Abstract
We conducted a multicenter study to determine whether Mycobacterium tuberculosis complex (MTBC) cultures in automated broth-based systems could reliably be considered negative sooner than 6 weeks. Laboratory sites used Bactec MGIT or BacT/Alert and tracked results of time to detection of all mycobacteria (TTD-all, n = 1547) and of MTBC (TTD-MTBC, n = 466) over 6-month periods from primarily (93%) respiratory specimens. Cumulative percentages by day detected and median TTD of initial and follow-up specimens were analyzed. The median TTD-MTBC for MGIT (n = 6 sites) was 14 days. For laboratories using standard processing procedures, 100% of MTBC were detected from initial and follow-up specimens in 28 and 35 days, respectively, and no yield of MTBC on solid or MGIT liquid media was observed after 5 weeks. The median TTD-MTBC for BacT/Alert (n = 3 sites) was 18 days, with 95% and 100% detected within 37 and 42 days, respectively. Analysis of TTD of positive MTBC cultures in broth can predict the probability of culture negativity at defined time points. Receipt of interim negative reports earlier than 6 weeks could assist clinicians in considering alternative diagnoses and could alter the timing and prioritization of public health interventions. Laboratories should analyze their own TTD data to inform protocol decisions. Laboratories using MGIT could issue reports of no growth of MTBC on initial specimens as early as 4 weeks and for patients undergoing treatment as early as 5 weeks postinoculation.
INTRODUCTION
Rapid diagnosis is critical for initiating effective treatment and establishing infection control measures to prevent ongoing transmission of tuberculosis. The laboratory plays an essential role in this diagnostic process and is responsible for providing rapid turnaround of accurate results (29). Although molecular methods to rapidly identify Mycobacterium tuberculosis complex (MTBC) directly from clinical specimens have become an accepted practice, current testing algorithms still require isolation and identification (ID) after growth in culture (9, 16). Culture remains the most sensitive method for identification of MTBC (33), and an isolate is required for phenotypic drug susceptibility testing and genotyping. However, due to the slow growth of MTBC in culture, patients suspected of having tuberculosis often need to be prescribed empirical treatment before culture and other microbiological results are known (14). Because MTBC grows faster in liquid than on solid media (3, 4, 13), the use of liquid media is the recommended practice (13, 18) for faster isolation and to meet the laboratory turnaround time objectives developed in 1993. These objectives were written to assist laboratories in avoiding testing delays and to help achieve the goal of reduced transmission by providing rapid diagnosis (29).
Although evidence exists that most cultures positive for MTBC occur in the first 1 to 3 weeks of incubation (28, 30, 32), current laboratory guidelines still require holding mycobacterial cultures for 6 weeks as indicated by commercial broth system protocols (5, 6) and up to 8 weeks for solid media (13, 17) before regarding results as negative. Standard practice indicates that when initial acid-fast-bacillus (AFB) smears and cultures are negative, a diagnosis other than tuberculosis should be considered and relevant evaluations undertaken (3). According to American Thoracic Society (ATS) and U.S. Centers for Disease Control and Prevention (CDC) diagnostic standards, patients for whom a diagnosis of tuberculosis disease is being considered should remain in the category “tuberculosis suspect, diagnosis pending” (i.e., class 5), until diagnostic procedures are complete (3). This process could take 2 months, delaying patient placement into a lesser category (e.g., class 2 [latent tuberculosis infection, no disease]). Additionally, the standard practice of waiting 6 to 8 weeks for such a result affects policies governing immigration (11) and foreign-born adoptions (10).
Negative reports could be issued earlier if shorter incubation times were sufficient. Prior studies have focused on the ability of liquid broth culture systems to rapidly detect MTBC (20, 21, 24, 26, 30), but few have addressed the corollary argument concerning the ability to dependably shorten incubation times and release earlier negative reports, thereby altering patient management and influencing public health interventions. We hypothesized that mycobacterial cultures in broth-based systems could reliably be considered negative at less than the historically recommended and accepted (13, 17) 6 to 8 weeks of incubation, especially for initial diagnostic specimens from those patients for whom a definitive diagnosis of tuberculosis has not yet been made and who are not undergoing treatment with anti-tuberculosis drugs. Changes in reporting protocols could result in considerable benefits to patients, clinicians, and laboratories.
MATERIALS AND METHODS
We conducted a multisite analysis of time to detection in days for cultures from specimens positive for any mycobacteria (TTD-all) and for cultures from specimens positive for MTBC (TTD-MTBC) by the use of Bactec MGIT 960 (Becton, Dickinson, Cockeysville, MD) (MGIT) and BacT/Alert (bioMérieux, Inc., Durham, NC) (BacT/Alert) commercial automated broth-based systems. The analyses included a review of incubation lengths to determine frequency and type of mycobacteria isolated and characterization of isolates that grew later than 28 days postinoculation.
Site selection.
To be considered for participation, laboratories had to perform AFB smears and mycobacterial culture using the MGIT or the BacT/Alert commercial automated broth system, perform species identification (ID) of mycobacteria, and perform an average of at least 10 mycobacterial cultures per week. Additional selection criteria included the ability to track or retrieve the date of inoculation and placement of broth cultures on the instrument, the date of positive signal by the instrument, the date a signal-positive broth culture was removed from the instrument, and the date of final ID, or disposition, of all signal-positive broth cultures. Site-specific information was collected, including testing volumes, specimen processing methods, protocols for reprocessing contaminated liquid broths, and standard lengths of incubation for solid and liquid media. Laboratories were excluded if they did not use liquid broth methods or were unable to collect data as described. Nine sites (five public health and four hospital-based laboratories) were included in the analysis. Six laboratories used the MGIT system for mycobacterial culture; three sites used BacT/Alert. For 6-month periods between January 2009 and February 2011, sites kept detailed culture records. Two sites recorded data prospectively, and seven sites retrieved data from laboratory records for all specimens received for mycobacterial culture that did not produce a “no-growth” result at the end of the incubation time (i.e., that produced a final positive culture result). Following data collection and preliminary analysis, a meeting of participants was held to discuss findings and their implications for laboratory practice.
Laboratory methods.
Specimens were processed in each participating laboratory following standard procedures for decontamination and digestion of specimens, AFB-smear microscopy, culture, and ID (13, 17, 18). For MGIT, one mycobacterial growth indicator tube was prepared for each resuspended sample pellet by inoculating 0.5 ml of the sample into the tube, which was then incubated at 37°C in an automated MGIT instrument and monitored continuously. For BacT/Alert, one bottle was prepared for each resuspended sample pellet by inoculating 0.5 ml of the processed sample into the bottle, which was then incubated at 37°C in the BacT/Alert instrument and monitored continuously. The nine laboratories used various smear-grading protocols. One of the laboratories (site K) did not use solid media in the initial culture setup. One site used a processing procedure that differed in the sequence of steps from the commonly accepted standard processing procedure (17) (site B). Liquid cultures were incubated in the respective automated instruments for the time period indicated in either the manufacturer's package inserts or the institution's validated protocols according to specimen type or until a positive signal was seen, whichever occurred first. Signal-positive broths were removed and the date was recorded, and then the tube or bottle was processed for identification according to each site's laboratory protocol. Signal-negative broths were removed, examined, and discarded at the end of the incubation period according to each site's protocol. Signal-positive broths that gave AFB smear-negative results were treated according to the policy in place for each site. Site characteristics are displayed in Table 1.
Table 1.
Site characteristics
| System | Site code | Site typea | No. of beds | Protocol for length of incubation of media |
No. (% of total) of specimens cultured | No. (% of total) of positive culture results |
MTBC recovery rate (%)b | MTBC smear sensitivity (%)c | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Broth (days) | Solid (wk) | All mycobacteria | MTBC | |||||||
| MGIT | ||||||||||
| B | PH | 42 | 8 | 2,793 (14.5) | 263 (17.0) | 123 (26.4) | 4.4 | 74.0 | ||
| D | Hosp | 477 | 42 | 6 | 1,143 (5.9) | 103 (6.7) | 51 (10.9) | 4.5 | 33.3 | |
| F | PH | 42 | 6 | 1,777 (9.2) | 91 (5.9) | 35 (7.5) | 2.0 | 60.0 | ||
| H | Hosp | 1,000 | 42 | 6 | 2,762 (14.3) | 146 (9.4) | 19 (4.1) | 0.7 | 31.6 | |
| K | PH | 42 | n/ae | 2,000 (10.4) | 171 (11.1) | 80 (17.2) | 4.0 | 48.8 | ||
| L | PH | 42 | 8 | 1,524 (7.9) | 135 (8.7) | 70 (15.0) | 4.6 | 78.6 | ||
| Total MGIT | 11,999 (62.3) | 909 (58.8) | 378 (81.8) | 3.2 | 60.6 | |||||
| BacT/Alert | ||||||||||
| C | PH | 42 | 6 | 471 (2.4) | 27 (1.7) | 10 (2.1) | 2.1 | 60.0 | ||
| G | Hosp | 1,000 | 42 | 5 | 3,963 (20.6) | 472 (30.5) | 74 (15.9) | 1.9 | 58.0 | |
| M | Hosp | 866 | 42+d | 8 | 2,842 (14.7) | 139 (9.0) | 4 (0.9) | 0.1 | 0.0 | |
| Total BacT/Alert | 7,276 (37.7) | 638 (41.2) | 88 (18.9) | 1.2 | 56.8 | |||||
| Overall total | 19,275 | 1,547 | 466 | 2.4 | 59.9 | |||||
PH, public health laboratory; Hosp, hospital-based laboratory.
Percent positive for MTBC of specimens cultured.
Percent of MTBC final culture results that were AFB smear positive.
Held 56 days for blood specimens (up to 62 days under special circumstances).
n/a, site does not use solid media.
Data collection.
Each participating laboratory was assigned a 6-month data collection period over which to retrieve or record data for all variables included in the study for all positive culture results. This time frame was determined to be sufficiently long to achieve a combined sample size of at least 10,000 total culture specimens and yet short enough for data collection to be manageable by each participating laboratory. Data collection periods were staggered to reduce potential seasonal variability in mycobacterial positivity rates. Accession numbers were removed prior to submission, and specimens were made anonymous by generation of a code number for each specimen. Personal identifying information was not collected for the study. Data were recorded for each specimen that did not generate a no-growth result (e.g., positive growth or contamination) by the end of incubation during the 6-month data collection period. Dates of specimen receipt, inoculation, positive signal on instrument of broth, recovery on solid media if known, and generation of the final report, as well as AFB smear result, specimen type, broth result (identification of growth from a signal-positive broth), and final culture result (final report of identification of growth from either broth or solid media), were collected. For any culture positive for MTBC that signaled in broth in more than 28 days, and for cultures positive for MTBC growing on solid media that were not detected in broth, further data were collected, including whether the specimen was an initial specimen (defined as the first known or one of a first set of patient specimens to be processed in the laboratory) or a follow-up specimen (defined as a subsequent specimen from a patient with a culture previously identified in the laboratory as positive for MTBC). Not all sites tracked dates of recovery on solid media for every specimen. Once data collection was completed at each site, data were transmitted to the principal investigator using a secure file transfer protocol. Details concerning the data collection instruments are available on request.
Data analysis.
SPSS version 18.0 (IBM, Armonk, NY) was used for all data analysis. Time to detection in broth in days (TTD) was calculated by subtracting the date of specimen inoculation from the date of broth signal positivity and determined for MTBC recovered (TTD-MTBC) and for all mycobacteria recovered (TTD-all). Median TTD-all and TTD-MTBC were determined for each site individually and overall for each culture system used. The cumulative percentages of TTD were determined for all mycobacterial species and for MTBC. For TTD-MTBC cumulative percent determinations, analysis included TTD of MTBC for all specimens and for MTBC with follow-up specimens excluded. Because TTD values were not normally distributed, nonparametric analysis was employed using an independent-sample median test for comparing TTD-MTBC and TTD-all between the two culture systems used, comparing TTD-MTBC and TTD-all specimens from pulmonary versus nonpulmonary sites, comparing TTD-MTBC and TTD-all smear-positive to smear-negative specimens, and for TTD comparisons among sites. P values of less than 0.05 were considered statistically significant. This study was determined to not be human subject research by the CDC National Center for HIV/AIDS, Viral Hepatitis, STD, and TB Prevention, as defined by Code of Federal Regulations (CFR) 45, part 46.
RESULTS
All sites.
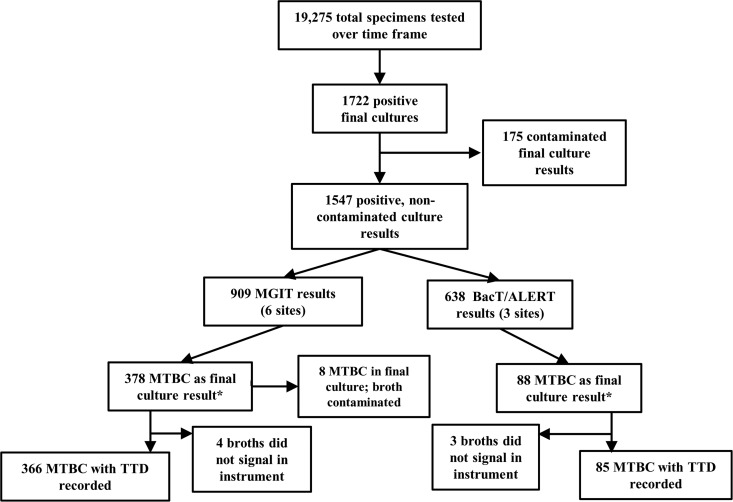
A total of 19,275 specimens for mycobacterial culture across the nine sites during the data collection period were received; of those specimens, 1,722 (8.9%) had a reported final positive or contaminated-culture result either in liquid or on solid media. Reported final culture results were divided into categories that consisted of 594 M. avium complex (34%), 466 MTBC (27%), 141 M. gordonae (8%), 329 other mycobacteria (19%), 175 contaminated (10%), 9 nonmycobacterial (e.g., Actinomyces sp.) (0.6%), and 8 nonviable (i.e., specimen AFB smear positive but no growth on media) (0.5%). Of the 1,722 culture-positive results, 175 had a final result of “contaminated,” defined either as any growth other than AFB or as a culture result from both liquid and solid media that was uninterpretable due to contaminating overgrowth. The majority of these (150 [86%]) were from one site, and because the TTD-all for overall final culture results was significantly different for this site when contaminated results were included, contaminated results were excluded from the overall analysis of TTD-all. A final sample size of 1,547 positive, noncontaminated culture results (8.0% overall mycobacterial recovery rate) was used for analysis of TTD-all and TTD-MTBC (Fig. 1). The overall MTBC recovery rate (n = 466) was 2.4%.
Fig 1.
Characterization of the numbers of specimens submitted for culture and results submitted to the study over a 6-month period by automated culture system used. *, final culture result, MTBC detected in broth or solid media.
(i) Specimen characteristics.
Personnel at each of the sites were asked to characterize each specimen by type (i.e., anatomic site) and to record the original AFB smear results. Specimens examined by culture included 1,234 sputa (80%), 188 bronchial washes (12%), 46 tissue samples (3%), 13 stool samples (0.8%), 11 sterile-fluid samples (0.7%), 5 urine specimens (0.5%), 5 wound specimens (0.5%), 21 other nonpulmonary specimens (1.4%), and 19 specimens from other pulmonary sources (1.2%). For TTD data analysis, specimens were categorized as either pulmonary or nonpulmonary; pulmonary specimens accounted for 93% of all specimens. Because the grading scales and subsequent assignments of results for AFB smears differed among the laboratories in this study, AFB smear results were categorized into smear-positive and smear-negative categories only. Equivocal or indeterminate smear results were categorized in the smear-negative category. Of the 1,547 final positive culture results, 1,071 were smear negative (69%) and 473 (31%) were smear positive (3 missing results). In our study, sensitivities of the AFB smear analysis for MTBC, defined as the proportion of MTBC with smear-positive results in the final culture (61% for MGIT and 57% for BacT/Alert [P = 0.465], for an overall smear sensitivity of 60%), were not statistically different between systems. However, smear sensitivities differed considerably across sites, ranging from 32% to 79% in sites using MGIT and 0% to 60% in those using BacT/Alert. Table 1 summarizes the site characteristics of participating laboratories by system used.
(ii) TTD differences.
Medians for TTD for all species (TTD-all) and TTD for MTBC (TTD-MTBC) were significantly different for smear-positive and smear-negative specimens, with 13 days for smear-positive versus 17 days for smear-negative specimens for TTD-MTBC (P < 0.01) and 11 days for smear-positive versus 16 days for smear-negative specimens for TTD-all (P < 0.01). TTD data were not significantly different for comparisons of specimen types (pulmonary versus nonpulmonary), with 14 days versus 15 days, respectively, for TTD-all (P = 0.166) and 14 days versus 16.5 days for TTD-MTBC (P = 0.070). Medians for TTD for all species (TTD-all) and TTD for MTBC (TTD-MTBC) were significantly different between the two culture systems, with 16 versus 13 days (P < 0.001) for TTD-all and 18 versus 14 days (P = 0.005) for TTD-MTBC in the BacT/Alert and MGIT systems, respectively. Because of this difference, all further TTD analyses were conducted separately for each culture system.
MGIT sites.
Table 2 displays a summary of results for time to detection of mycobacteria and cumulative percentage of MTBC detected in MGIT. For sites using the MGIT system, overall median TTD-all was 13 days (interquartile range [IQR], 7 to 19), and for MTBC, the median TTD-MTBC was 14 days (IQR, 10 to 18). At the end of 3, 4, and 5 weeks of incubation, MTBC had been detected in broth from all specimens at levels of 88%, 94%, and 99%, respectively (n = 366). With follow-up specimens excluded, median TTD-MTBC for initial specimens was 13 days (IQR, 9 to 17), and at the end of 4 and 5 weeks of incubation, MTBC had been detected in 99.4% and 100%, respectively (n = 348) (Fig. 2). Of the 366 specimens that were culture positive for MTBC in MGIT, only 21 (5.7%) (13 of which were from site B) were detected beyond 28 days and only 2 were initial specimens, detected in broth in 31 and 32 days. Both of those two specimens had MTBC recovered from solid media in less than 28 days, yielding 100% detection of MTBC from initial specimens within 28 days.
Table 2.
Summary of results of determinations of time to detection in days and percent detected by week for MTBC and all mycobacteria and late-growing MTBC by culture system used
| Organism | System | Median TTD (IQR)a |
% detected by end of wkb: |
No. (%) recovered |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 4 |
5 |
|||||||||
| All | Smear positive | Smear negative | All specimens | Initial specimens | All specimens | Initial specimens | >28 days | >35 days | ||
| MTBC | MGIT (366) | 14 (10–18) | 12 (8–16) | 16 (13–21) | 94.4 | 99.4 | 98.9 | 100 | 21(5.7) | 4 (1.1) |
| BacT/Alert (85) | 18 (11–27.5) | 14 (9–19) | 26 (15.75–32) | 80.0 | 89.2 | 90.6 | 94.6 | 17 (20.0) | 8 (9.0) | |
| All mycobacteria | MGIT (852) | 13(7–19) | 11 (6–15) | 14 (8–21) | 91.6 | 97.6 | ||||
| BacT/Alert (580) | 16 (13–22.75) | 11 (8–17) | 17 (14–24) | 87.6 | 95.2 | |||||
TDD, time to detection in days; IQR, interquartile range.
For “All specimens” data, follow-up specimens, initial specimens, and undifferentiated specimens (designated for MTBC only) were combined. For “Initial specimens” data, known follow-up specimens (designated only for MTBC detected at >28 days) were excluded from analysis.
Fig 2.
Cumulative percentages of TTD-MTBC with and without the inclusion of follow-up specimens for each culture system. At the end of 4 weeks of incubation, using the MGIT system, 99.4% of MTBC from initial specimens had been detected. In contrast, only 89% of MTBC had been detected by this time in the BacT/Alert system.
Overall, in MGIT, a total of 20 cultures were positive after 5 weeks (35 days) of incubation; 9 grew M. gordonae, 6 grew other mycobacteria, 4 grew MTBC, and 1 grew M. avium. Of the four cultures that grew MTBC in greater than 5 weeks in MGIT, all were from follow-up specimens from one laboratory site (site B), three were smear positive, and three grew on solid media in less than 5 weeks. In MGIT, the median TTD-MTBC for smear-positive specimens (n = 229) was 12 days (IQR, 8 to 15.5) compared to 16 days (IQR, 13 to 20) for smear-negative specimens (n = 149, P = 0.005). Only six initial specimens that were AFB smear positive grew in greater than 28 days in MGIT, and only one of these grew MTBC, which was detected using solid media in less than 28 days (Table 3).
Table 3.
Smear status and number of MTBC and NTM detected from initial specimens in ≤28 days and >28 days by MGIT and BacT/Alerta
| System used | Smear status | No. of specimens detected |
|||
|---|---|---|---|---|---|
| All |
>28 days, NTM | >28 days, MTBC | |||
| ≤28 days | >28 days | ||||
| MGIT | |||||
| + (n = 315) | 309 | 6 | 5 | 1b | |
| − (n = 510) | 464 | 46 | 45 | 1 | |
| BacT/Alert | |||||
| + (n = 117) | 114 | 3 | 2 | 1 | |
| − (n = 444) | 386 | 58 | 51 | 7 | |
“Initial specimens” refers to known follow-up specimens excluded from analysis.
This MTBC was recovered in <28 days on solid media.
Among the six laboratory sites using MGIT, the range of medians for TTD-MTBC was 11 to 16 days (Table 4). Site D had the lowest overall median TTD of MTBC, site B had the highest, and all others were at or lower than the overall median. There was a significant difference in median TTD across laboratory sites (P = 0.024) but only between sites D and B after post hoc pairwise analysis (P = 0.044).
Table 4.
Comparison of results of determinations of time to detection in days (all, MTBC, late-growing MTBC, and MTBC recovered on solid media only by laboratory site)
| System and site code | Median TTD (IQR)a |
No. MTBC recovered in: |
||||
|---|---|---|---|---|---|---|
| All mycobacteria | MTBC |
|||||
| Smear positive | Smear negative | All | Solid media onlyc | Broth (>5 wk) | ||
| MGIT | ||||||
| B | 13 (7.5–20.5) | 15 (11–19) | 21 (15–28) | 16 (12–20) | 3 | 4 |
| D | 9 (4–15) | 6 (4–9) | 13.5 (10.25–18) | 11 (5–15) | 0 | 0 |
| F | 14 (7.5–20) | 13 (9–15.5) | 15 (12.5–21.5) | 14 (10.75–16.5) | 1 | 0 |
| H | 11 (7–16.5) | 8 (7.75–9.25) | 15 (11.5–20.5) | 13 (9–17) | 0 | 0 |
| K | 14 (9–20) | 11 (9–14) | 16 (14–21) | 14 (11–18) | 0 | 0 |
| L | 12 (6–16) | 12 (7–15) | 16 (13–21) | 12.5 (7.75–16) | 0 | 0 |
| Total MGIT | 13 (7–19) | 12 (8–16) | 16 (13–21) | 14 (10–18) | 4 | 4 |
| BacT/Alert | ||||||
| C | 21.5 (6–35) | 30 (20.5–32) | 28 (25–31) | 28 (23.5–31.5) | 1 | 0 |
| G | 17.5 (4–47) | 14 (8.75–18.25) | 25 (14–34.5) | 16 (10–25.75) | 2 | 8 |
| M | 19.5 (2–62) | n/ab | 23 (16.5–30.5) | 23 (16.25–30.5) | 0 | 0 |
| Total BacT/Alert | 16 (13–22.75) | 14 (9–19) | 26 (15.75–32) | 18 (11–27.5) | 3 | 8 |
TDD, time to detection in days; IQR, interquartile range.
n/a, site had no smear-positive MTBC.
Broth did not signal by end of incubation period.
Inoculation of solid media in the culture setup process was practiced by five of the six MGIT sites. In addition to the 366 specimens culture positive for MTBC in broth, 12 additional specimens resulted in an MTBC-positive culture only after growth on solid media, yielding an overall MGIT sensitivity for MTBC of 96.8%. Four of the 12 (3 from site B, 1 from site F) were follow-up specimens that did not signal in MGIT broth by the end of the incubation period but grew on solid media only; all were smear negative. The presence of MTBC in the specimen from site F was detected on Lowenstein-Jensen (LJ) medium at 24 days; the three from site B were detected between 40 and 43 days on LJ media. Additionally, eight MTBC specimens (all from site D) reported in a final culture were not detected in broth; these specimens gave positive signals but grew only nonmycobacteria (i.e., contamination) in broth and later grew MTBC on solid media. The range of time to recovery of MTBC on solid media for these eight was 16 to 28 days. Because MTBC was not detected in broth, these 12 specimens were not included in the analysis of TTD-MTBC. Among MGIT sites, of the 378 MTBC in final culture, two (0.5%) MTBC-positive cultures which did not signal in MGIT were recovered after 6 weeks of incubation on solid media. Of 516 final cultures identified as harboring nontuberculous mycobacteria (NTM) from MGIT sites, 4 (0.75%) that had not already been recovered in MGIT broth grew on solid media beyond 6 weeks. All of these were from site B (Table 5).
Table 5.
Number and percent of MTBC and NTM recovered on solid media in greater than 6 weeks
| Organism and system used (no. of specimens) | No. (%) of specimens detected in indicated media at: |
||
|---|---|---|---|
| >6 wk (solid) | ≤6 wk (broth) | >6 wka | |
| MTBC (466) | |||
| MGIT (378) | 9 (2.4) | 7 (1.8) | 2b (0.5) |
| BacT/Alert (88) | 2 (2.2) | 1 (1.1) | 1 (1.1) |
| Total MTBC | 11 (2.3) | 8 (1.7) | 3 (0.6) |
| NTM (1,064) | |||
| M. avium (594) | |||
| MGIT (216) | 15 (6.9) | 11 (5.1) | 4b (1.9) |
| BacT/Alert (378) | 1 (0.3) | 1 (0.3) | 0 |
| Other mycobacteria (470) | |||
| MGIT (300) | 3 (1.0) | 3 (1.0) | 0 |
| BacT/Alert (170) | 0 | 0 | 0 |
| Total NTM | 19 (1.8) | 15 (1.4) | 4 (0.1) |
Data represent specimens not detected in broth.
Data are from site B.
BacT/Alert sites.
For sites using BacT/Alert, the overall median TTD-all was 16 days (IQR, 13 to 22.75; n = 580) and the median TTD-MTBC was 18 days (IQR, 11 to 27.5). At the end of 3, 4, and 5 weeks of incubation, MTBC in broth had been detected from all specimens at levels of 60%, 80%, and 91%, respectively (n = 85) (Table 2). With follow-up specimens excluded, the median TTD-MTBC for initial specimens was 16.5 days, and at the end of 4 and 5 weeks of incubation, MTBC had been detected at levels of 89% and 95%, respectively (n = 74). (Fig. 2). Of the 85 specimens culture positive for MTBC in BacT/Alert, 17 (20%) were detected in greater than 28 days. Four of these 17 were initial specimens, 11 were follow-up specimens, and the status of 2 was unknown. The median TTD of these was 34 days; the range was 29 to 43, and 8 signaled positive beyond 35 days.
Growth after 5 weeks (35 days) of incubation in BacT/Alert yielded 28 isolates, including 8 isolates of MTBC, 11 isolates of M. avium, 6 isolates of M. gordonae, and 3 isolates of other mycobacteria. Of the eight MTBC isolates detected in greater than 5 weeks, 100% were from site G, which accounted for 84% of all MTBC isolates collected in BacT/Alert sites. In the BacT/Alert results, the median TTD-MTBC for smear-positive specimens was 13 days (IQR, 8.5 to 18; n = 49) compared to 24 days for smear-negative specimens (IQR, 15.25 to 28; n = 38; P = 0.005). Only three initial specimens that were AFB smear positive grew in greater than 28 days, and only one of these grew MTBC (Table 3).
Among the three BacT/Alert sites, the range of medians for TTD-MTBC was 16 to 28 days (Table 4). Overall, across the BacT/Alert sites, there was a significant difference in median TTD among laboratories (P = 0.028) but only between sites C and G after post hoc pairwise analysis (P = 0.022). Inoculation of solid media in the culture setup process was practiced by all BacT/Alert users. Three specimens resulted in an MTBC-positive culture only after growth on solid media, yielding an overall sensitivity of 96.5% for MTBC detection by BacT/Alert (85/88). Of the three MTBC specimens that never signaled in the BacT/Alert system, one was a urine specimen detected on solid media in 33 days and two were from sputum: one of the two was a follow-up specimen which grew on solid media in 52 days and the other was a specimen which grew on solid media in 10 days (not identified as a follow-up or initial specimen [data missing]). Of the 88 MTBC final cultures in BacT/Alert sites, 2 (1 of which had already been detected in broth) grew on solid media beyond 6 weeks, leaving only 1 MTBC culture recovered after 6 weeks of incubation (1.1%). Of 548 NTM final cultures, only 1 grew on solid media in greater than 6 weeks, and that had already been detected in broth at 36 days (Table 5).
DISCUSSION
In this multicenter study, we determined that mycobacterial cultures in automated broth systems could reliably be identified as negative for MTBC earlier than 6 weeks. Based on data from MGIT users in this study, this determination can be made with 100% confidence for both initial and follow-up specimens at the end of 5 weeks of incubation and with 100% confidence for initial specimens at the end of 4 weeks, provided standard processing procedures (17) are used. Using data from BacT/Alert system users in this study, this determination can be made with 95% confidence at the end of 5 weeks of incubation and with 89% confidence at the end of 4 weeks of incubation for initial diagnostic specimens. To our knowledge, this work represents the first evaluation of the probability of culture negativity based on TTD of positive cultures for the MGIT and BacT/Alert systems, which currently are the two automated broth-based mycobacterial culture systems most commonly used in the United States (Association of Public Health Laboratories TB Laboratory Services Survey, 2011, unpublished data). Our results correspond to previous studies examining TTD of MTBC using the Bactec 12B radiometric system and indicating that incubation lengths in liquid broth media could be shortened to 5 weeks (28; B. Metchock, meeting, Grady Memorial Hospital and Emory Medical School, 1995). We obtained similar results in our study, as the only MTBC species recovered in MGIT after 5 weeks of incubation were from follow-up specimens from one laboratory site which used a processing procedure that differed from those used by the other sites. Our study also looked at potential differences in TTD between specimens considered to be initial diagnostic specimens and those considered to be follow-up specimens, based on the laboratory's knowledge of previous specimens. In reality, and possibly in our study, the laboratory may be unaware of the status of previous specimens processed elsewhere, so the designation we apply here as “initial” may not always correspond to the initial diagnostic specimen for a given patient. However, even though excluding those specimens designated follow-up specimens yielded a subset of data (designated initial specimens) that may have included both initial diagnostic and follow-up specimens from patients, the potential inclusion of specimens from patients on treatment would not detract from the strength of the probability of detecting virtually all MTBC-positive isolates from nontreated patients in 28 days or less.
Among MGIT sites, the TTD-MTBC for site B was the longest and was longer than that for sites in previous MGIT system studies (13, 16, 17, 29, 35), and site B was the only site to recover MTBC at more than 5 weeks. This may be explained by the different processing procedure used by site B, in which a portion of the processed pellet was removed for performing the AFB smear, prior to reconstituting and mixing the pellet with buffer for inoculation of media, which may have led to fewer organisms being available for culture. All other sites reconstituted and mixed the entire pellet for smear and inoculation. However, other factors may have contributed to this site's longer TTD-MTBC, such as not rejecting sputum specimens based on volume and the proportion of specimens from patients on anti-tuberculosis treatment for extended periods of time. Site D had the shortest TTD-MTBC and yet recovered more MTBC exclusively on solid media. This site reports that it experiences high contamination rates possibly due to specimen delivery delays. This could have caused some MGIT tubes to signal positive due to bacterial contamination, with the MTBC in these subsequently being recovered on solid media only. It is plausible that MTBC was growing in those tubes with its presence masked by the contaminating bacteria. Since patient data were not collected in this study, there is insufficient information to definitively explain the TTD differences in these two sites. Additional studies incorporating patient characteristics with TTD data would be of value in elucidating TTD disparities.
Our findings of the ability of the MGIT system to more rapidly detect mycobacteria than the BacT/Alert system correspond to those in other studies that compared the two (19, 27, 31, 34). Other investigators have suggested that the addition of vancomycin to the BacT/Alert antibiotic supplement (19) or differences in the detection mechanisms (34) may be partially responsible. Additional findings from our study revealed the high sensitivities of the two broth systems for detection of MTBC, which were comparable to those reported by others evaluating these systems (1, 12, 23, 24, 27, 34). However, none of those studies differentiated initial diagnostic from follow-up specimens. In our study, among MGIT sites, only follow-up specimens were found to have grown on solid media exclusively, and in BacT/Alert sites, only one initial specimen grew MTBC exclusively on solid media. While the high sensitivity of both culture systems in our study revealed the relatively low value of using solid media in the initial setup of mycobacterial cultures to detect MTBC in specimens from pulmonary sites, a decision on whether to exclude solid media from the setup process should be made by a laboratory only after analysis of its own data and careful consideration of several criteria, such as specimen type, contamination rates, and incidence and clinical significance of NTM infections. Our study also demonstrates the lack of evidence to support the practice of holding solid media for mycobacterial cultures beyond 6 weeks. Laboratories that routinely hold mycobacterial cultures for 8 weeks before issuing negative results should examine their own data to determine if there is a compelling reason to continue this practice.
Our data indicate that, using the MGIT system, and standard processing procedures, for a given initial diagnostic specimen, there is 100% probability that if the culture is negative at 28 days, the specimen will be final culture negative for MTBC (at 6 weeks). For a clinician considering a differential diagnosis for a new patient being evaluated for tuberculosis, receiving a report with this information could help to guide patient management decisions, as treatment may be begun prior to culture results being completed (2). One benefit to rapidly excluding tuberculosis in evaluation of these patients is the potential to lessen possible adverse anti-tuberculosis treatment effects, which include hepatotoxicity (15, 22, 25, 35). Using ATS/CDC guidelines (3), patients may be considered tuberculosis suspects on the basis of a positive AFB smear result, and such patients are subsequently started on a four-drug regimen while awaiting final culture results. In our study, among MGIT sites, all smear positives from initial specimens detected beyond 28 days of incubation were NTM. These data may also be applicable to tuberculosis programs to decide when to conduct contact investigations for tuberculosis suspects (7). This decision is often contingent on pending culture results, as recommended by CDC instructional materials (8), and if the culture is not MTBC and clinical tuberculosis is ruled out, the investigation can be stopped, allowing a prioritization of scarce public health resources. While our data suggest that a shortened routine incubation length in liquid media may be sufficient to recover MTBC, there are other considerations in determining whether to hold broth cultures up to 6 weeks. These may include cultures from patients who are on antimycobacterial therapy (i.e., follow-up specimens), cultures from patients who may be suspected of having disease caused by a clinically significant NTM species, cultures from sites other than pulmonary sites (28), cultures from patients who have drug-resistant disease or from geographical areas experiencing high levels of drug-resistant tuberculosis, or cultures incubated using systems other than MGIT (e.g., BacT/Alert). Importantly, because nonpulmonary sites accounted for less than 7% of our overall sample size, there are not enough data to apply our findings to body sites other than pulmonary sites. Because we did not collect patient data, the added value of the increased probability of a patient being shown to be negative for MTBC with two or three additional specimens or as a consequence of using combined testing methods such as nucleic acid amplification assays was not determined in our study and warrants further investigation. Additional studies incorporating TTD with patient information, drug resistance rates, and clinical outcome data could contribute greatly to the utility of analyzing TTD for patient management and are necessary for the potential application of these findings to clinical practice.
In light of these limitations, and because changing the protocols for length of incubation may not be practical for laboratories due to regulatory and validation issues, it may be more feasible for laboratories to use these findings to alter reporting practices. Many laboratories do not issue preliminary “no-growth” results for mycobacterial cultures. Our data demonstrate the potential value of issuing such reports. Preliminary no-growth reports after 4 and 5 weeks of incubation with wording to inform providers of the high probability of a final culture-negative result could be issued, and at the discretion of each laboratory, final reports could then be issued at a later date. In the event that subsequent growth occurred, a laboratory could issue a “critical value” report.
In conclusion, based on our findings, for a laboratory following standard processing procedures using the MGIT system, a preliminary report of no growth of MTBC could be generated at the end of 4 weeks of incubation for specimens from untreated patients and at 5 weeks for follow-up specimens from treated patients, with 100% confidence of a final negative result. Laboratories should collect and analyze TTD data in their own facilities to determine the optimal date to issue preliminary reports and should tailor their reporting language to indicate the probability of a final negative result based on their TTD data and on the designation of initial or follow-up specimens. Time to detection of mycobacterial cultures is a straightforward measurement that laboratories could undertake with little added burden to workload. Further evaluation of the utility of TTD data, along with its analysis and application to laboratory practices, could contribute to substantial clinical and public health impacts.
ACKNOWLEDGMENTS
We acknowledge Beverly Metchock for assistance with study design and manuscript review, Michael P. Chen for statistical analysis, and Kim Dionne, Tim Drake, Amy Fothergill, Hiba Ahmad, and Wan-hsuan Kou for data collection assistance.
The findings and conclusions in this report are ours and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Footnotes
Published ahead of print 25 July 2012
REFERENCES
- 1. Alcaide F, Benitez MA, Escriba JM, Martin R. 2000. Evaluation of the BACTEC MGIT 960 and the MB/BacT Systems for recovery of mycobacteria from clinical specimens and for species identification by DNA AccuProbe. J. Clin. Microbiol. 38:398–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. American Thoracic Society Centers for Disease Control and Infectious Diseases Society of America 2003. Treatment of tuberculosis. Morb. Mortal. Wkly. Rep. 52(RR11):1–77 [Google Scholar]
- 3. American Thoracic Society Centers for Disease Control and Prevention and Infectious Diseases Society of America 2000. Diagnostic standards and classification of tuberculosis in adults and children. Am. J. Respir. Crit. Care Med. 161(Pt 1):1376–1395 [DOI] [PubMed] [Google Scholar]
- 4. Association of Public Health Laboratories 2009. Mycobacterium tuberculosis: assessing your laboratory. Association of Public Health Laboratories, Silver Spring, MD [Google Scholar]
- 5. Becton Dickinson 2010. BBL MGIT 7-ml tube package insert. Becton Dickinson and Company, Sparks, MD [Google Scholar]
- 6. bioMerieux 2008. BacT/ALERT MP package insert, p 1 bioMerieux, Durham, NC [Google Scholar]
- 7. Centers for Disease Control and Prevention 2011. Chapter 8, community tuberculosis control, p 227 Core curriculum on tuberculosis: what the clinician should know, 5th ed CDC, Atlanta, GA [Google Scholar]
- 8. Centers for Disease Control and Prevention 1999. Self study module 6: contact investigations for tuberculosis. CDC, Atlanta, GA [Google Scholar]
- 9. Centers for Disease Control and Prevention 2009. Updated guidelines for the use of nucleic acid amplification tests in the diagnosis of tuberculosis. MMWR Morb. Mortal. Wkly. Rep. 58:7–10 [PubMed] [Google Scholar]
- 10. Centers for Disease Control and Prevention Division of Global Migration and Quarantine 2009. International adoption: health guidance and the immigration process. CDC, Atlanta, GA [Google Scholar]
- 11. Centers for Disease Control and Prevention Division of Global Migration and Quarantine 2009. Technical instructions for tuberculosis screening and treatment, p 7 In Technical instructions for panel physicians. CDC, Atlanta, GA [Google Scholar]
- 12. Chien HP, Yu MC, Wu MH, Lin TP, Luh KT. 2000. Comparison of the BACTEC MGIT 960 with Löwenstein-Jensen medium for recovery of mycobacteria from clinical specimens. Int. J. Tuberc. Lung Dis. 4:866–870 [PubMed] [Google Scholar]
- 13. CLSI 2008. Laboratory detection and identification of mycobacteria; approved guideline. CLSI document 48-A. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 14. Davis JL, et al. 2011. The clinical and public health impact of automated nucleic acid testing for tuberculosis evaluation in San Francisco. Am. J. Respir. Crit. Care Med. 183:A5314 http://ajrccm.atsjournals.org/search?submit=yes&fulltext=A5314&sendit=Enter&volume=183&issue=1+MeetingAbstracts&flag=MTG_ABSTRACT_ARTICLE&journalcode=ajrccm [Google Scholar]
- 15. Forget EJ, Menzies D. 2006. Anti-infectives—adverse reactions to first-line antituberculosis drugs. Expert Opin. Drug Saf. 5:231–249 [DOI] [PubMed] [Google Scholar]
- 16. Gen-Probe 2001. Amplified Mycobacterium Tuberculosis Direct Test package insert, p 1 Gen-Probe, San Diego, CA [Google Scholar]
- 17. Kent P, Kubica G. 1985. Public health mycobacteriology: a guide for the level III laboratory. U.S. Department of Health and Human Services, Atlanta, GA [Google Scholar]
- 17a. Metchock B. 1995. Evaluation of length of incubation of negative BACTEC 12B cultures. Meeting of the American Society for Microbiology, 21–25 May 1995, Washington, DC http://www.newsrx.com/newsletters/TB-and-Outbreaks-Week/1995-07-24/072495767124292TW.html [Google Scholar]
- 18. Murray PR, et al. (ed). 2007. Manual of clinical microbiology, 9th ed, vol 1 ASM Press, Washington, DC [Google Scholar]
- 19. Parrish N, Dionne K, Sweeney A, Hedgepeth A, Carroll K. 2009. Differences in time to detection and recovery of Mycobacterium spp. between the MGIT 960 and the BacT/ALERT MB automated culture systems. Diagn. Microbiol. Infect. Dis. 63:342–345 [DOI] [PubMed] [Google Scholar]
- 20. Pfyffer G, Cieslak C, Welscher H, Kissling P, Rusch-Gerdes S. 1997. Rapid detection of mycobacteria in clinical specimens by using the automated BACTEC 9000 MB system and comparison with radiometric and solid-culture systems. J. Clin. Microbiol. 35:2229–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Piersimoni C, et al. 2001. Comparison of MB/BacT ALERT 3D system with radiometric BACTEC system and Lowenstein-Jensen medium for recovery and identification of mycobacteria from clinical specimens: a multicenter study. J. Clin. Microbiol. 39:651–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Resi D, Gagliotti C, Moro ML. 2004. Side effects of antituberculosis therapy. Am. J. Respir. Crit. Care Med. 169:542. [DOI] [PubMed] [Google Scholar]
- 23. Rishi S, Sinha P, Malhotra B, Pal N. 2007. A comparative study for the detection of Mycobacteria by BACTEC MGIT 960, Lowenstein Jensen media and direct AFB smear examination. Indian J. Med. Microbiol. 25:383–386 [DOI] [PubMed] [Google Scholar]
- 24. Rohner P, Ninet B, Metral C, Emler S, Auckenthaler R. 1997. Evaluation of the MB/BacT system and comparison to the BACTEC 460 system and solid media for isolation of mycobacteria from clinical specimens. J. Clin. Microbiol. 35:3127–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Saukkonen JJ, et al. 2006. An official ATS statement: hepatotoxicity of antituberculosis therapy. Am. J. Respir. Crit. Care Med. 174:935–952 [DOI] [PubMed] [Google Scholar]
- 26. Scarparo C, et al. 2002. Evaluation of the BACTEC MGIT 960 in comparison with BACTEC 460 TB for detection and recovery of mycobacteria from clinical specimens. Diagn. Microbiol. Infect. Dis. 44:157–161 [DOI] [PubMed] [Google Scholar]
- 27. Sorlozano A, et al. 2009. Comparative evaluation of three culture methods for the isolation of mycobacteria from clinical samples. J. Microbiol. Biotechnol. 19:1259–1264 [DOI] [PubMed] [Google Scholar]
- 28. Spark RP, Fried ML. 1988. Negative BACTEC 460-TB cultures. How long to incubate? Am. J. Clin. Pathol. 90:213–215 [DOI] [PubMed] [Google Scholar]
- 29. Tenover FC, et al. 1993. The resurgence of tuberculosis: is your laboratory ready? J. Clin. Microbiol. 31:767–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tortoli E, et al. 1999. Use of BACTEC MGIT 960 for recovery of mycobacteria from clinical specimens: multicenter study. J. Clin. Microbiol. 37:3578–3582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Whyte T, Hanahoe B, Collins T, Corbett-Feeney G, Cormican M. 2000. Evaluation of the BACTEC MGIT 960 and MB BAC/T systems for routine detection of Mycobacterium tuberculosis. J. Clin. Microbiol. 38:3131–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Williams-Bouyer N, Yorke R, Lee HI, Woods GL. 2000. Comparison of the BACTEC MGIT 960 and ESP culture system II for growth and detection of mycobacteria. J. Clin. Microbiol. 38:4167–4170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. World Health Organization Stop TB Department 2009. Treatment of tuberculosis guidelines, 4th ed World Health Organization, Geneva, Switzerland [Google Scholar]
- 34. Yan JJ, et al. 2000. Comparison of the MB/BacT and BACTEC MGIT 960 system for recovery of mycobacteria from clinical specimens. Diagn. Microbiol. Infect. Dis. 37:25–30 [DOI] [PubMed] [Google Scholar]
- 35. Yee D, et al. 2003. Incidence of serious side effects from first-line antituberculosis drugs among patients treated for active tuberculosis. Am. J. Respir. Crit. Care Med. 167:1472–1477 [DOI] [PubMed] [Google Scholar]