxycycline; FEP, cefepime; TGC, tigecycline; AUG, amoxicillin-clavulanic acid.
We recently reviewed 16S sequences of clinical isolates that belonged to as-yet-undescribed bacterial taxa (8). Among these were 12 of the 14 isolates presented here, which could retrospectively be identified as K. eburnea. Two additional isolates were subsequently identified. Members of the Thermoactinomycetaceae are rarely isolated from patient specimens and are difficult to identify by conventional phenotypic testing. Their recognition has been facilitated by molecular identification methods, and several novel species have recently been described (1, 2, 5, 12, 14, 15). Here we showed that K. eburnea has been repeatedly isolated from clinical specimens in different geographic regions of the United States. Isolates were readily identified by 16S sequencing and MALDI-TOF mass spectrometry.
Of the 14 isolates, 13 (93%) were recovered from normally sterile body sites (blood, CSF, peritoneal fluid), thus warranting definitive identification. Limited medical histories were available for only 3 patients. Repeated blood cultures in the 2 cases with blood culture isolates failed to reisolate this organism, and antimicrobial treatment was not initiated for any of the 3 patients. Establishing the pathogenic role of the organism in these cases was beyond the scope of this study, and the pathogenic role of these isolates in the three cases presented is unclear.
Little is known about the environmental distribution of K. eburnea. It was first identified by surface sampling in an air grade C area of a contract manufacturing organization in Germany. K. eburnea 16S sequences do not appear as part of human microbiome sequencing surveys in GenBank (data not shown), suggesting that they are unlikely to be common commensals. Thus, their significance and origin in these clinical cases remain unclear. By providing information on isolation and rapid identification of K. eburnea, this report can facilitate its identification in clinical microbiology laboratories and help determine the environmental distribution and pathogenicity of this recently described organism.
ACKNOWLEDGMENTS
This work was supported by the ARUP Institute for Experimental and Clinical Pathology.
We thank the ARUP bacteriology and special microbiology laboratories for technical assistance.
Footnotes
Published ahead of print 1 August 2012
REFERENCES
- 1. Addou AN, et al. 19 August 2011, posting date Melghirimyces algeriensis gen. nov., sp. nov., a member of the family Thermoactinomycetaceae, isolated from a salt lake. Int. J. Syst. Evol. Microbiol. 62(Pt 7):1491–1498 [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 2. Chen JJ, et al. 2012. Laceyella sediminis sp. nov., a thermophilic bacterium isolated from a hot spring in China. Int. J. Syst. Evol. Microbiol. 62:38–42 [DOI] [PubMed] [Google Scholar]
- 3. Hatayama K, Shoun H, Ueda Y, Nakamura A. 2005. Planifilum fimeticola gen. nov., sp. nov. and Planifilum fulgidum sp. nov., novel members of the family “Thermoactinomycetaceae” isolated from compost. Int. J. Syst. Evol. Microbiol. 55(Pt 5):2101–2104 [DOI] [PubMed] [Google Scholar]
- 4. Ismail T, McSharry C, Boyd G. 2006. Extrinsic allergic alveolitis. Respirology 11:262–268 [DOI] [PubMed] [Google Scholar]
- 5. Li J, et al. 2011. Marina mesophila gen. nov., sp. nov., a novel thermoactinomycete isolated from a deep sea sediment, and emended description of the family Thermoactinomycetaceae Matsuo et al. 2006 emend. Yassin et al. 2009, emend. Von Jan et al. Int. J. Syst. Evol. Microbiol. 62:1383–1388 [DOI] [PubMed] [Google Scholar]
- 6. Petti CA, et al. 2008. Interpretive criteria for identification of bacteria and fungi by DNA target sequencing; approved guidelines, vol 28. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 7. Pinto A, et al. 2011. Matrix-assisted laser desorption ionization-time of flight mass spectrometry identification of yeasts is contingent on robust reference spectra. PLoS One 6:e25712 doi:10.1371/journal.pone.0025712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schlaberg R, Simmon KE, Fisher MA. 2012. A systematic approach for discovering novel, clinically relevant bacteria. Emerg. Infect. Dis. 18:422–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Simmon KE, et al. 2008. Phylogenetic analysis of viridans group streptococci causing endocarditis. J. Clin. Microbiol. 46:3087–3090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Suihko ML, et al. 2009. Detection and molecular characterization of filamentous actinobacteria and thermoactinomycetes present in water-damaged building materials. Indoor Air 19:268–277 [DOI] [PubMed] [Google Scholar]
- 11. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 12. von Jan M, et al. 2011. Kroppenstedtia eburnea gen. nov., sp. nov., a thermoactinomycete isolated by environmental screening, and emended description of the family Thermoactinomycetaceae Matsuo et al. 2006 emend. Yassin et al. 2009. Int. J. Syst. Evol. Microbiol. 61(Pt 9):2304–2310 [DOI] [PubMed] [Google Scholar]
- 13. Woods GL, et al. 2011. Susceptibility testing of Mycobacteria, Nocardiae, and other aerobic actinomycetes; approved standard—2nd ed, vol 31. Clinical and Laboratory Standards Institute, Wayne, PA: [PubMed] [Google Scholar]
- 14. Yassin AF, Hupfer H, Klenk HP, Siering C. 2006. 2009 Desmospora activa gen. nov., sp. nov., a thermoactinomycete isolated from sputum of a patient with suspected pulmonary tuberculosis, and emended description of the family Thermoactinomycetaceae Matsuo et al. Int. J. Syst. Evol. Microbiol. 59:454–459 [DOI] [PubMed] [Google Scholar]
- 15. Zhang J, et al. 2010. Laceyella tengchongensis sp. nov., a thermophile isolated from soil of a volcano. Int. J. Syst. Evol. Microbiol. 60:2226–2230 [DOI] [PubMed] [Google Scholar]