Abstract
The most common cause of invasive aspergillosis (IA) in patients with chronic granulomatous disease (CGD) is Aspergillus fumigatus followed by A. nidulans; other aspergilli rarely cause the disease. Here we review two clinical cases of fatal IA in CGD patients and describe a new etiologic agent of IA refractory to antifungal therapy. Unlike typical IA caused by A. fumigatus, the disease caused by the new species was chronic and spread from the lung to multiple adjacent organs. Mycological characteristics and the phylogenetic relationship with other aspergilli based on the sequence analysis of Mcm7, RPB2, and Tsr1 indicated that the new species, which we named as A. tanneri, belongs to Aspergillus section Circumdati. The species has a higher amphotericin B, voriconazole, and itraconazole MIC and causes more chronic infection in CGD mice than A. fumigatus. This is the first report documenting IA in CGD patients caused by a species belonging to the Aspergillus section Circumdati that is inherently resistant to azoles and amphotericin B. Unlike the results seen with many members of Aspergillus section Circumdati, ochratoxin was not detected in filtrates of cultures grown in various media. Our phenotypic and genetic characterization of the new species and the case reports will assist future diagnosis of infection caused by A. tanneri and lead to more appropriate patient management.
INTRODUCTION
Patients with chronic granulomatous disease (CGD) suffer from recurrent life-threatening bacterial and fungal infections due to dysfunctional phagocytic cells resulting from genetic defects in structural components of the NADPH oxidase complex (13, 31, 42). The most common fungal infection encountered by CGD patients is invasive aspergillosis (IA), and the disease has been the most important cause of mortality in these patients (9, 23, 42). The genus Aspergillus contains over 200 species (15), of which 9, Aspergillus fumigatus, A. nidulans, A. udagawae (Neosartorya udagawae), A. viridinutans, A. flavus, A. terreus, A. quadrilineatus (Emericella quadrilineatus), A. niger,and A. calidoustus, have been documented from clinical cases of IA in CGD patients (9, 17, 30, 37–39). Among these, A. fumigatus is by far the most common cause of IA, followed by A. nidulans, while the remaining seven species have been reported as a cause of IA only rarely (4, 9, 10, 17, 30, 38, 39). The susceptibilities of these species to antifungals in vitro differ, regardless of their morphological similarities. For example, most strains of A. fumigatus are considered susceptible to amphotericin B, triazoles, and caspofungin (40), while a morphologically similar and phylogenetically related species, A. udagawae, has higher resistance to these antifungals and tends to cause refractory infection (39). Aspergillus nidulans is reported to be more resistant to amphotericin B but more susceptible to caspofungin than the similar-appearing and phylogenetically related species A. quadrilineatus (37). Correct identification of Aspergillus species, therefore, is essential for appropriate disease management. We describe a new pathogenic species of Aspergillus that caused refractory invasive disease in two young male patients with X-linked CGD. The new species, named A. tanneri, is closely related to the members of Aspergillus section Circumdati. Unlike other members of Circumdati, the species failed to produce conidiophores on most of the fungal culture media routinely used in clinical laboratories, such as Sabouraud dextrose and potato dextrose agar media. Because of its sterility on routine culture media and lack of the DNA sequence in the database, this species had been treated as a “sterile mold” to date.
MATERIALS AND METHODS
Morphological diagnosis.
Primary cultures from biopsy specimens or bronchoalveolar lavage were grown on Sabouraud dextrose agar (SDA) and potato dextrose agar. Isolates were then subcultured on the following mycological agar media: Czapeck's, malt extract agar (MEA), corn meal (CM), V8 juice, and oatmeal (33). Culture plates were incubated for 2 to 3 weeks at 25, 30, 37, and 42°C. Morphologies of fruiting structures were studied under light microscopy and surface markings of conidia were examined under scanning electron microscopy according to methods described previously (34).
DNA sequencing and construction of a phylogenetic tree.
DNA was isolated from mycelium grown in yeast nitrogen broth (Becton, Dickinson and Co., Sparks, MD) on a rotary platform (200 rpm) for 5 days at 30°C using the cetyltrimethylammonium bromide (CTAB) method (19) and kept at −20°C until used in PCR amplifications. Loci were amplified using primers ITS5 and ITS4 (41) for ITS (for internal transcribed spacer [ITS] 1, 5.8S ribosomal DNA [rDNA]; for internal transcribed spacer 2, contiguous region), primers Mcm7-709for and Mcm7-1348rev for Mcm7 (partial minichromosome maintenance protein), primers fRPB2-5F and fRPB2-7cR for RPB2 (partial second-largest subunit of RNA polymerase II), and primers Tsr1-1453for and Tsr1-2308rev for Tsr1 (partial preribosomal processing protein) as described elsewhere (21, 29). Amplicons were prepared for sequencing using ExoSapIt. DNA sequences were determined on each complementary strand, and discrepancies between the two were resolved by comparisons of the complements using Sequencher. ITS sequences were used for BLASTn queries of the GenBank nucleotide database. Verified Mcm7, RPB2,and Tsr1 sequences along with homologous Aspergillus species sequences (25) from GenBank were aligned into data sets using CLUSTALW (5). PAUP* (Phylogenetic Analysis Using Parsimony* and other methods, version 4.0b10; D. L. Swofford, Sinauer Associates, Sunderland, MD) was used to conduct parsimony analysis and to generate phylogenetic trees for single gene alignments as well as for concatenated data sets. Parsimony analysis was conducted in two phases, with the first phase using random order addition (5,000 replicate experiments), NNI (nearest-neighbor-interchange) branch swapping, and maximum trees set at 10,000. Results from the random addition runs were used as starting trees for maximum parsimony analysis using as-is addition order and TBR (tree bisection-reconnection) branch-swapping analysis. Bootstrapping (bs) was performed in PAUP* with maximum parsimony criteria and TBR branch swapping for 1,000 replicates. MrBayes 3.1.2 (18, 28) was used to calculate Bayesian posterior probabilities (pp) of branches using the GTR+G+I model chosen using ModelTest 3.7 (26). Mcm7, Tsr1, and RPB2 data sets included only protein-coding DNA, and the combined data set was partitioned into codon positions 1, 2, and 3, where the G and I parameters are independent in each partition during the analysis. Tree diagrams generated by PAUP* were viewed and transformed to a transportable format using TREEVIEW (24), formatted, and annotated for publication using CorelDraw.
Test of ochratoxin in culture filtrates.
Culture filtrates of 10-day cultures in potato dextrose and YES (2% yeast extract, 15% sucrose) broth were analyzed by high-performance liquid chromatography (HPLC) (1, 34) for the presence of ochratoxin. Chromatography was conducted on a C18 Ultrasphere column (Beckman Coulter) developed in water/acetonitrile/acetic acid (99:99:2). Online fluorescence detection was used. Culture filtrates of a known ochratoxin producer, A. westerdijkiae NRRL 5175 (22), belonging to Aspergillus section Circumdati, as well as a solution of commercially obtained ochratoxin A (Sigma-Aldrich) were used as positive controls.
Antifungal susceptibility testing.
MICs were assayed according to the Clinical and Laboratory Standards Institute reference method for broth dilution (M38-A2) (8). The drugs tested were voriconazole (Pfizer), itraconazole (Sigma, St. Louis, MO), and amphotericin B (Sigma, St. Louis, MO). An Etest for voriconazole, posaconazole, itraconazole, and amphotericin B was carried out using test strips from bioMérieux S. A., France. Tests were performed on MEA and RPMI 1640 (catalog no. 31800-022; Invitrogen, Grand Island, NY) plates. Conidia were harvested with 0.01% Tween 20–phosphate-buffered saline (PBS) (TP), filtered through a cell strainer (40 μm pore size), washed twice with TP, and counted with a hemocytometer. Conidia were evenly spread (3 × 106/plate) using sterile glass beads, and test strips were laid in the center of the plate before incubation at 37°C for 72 h.
Experimental infection.
The animal experiments were carried out with the approval and oversight of the Animal Care and Use Committee of the National Institute of Allergy and Infectious Diseases at the U.S. National Institutes of Health (NIH). Virulence of A. tanneri NIH1004 was compared with that of A. fumigatus strain B-5233 in two CGD animal models: p47phox−/− mice (B6.129S2-Ncf1tm1Shl N14) (Taconic Farms, Inc.) and gp91phox-deficient mice (B6.129S6-Cybbtm1Din/J) (The Jackson Laboratory). Mice were inoculated with freshly harvested conidia via pharyngeal aspiration (27). CGD p47phox−/− and gp91phox-deficient mice received 2 × 104 and 5 × 103 CFU/mouse, respectively. These inoculum sizes were observed to cause 80% to 100% mortality within 10 to 15 days in the CGD mice inoculated with A. fumigatus B-5233 (J. A. Sugui and K. J. Kwon-Chung, unpublished data). Survival was monitored for up to 80 days postinoculation. Survival data were analyzed using the log rank test. Lungs from mice were subjected to histopathologic staining with hematoxylin and eosin (H&E) and Gomori methenamine silver (GMS).
Nucleotide sequence accession numbers.
A. tanneri isolates NIH1004 and NIH1005 were deposited at the Agricultural Research Service Culture Collection, Peoria, IL, under accession numbers NRRL 62425 and NRRL 62426, respectively. The holotype of NIH1005 (NRRL 62426), a dried colony grown on MEA, was deposited at the National Fungus Collection, U.S. Department of Agriculture, Beltsville, MD, under accession number BPI 882529. DNA sequences from NIH1004 were deposited at GenBank under the following accession numbers: JN853798, JN896584, JN896585, and JN896586 for ITS, Mcm7, RPB2, and Tsr1, respectively. Accession numbers for NIH1005 are JN896579, JN896580, and JN896581 for Mcm7, RPB2, and Tsr1, respectively.
RESULTS
Case reports.
Patients 1 and 2 were seen using institutional review board (IRB)-approved protocols of the National Institute of Allergy and Infectious Diseases, NIH.
Patient 1.
A 17-year-old male had been diagnosed with X-CGD at 7 months of age because of Staphylococcus aureus osteomyelitis (splice mutation exon 6, absent superoxide production). He was maintained on antibiotic and gamma interferon prophylaxis. At 8 years of age, he had experienced bacteremia due to a Klebsiella sp. after an appendectomy. Recurrent pneumonias at ages 12, 13, and 15 all responded to empirical antibiotics. At 16 years of age, he was diagnosed with mold pneumonia, with pleural effusion, mediastinal abscess, and extension to liver and spleen. Excision of his liver abscess and splenectomy were followed by aggressive treatment with amphotericin B, itraconazole, voriconazole, and caspofungin. Gamma interferon and granulocyte transfusions were added. Despite intensive therapy, infection progressed and disseminated further. Fever, dyspnea, and edema culminated in respiratory failure due to massive pulmonary infection. The fungal isolates grown from biopsy specimens of lung, liver, spleen, and gastric fistula and from bronchoalveolar lavage fluid produced identical colonies of a nonsporulating white mold. An autopsy showed innumerable abscesses in lung, liver, mediastinum, and abdominal cavity specimens, with multiple adhesions, intense inflammatory infiltrates, and necrosis. GMS staining of tissue sections showed septate hyphal strands with or without branching.
Patient 2.
A 19-year-old male had been diagnosed with CGD at 2 months of age (large interstitial deletion of the entire CYBB and XK genes, yielding the McLeod phenotype, absent superoxide production). At the age of 2 years, he received antituberculous drugs for mediastinal adenopathy and presumptive tuberculosis. At 8 years of age, he presented with pneumonia of the right upper lobe, which grew Phaeoacremonium inflatipes that responded to amphotericin B and itraconazole therapy. At 13 years of age, he required dilation of a ureter and stent placement. He also had cervical adenopathy due to Staphylococcus aureus and hepatosplenic abscesses caused by Serratia marcescens. At age 17 years, a pneumonia of the right upper lobe caused hemoptysis, for which he received voriconazole, with gradual clinical and radiographic improvement. At 18 years of age, while still on voriconazole therapy, he had a new episode of pneumonia (Fig. 1A and B), which led to the addition of treatment with broad-spectrum antibiotics. However, cough, intermittent hemoptysis, weight loss, and fever continued. Posaconazole treatment was added. A bronchoalveolar lavage fluid culture yielded a strain of white mold resembling those isolated from patient 1. Micafungin, ambisome, voriconazole, terbinafine, and albendazole were added to the treatment regimen without sustained improvement. Moderate to severe pulmonary restrictions on pulmonary function matched the extensive infiltration observed upon computed tomography (CT) (Fig. 1C and D). A percutaneous needle biopsy specimen of the lung grew the same mold that had been isolated from the bronchoalveolar lavage fluid. A trial treatment using intravenous pentamidine, based on its activity in murine models of fusariosis (20), resulted in transiently altered mental status. Lung and chest wall biopsy specimens showed numerous hyphae (Fig. 1E and F) and grew a mold which was confirmed to be identical to the one isolated from bronchoalveolar lavage. Despite intensive therapy, infection was progressive and fatal. An autopsy found bilateral fungal pneumonia with cavities, ocular vein involvement, hepatomegaly, and periaortic and mesenteric lymphadenopathy.
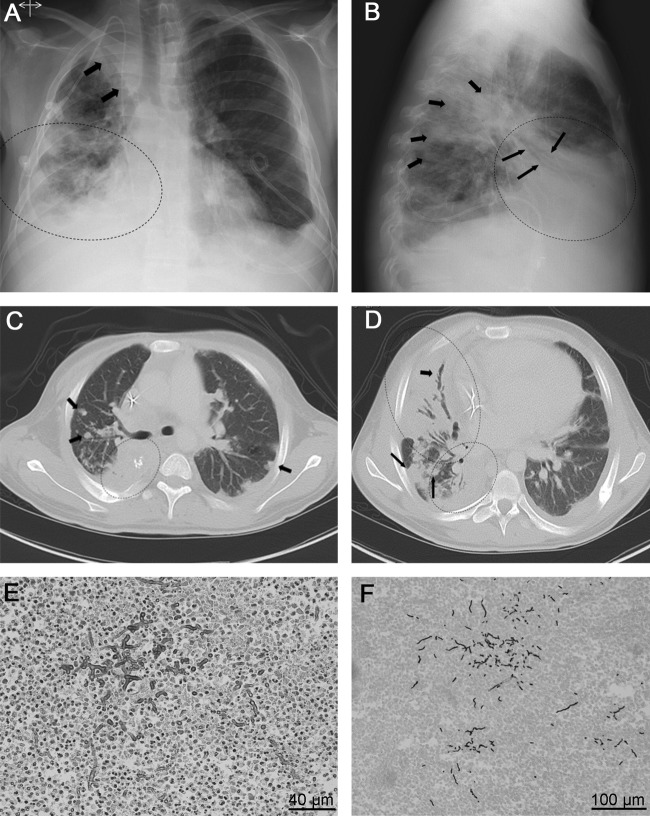
Fig 1.
Radiological findings of patient 2. (A) Posteroanterior view of chest X-ray, demonstrating reticulonodular opacities (dotted oval) at the right lower lung field. The right paratracheal line and superior vena caval edge are obliterated by an opacity with a dual edge confirmed on CT to be right upper lobe atelectasis, loculated fluid, and masses-nodules (arrows). The costophrenic angles are blunted bilaterally and are worse on the right periphery. A pleural drain is noted on the left, and a central line is in the superior vena cava. (B) In the lateral view, air-bronchograms (long arrows) overlying the cardiac silhouette are confirmed, supporting right middle lobe consolidation (dotted oval). The short arrows demonstrate nodules confirmed on CT. (C) Axial CT in lung windows just below the level of the carina demonstrates right lower lobe consolidation (dotted oval) along with nodular opacities (arrows). (D) Axial CT of the more inferior level demonstrates extensive consolidations with bronchioectatic air-bronchograms (short arrow) in the right middle lobe consolidation (large dotted oval) and right lower lobe consolidation (small dotted oval). The long arrows show multiple large nodules compatible with fungal lesions. (E and F) Histological sections of lung biopsy specimens, showing fungal hyphae stained with H&E and GMS, respectively.
Fungal diagnosis.
Initial culture of biopsy specimens or bronchoalveolar lavage fluid from both patients on agar media commonly used in the clinical laboratory produced morphologically similar white colonies that lacked fruiting structures (Fig. 2). The ITS sequences of the two fungi were identical, and, as shown by alignment with sequences in the GenBank database (BLASTn; May 2012), the species with the highest (94%) identity was Aspergillus robustus (Aspergillus section Circumdati). However, with an ITS sequence identity of less than 95%, which is too low to represent conspecificity, this fungus was considered hitherto unreported (16). Among the subcultures on various agar media made from the nonsporulating colonies, conidial structures typical of the genus Aspergillus were produced only on corn meal agar after 3 weeks of incubation at 30°C, when the agar plates were nearly dehydrated. Subcultures on MEA from the sporulating mycelium grown on cornmeal agar were fertile, but the morphological characteristics of colonies as well as conidial structures did not resemble those of any of the existing species in Aspergillus section Circumdati or those of species in any other section. Subsequent subcultures on V8 juice agar and oatmeal agar from sporulating mycelium that had been grown on MEA were also fertile. Partial sequences of three phylogenetically informative genes, Mcm7, Tsr1, and RPB2, of the two isolates were identical, indicating that they are conspecific. Phylogenetic analysis of the three catenated genes indicated that the isolates represented new species belonging to the Circumdati clade (Fig. 3). The new species was named Aspergillus tanneri.
Fig 2.
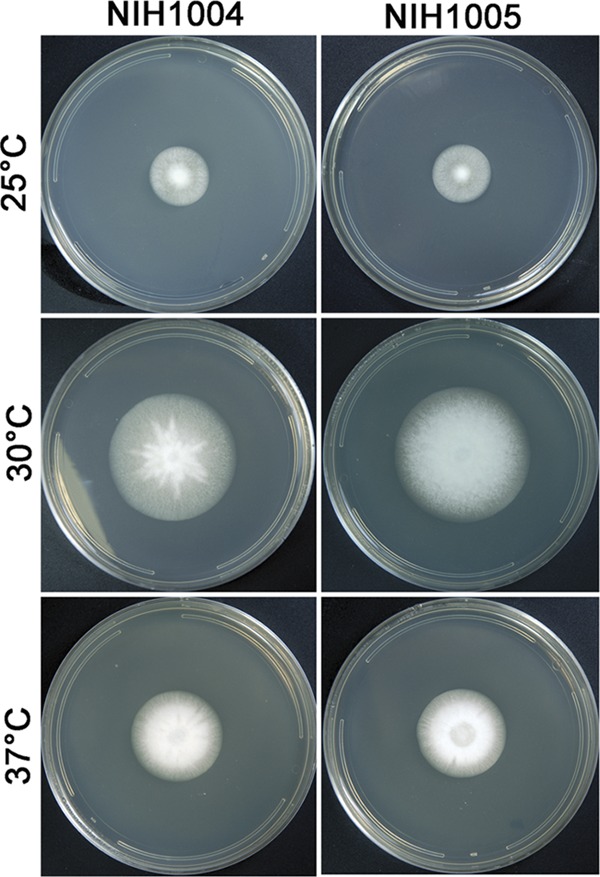
Colony morphology of A. tanneri strains NIH1004 and NIH1005 grown on malt extract agar (MEA) for 2 weeks at 25, 30, and 37°C.
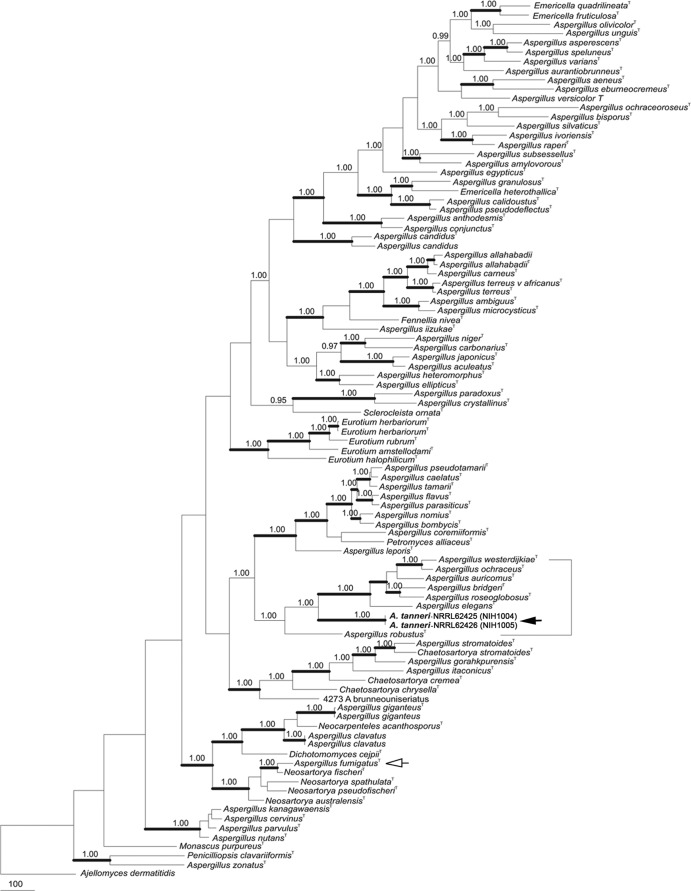
Fig 3.
Phylogenetic tree inferred from combined Mcm7, RPB2, and Tsr1 data set. A white arrow identifies A. fumigatus (section Fumigati), and a black arrow identifies A. tanneri (section Circumdati). Thick branches indicate greater than 95% bootstrap support; numbers above nodes are Bayesian probabilities greater than 0.90. Strong statistical support places A. tanneri in section Circumdati.
Description of the new species.
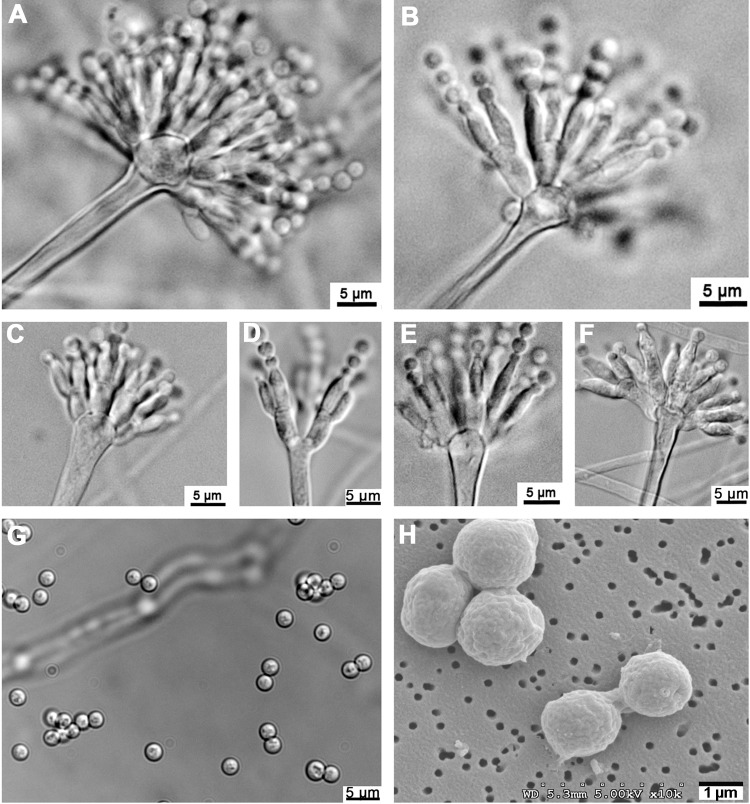
Aspergillus tanneri Kwon-Chung, Sugui & S. W. Peterson, sp. nov. Colonies grew slowly on Czapek's solution agar and were white in color, attaining a diameter of 1.5 to 2 cm in 2 weeks at 25°C, 4 to 4.5 cm at 30°C, and up to 3 cm at 37°C; substrate mycelium, thin and hyaline. Colonies on MEA grew faster and were 2.9 to 3.0 cm in diameter in 2 weeks at 25°C, 6 to 7 cm at 30°C, and 4 to 5 cm at 37°C (Fig. 2). Substrate mycelium was raised, and the colony became faintly tinged with yellowish pigment as it aged. The colony reverse remained colorless. Conidiophores sparsely formed in the dark at 30°C, and conidiophore formation was enhanced as the culture aged. Conidiophore hyaline, smooth, long, and slender, 2 to 4 μm wide, bearing hyaline pear-shaped or clavate vesicles of 5 to 7 by 7 to 10 μm (Fig. 4A, C, E, and F); metulae, 1.5 to 3 by 4 to 8 μm, covering one-half to two-thirds of the surface of the vesicle (Fig. 4A, B, C, and E), each bearing two or more phialides, sometimes becoming septate, 1 to 2 by 4 to 10 μm (Fig. 4A to F). Conidia globose, hyaline, smooth, 2 to 3 μm in diameter (Fig. 4G and H). Diminutive conidial heads with uniseriate or biseriate phialides of reduced size are often found on trailing hyphae. These diminutive heads lack vesicles, and the conidial structures appear almost penicillate (Fig. 4D). A teleomorph was not observed.
Fig 4.
Conidiophores and conidia of A. tanneri NIH1004 and NIH1005. Conidial heads were produced in NIH1004 (A, C, and D) and NIH1005 (B, E, and F) on malt extract agar (MEA) and corn meal agar, respectively. Globose conidia with smooth walls viewed under light microscopy (G) and by scanning electron microscopy (H) are shown.
Etymology.
The species epithet tanneri was given with permission of the family in memory of CGD patient 2, who was infected by strain NIH1004. The NIH1005 type strain was isolated from a gastric fistula abscess of patient 1, and strain NIH1004 was isolated from a lung biopsy specimen from patient 2.
Susceptibility to antifungal drugs.
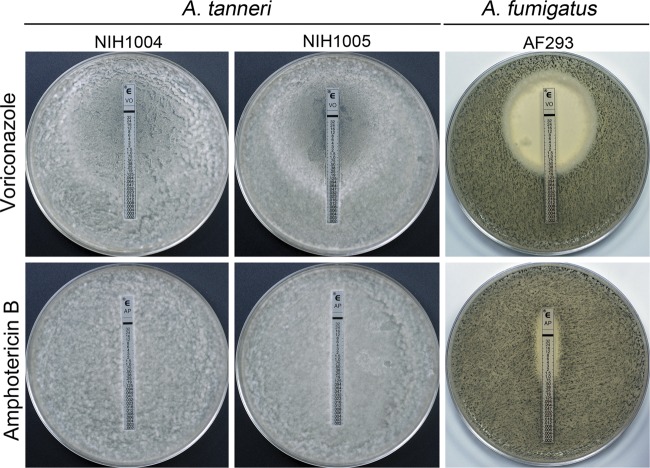
Failure of both patients to respond to aggressive antifungal therapy suggested that the new species could be inherently resistant to existing antifungals. To test if A. tanneri is highly resistant to antifungals, we compared the voriconazole, itraconazole, and amphotericin B MIC values for A. tanneri with the values for the two A. fumigatus reference strains, which are widely used for pathobiological studies. A. tanneri had significantly higher amphotericin B MICs and generally higher azole MICs than A. fumigatus (Table 1). Of note, A. tanneri was remarkably resistant to amphotericin B, showing extensive mycelial growth at 16 μg/ml, the highest concentration tested. The higher resistance of A. tanneri to antifungal drugs was also confirmed by the Etest. Figure 5 shows the susceptibility of A. tanneri and A. fumigatus to voriconazole and amphotericin B on MEA media. Conidial plates with the test strip were incubated for 72 h at 37°C. Under these conditions, A. fumigatus produced a green lawn of sporulating culture whereas A. tanneri grew mostly as thin white mycelia without sporulation. Aspergillus fumigatus strain AF293 showed a large clear zone of growth inhibition for voriconazole (MIC, 0.5 μg/ml) and a smaller zone of growth inhibition for amphotericin B (MIC, 1.5 μg/ml). In contrast, the zone of growth inhibition for voriconazole as well as for amphotericin B was absent in plates inoculated with A. tanneri. Similar results were obtained with RPMI 1640 as a growth medium. The Etest with posaconazole and itraconazole performed using both MEA and RPMI 1640 showed the same trend of higher resistance of A. tanneri compared to A. fumigatus. Our findings suggest that the high resistance of A. tanneri to antifungal drugs may have contributed to therapy failure.
Table 1.
In vitro susceptibility of A. fumigatus and A. tanneri to voriconazole, itraconazole, and amphotericin B determined by CLSI microdilution method (M38-A2)
| Strain | MICa (μg/ml) |
||
|---|---|---|---|
| Voriconazole | Itraconazole | Amphotericin B | |
| A. tanneri (NIH1005) | 4 | 4 | >16 |
| A. fumigatus (B-5233) | 1 | 1 | 2 |
| A. fumigatus (AF293) | 2 | 1–4 | 2 |
The MIC (minimal inhibitory concentration) was assayed with four replicate experiments.
Fig 5.
Susceptibility of A. tanneri and A. fumigatus to voriconazole and amphotericin B assayed by the Etest. Conidia (3 × 106/plate) were evenly spread on malt extract agar (MEA) plates, and test strips were laid at the center of the plate before incubation at 37°C for 72 h.
Ochratoxin analysis.
Analysis of culture filtrates harvested from A. tanneri strains NIH1004 and NIH1005 showed no evidence of ochratoxin production, whereas the culture filtrate of the known ochratoxin producer A. westerdijkiae strain NRRL 5175 (22) showed the typical HPLC profile of ochratoxin that matched the profile of the commercially available pure ochratoxin standard (see Fig. S1 in the supplemental material).
Experimental infection.
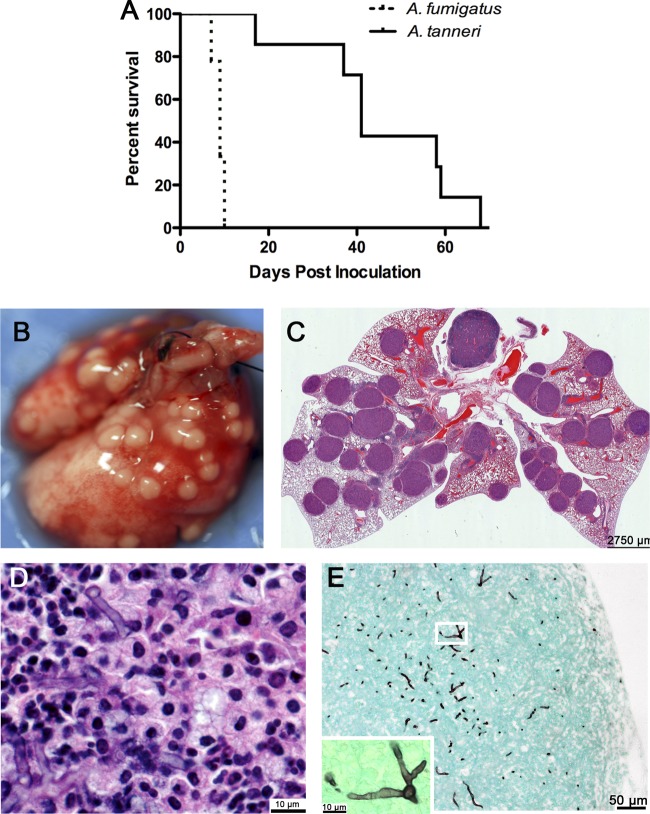
A. fumigatus produced acute infection in CGD mice (p47phox−/−), resulting in 100% mortality within 10 days, while A. tanneri caused chronic disease and required over 60 days to cause 100% death (P = 0.0001 for survival difference) (Fig. 6A). The results were similar with gp91phox-deficient mice (data not shown). Histological sections revealed that the degrees of granuloma formation in lung parenchyma seen with A. fumigatus and A. tanneri were similar, although mice died faster from IA due to A. fumigatus. The lung specimens isolated from CGD mice that succumbed to A. tanneri infection showed numerous granulomatous lesions protruding from the lung surface that were visible to the naked eye (Fig. 6B). Histology showed that the lungs were crowded with large granulomas containing hyphae (Fig. 6C to E). Severe splenomegaly was observed in CGD mice that succumbed to A. tanneri infection (data not shown).
Fig 6.
Animal experiment. (A) Survival rates of CGD p47phox−/− mice inoculated with 2 × 104 CFU per mouse. A survival study performed with CGD gp91phox-deficient mouse showed similar results. (B) Lungs of a CGD gp91phox-deficient mouse that succumbed to A. tanneri infection 63 days postinoculation. (C) Histological section of the lung shown in panel B stained with H&E. Various granulomatous lesions are seen in the parenchyma. (D) Higher magnification of one of the granulomatous lesions shown in panel C, revealing hyphae at the center of the lesion. (E) GMS-stained section of the lung shown in panel B, showing hyphae near the periphery where granulomatous lesions protruded to the lung surface. The inset at lower left shows branching hyphae at higher magnification (white box).
DISCUSSION
The two cases of infection caused by A. tanneri reported here represent examples of the most refractory cases of mold infection. The azole and amphotericin B MIC values for A. tanneri analyzed in comparison with the values seen with the A. fumigatus strain attested to the fungal resistance to various antifungals the patients have received. Cases of IA refractory to azoles and other antifungals similar to the cases reported here have been described in CGD patients infected by Neosartorya udagawae (39). Unlike IA caused by A. fumigatus, which is typically localized to the lungs, infection due to N. udagawae and A. tanneri showed primary pulmonary foci which subsequently progressed to adjacent organs. The propensity for contiguous and distant spread and elevated drug resistance appears to be inherent in both N. udagawae and A. tanneri. In addition to the pattern of invasive growth and elevated resistance to antifungals, A. tanneri appears to differ from A. fumigatus in the manner of the disease progression. Our survival studies with CGD mice revealed that, while A. fumigatus killed 100% of the infected mice in the first 10 days postinfection, A. tanneri-infected mice succumbed to the infection at a markedly lower rate. Whether the slower pace of the disease progression is because of the slow growth of A. tanneri compared to A. fumigatus in the host tissue or because A. tanneri remains mostly contained inside the granulomas and does not expand until changes in the environmental conditions of the host tissue trigger further growth is unknown. Another factor that can play a role in disease development is the secretion of secondary metabolites detrimental to host tissue. Although ochratoxin was not detected in the culture filtrates, it is possible that A. tanneri produces other secondary metabolites, which may contribute to virulence of this fungus. Sequencing of A. tanneri genome is under way, which should allow us to address this hypothesis.
A. tanneri may well have been responsible for more cases of refractory IA that have gone undiagnosed, since the species fails to sporulate on media routinely used by most clinical laboratories for a mycological diagnosis and because of the lack of available DNA sequence data for a molecular diagnosis. Although sporulation was lacking in the primary cultures, we suspected that both strains from our CGD patients were of the same species belonging to the genus Aspergillus. Not only did the two strains show similar growth characteristics, such as identical colony colors and slow growth at 37°C, but their ITS sequences matched 100%. When the ITS sequence of A. tanneri was used for a BLAST search of the GenBank database, the highest percent identity (94%) was found with A. robustus in the section Circumdati. A phylogenetic analysis based on the combined Mcm7, RPB2, and Tsr1 data set indeed placed A. tanneri in the Circumdati clade. These three genes combined are more informative for phylogeny reconstruction or genotyping aspergilli than benA and the ITS gene. The benA and ITS gene sequences may be suitable for phylogenetics analysis within the section Fumigati (2, 14) but not for a genus-wide analysis. Peterson (25) has shown that the beta tubulin gene in the genus Aspergillus is paralogous rather than homologous, rendering it unsuitable for broad phylogenetics analysis, and Galagan et al. (12) have shown that ITS analysis may yield misleading phylogenies in Aspergillus. In contrast, RPB2 was used in the “Assembling the fungal tree of life” (AFTOL) project and in a study examining the phylogeny of Aspergillus (25), and Tsr1 and Mcm7 have been used with success in several broad phylogenetic studies in Aspergillus as well as in other genera (29). One of the characteristics of various species belonging to Aspergillus section Circumdati is the production of ochratoxin, which has been shown to be nephrotoxic, immunosuppressive, teratogenic, and carcinogenic (32). Although A. tanneri belongs to section Circumdati, the toxin was not detected in culture filtrates, suggesting that A. tanneri may not be an ochratoxin producer. The other ochratoxin nonproducers within section Circumdati are A. bridgeri and A. elegans (3, 35). A. tanneri can be distinguished phenotypically from the other members of section Circumdati (A. westerdijkiae, A. cretensis, A. steynii, A. auricomus, A. flocculosus, A. ochraceus, A. petrakii, A. surphureus, A. persii, A. insulicola, A. ostianus, A. robustus, A. neobridgeri, A. pseudoelegans, A. sclerotiorum, A. melleus, A. roseoglobulosus) by its small pear[hyphen] to club-shaped vesicles (5 to 7 by 7 to 10 μm), lack of sclerotia, and better growth at 37°C than at 25°C. Most of the other members of the section Circumdati produce sclerotia, and their vesicles are globose and sometimes oblong and large (15 to 60 μm in diameter) (11). Aspergillus petrakii is most similar to A. tanneri in shape and size (15 to 30 by 20 to 35 μm) of the vesicles and absence of sclerotia, but its growth on Czapek's agar at 37°C is negligible (36). The colony reverse (Czapek agar) of species from section Circumdati is brightly colored (yellow, yellow brown, pink, vinaceous, or rose red), while the colony reverse of A. tanneri is uncolored. No member of section Circumdati other than A. tanneri has been reported as zoopathogenic. Since some species in this section, such as A. neobridgeri and A. roseoglobulosus (11), have growth rates comparable to that of A. tanneri at 37°C, it is unlikely that thermotolerance alone is the reason for A. tanneri pathogenicity. It is possible that a combination of thermotolerance and production of secondary metabolites yet to be identified enables this fungus to be a successful pathogen.
The environmental source of A. tanneri is not clear. It is noteworthy that both patients resided in the southwestern region of the United States: the first patient came from Demin, New Mexico, and the second patient from Los Angeles. It is possible that A. tanneri thrives in the environment of the southwestern region. Other members of section Circumdati have been isolated from soil (6, 7, 11), wild crabapple (Malus sylvestris), coffee bean, and decaying leaves of red mangrove (Rhizophora mangle) (11). Interestingly, A. tanneri sporulated on agar media containing sterilized carnation leaf (data not shown) in addition to cornmeal, malt extract, V8 juice, and oatmeal, suggesting a possible affinity of A. tanneri for plant materials.
In summary, IA caused by A. tanneri is distinct from typical infections caused by A. fumigatus. In CGD patients, the disease is chronic and has a tendency to spread contiguously from the lung in addition to being refractory to antifungals. Identification of the optimum growth conditions for sporulation in A. tanneri and the DNA sequences deposited in GenBank enable future diagnosis of the species and offer better-informed patient management. In view of the chronic, progressive disease course and poor response of A. tanneri to aggressive antifungal therapy, identification of the species should lead to an early consideration of definitive surgery.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the Intramural program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
Published ahead of print 1 August 2012
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Aboul-Enein HY, Kutluk OB, Altiokka G, Tuncel M. 2002. A modified HPLC method for the determination of ochratoxin A by fluorescence detection. Biomed. Chromatogr. 16:470–474 [DOI] [PubMed] [Google Scholar]
- 2. Balajee SA, et al. 2009. Molecular identification of Aspergillus species collected for the transplant-associated infection surveillance network. J. Clin. Microbiol. 47:3138–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bayman P, Baker JL, Doster MA, Michailides TJ, Mahoney NE. 2002. Ochratoxin production by the Aspergillus ochraceus group and Aspergillus alliaceus. Appl. Environ. Microbiol. 68:2326–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blumental S, et al. 2011. Invasive mold infections in chronic granulomatous disease: a 25-year retrospective survey. Clin. Infect. Dis. 53:e159–e169 [DOI] [PubMed] [Google Scholar]
- 5. Chenna R, et al. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Christensen M. 1982. The Aspergillus ochraceus group: two new species from Western soils and a synoptic key. Mycologia 74:210–225 [Google Scholar]
- 7. Christensen M, Raper KB. 1978. Aspergillus robustus, a new species in the A. ochraceus group. Mycologia 70:200–205 [Google Scholar]
- 8. Clinical and Laboratory Standards Institute 2008. Reference method for broth dilution antifungal susceptibility testing of filamentous fungi: approved standard—2nd ed (M38–A2). CLSI, Wayne, PA [Google Scholar]
- 9. Cohen MS, et al. 1981. Fungal infection in chronic granulomatous disease. The importance of the phagocyte in defense against fungi. Am. J. Med. 71:59–66 [DOI] [PubMed] [Google Scholar]
- 10. Dotis J, Roilides E. 2004. Osteomyelitis due to Aspergillus spp. in patients with chronic granulomatous disease: comparison of Aspergillus nidulans and Aspergillus fumigatus. Int. J. Infect. Dis. 8:103–110 [DOI] [PubMed] [Google Scholar]
- 11. Frisvad JC, Frank JM, Houbraken JAMP, Kuijpers AFA, Samson RA. 2004. New ochratoxin A producing species of Aspergillus section Circumdati. Stud. Mycol. 50:23–43 [Google Scholar]
- 12. Galagan JE, et al. 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438:1105–1115 [DOI] [PubMed] [Google Scholar]
- 13. Gallin JI, Zarember K. 2007. Lessons about the pathogenesis and management of aspergillosis from studies in chronic granulomatous disease. Trans. Am. Clin. Climatol. Assoc. 118:175–185 [PMC free article] [PubMed] [Google Scholar]
- 14. Geiser DM, Frisvad JC, Taylor JW. 1998. Evolutionary relationships in Aspergillus section Fumigati inferred from partial β-tubulin and hydrophobin DNA sequences. Mycologia 90:831–845 [Google Scholar]
- 15. Geiser DM, et al. 2007. The current status of species recognition and identification in Aspergillus. Stud. Mycol. 59:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Henry T, Iwen PC, Hinrichs SH. 2000. Identification of Aspergillus species using internal transcribed spacer regions 1 and 2. J. Clin. Microbiol. 38:1510–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hubka V, et al. 2012. Rare and new etiological agents revealed among 178 clinical Aspergillus strains obtained from Czech patients and characterized by molecular sequencing. Med. Mycol. 50:601–610 [DOI] [PubMed] [Google Scholar]
- 18. Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755 [DOI] [PubMed] [Google Scholar]
- 19. Jurjevic Z, Peterson SW, Horn BW. 2012. Aspergillus section Versicolores: nine new species and multilocus DNA sequence based phylogeny. IMA Fungus 3:59–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lionakis MS, et al. 2006. Pentamidine is active in a neutropenic murine model of acute invasive pulmonary fusariosis. Antimicrob. Agents Chemother. 50:294–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu YJ, Whelen S, Hall BD. 1999. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 16:1799–1808 [DOI] [PubMed] [Google Scholar]
- 22. Marino A, Nostro A, Fiorentino C. 2009. Ochratoxin A production by Aspergillus westerdijkiae in orange fruit and juice. Int. J. Food Microbiol. 132:185–189 [DOI] [PubMed] [Google Scholar]
- 23. Mouy R, et al. 1994. Long-term itraconazole prophylaxis against Aspergillus infections in thirty-two patients with chronic granulomatous disease. J. Pediatr. 125:998–1003 [DOI] [PubMed] [Google Scholar]
- 24. Page RD. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357–358 [DOI] [PubMed] [Google Scholar]
- 25. Peterson SW. 2008. Phylogenetic analysis of Aspergillus species using DNA sequences from four loci. Mycologia 100:205–226 [DOI] [PubMed] [Google Scholar]
- 26. Posada D, Crandall KA. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818 [DOI] [PubMed] [Google Scholar]
- 27. Rao GV, et al. 2003. Efficacy of a technique for exposing the mouse lung to particles aspirated from the pharynx. J. Toxicol. Environ. Health A 66:1441–1452 [DOI] [PubMed] [Google Scholar]
- 28. Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574 [DOI] [PubMed] [Google Scholar]
- 29. Schmitt I, et al. 2009. New primers for promising single-copy genes in fungal phylogenetics and systematics. Persoonia 23:35–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Segal BH, et al. 1998. Aspergillus nidulans infection in chronic granulomatous disease. Medicine (Baltimore) 77:345–354 [DOI] [PubMed] [Google Scholar]
- 31. Segal BH, Romani L, Puccetti P. 2009. Chronic granulomatous disease. Cell Mol. Life Sci. 66:553–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Smith JE, Moss MO. 1985. Mycotoxins. Formation, analysis and significance.John Wiley & Sons, Chichester, England [Google Scholar]
- 33. Sugui JA, et al. 2011. Identification and characterization of Aspergillus fumigatus “supermater” pair. mBio 2:e00234–11 doi:10.1128/mBio.00234-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sugui JA, et al. 2007. Role of laeA in the regulation of alb1, gliP, conidial morphology, and virulence in Aspergillus fumigatus. Eukaryot. Cell 6:1552–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Varga J, Kevei E, Rinyu E, Teren J, Kozakiewicz Z. 1996. Ochratoxin production by Aspergillus species. Appl. Environ. Microbiol. 62:4461–4464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Varga J, et al. 2000. Phylogenetic analysis of Aspergillus section Circumdati based on sequences of the internal transcribed spacer regions and the 5.8 S rRNA gene. Fungal Genet. Biol. 30:71–80 [DOI] [PubMed] [Google Scholar]
- 37. Verweij PE, et al. 2008. Emericella quadrilineata as cause of invasive aspergillosis. Emerg. Infect. Dis. 14:566–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vinh DC, et al. 2009. Chronic invasive aspergillosis caused by Aspergillus viridinutans. Emerg. Infect. Dis. 15:1292–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vinh DC, et al. 2009. Invasive aspergillosis due to Neosartorya udagawae. Clin. Infect. Dis. 49:102–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Walsh TJ, et al. 2008. Treatment of aspergillosis: clinical practice guidelines of the Infectious Diseases Society of America. Clin. Infect. Dis. 46:327–360 [DOI] [PubMed] [Google Scholar]
- 41. White TJ, Bruns T, Lee S, Taylor JW. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p 315–322 In Innis MA, Gelfand DH, Sninsky JJ, White TJ. (ed), PCR protocols: a guide to methods and applications. Academic Press, Inc., New York, NY [Google Scholar]
- 42. Winkelstein JA, et al. 2000. Chronic granulomatous disease. Report on a national registry of 368 patients. Medicine (Baltimore) 79:155–169 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.