Abstract
A real-time PCR assay to detect Histoplasma capsulatum in formalin-fixed, paraffin-embedded (FFPE) tissue is described. The assay had an analytical sensitivity of 6 pg/μl of fungal DNA, analytical specificity of 100%, and clinical sensitivity of 88.9%. This proof-of-concept study may aid in the diagnosis of histoplasmosis from FFPE tissue.
TEXT
Histoplasma capsulatum is a thermally dimorphic fungus that is endemic across the Americas. Exposure to H. capsulatum conidia causes disease states ranging from fatal disseminated fungemia in immunosuppressed patients to an asymptomatic infection in immunocompetent patients (6). A minimally symptomatic infection often results in lung or mediastinal nodules or calcifications that may be detected only incidentally years later (10).
Granulomatous pulmonary nodules related to histoplasmosis radiographically mimic malignancy, and biopsies are performed to resolve the differential diagnosis (7). Ideally, following biopsy, fresh tissue should be submitted for culture; however, the specimen is often evaluated histologically only. Lesions that consist of granulomatous inflammation are generally stained using Gomori's methenamine silver (GMS) stain or periodic acid-Schiff stain to look for fungal elements. In a patient with an illness consistent with an endemic mycosis where culture was not performed, the EORT/MSG Consensus Group has suggested that histoplasmosis could be diagnosed with the histopathologic demonstration of distinctive 2- to 4-μm oval, narrow-based budding intracellular yeast forms in tissue macrophages (3). However, the yeast form of Histoplasma may mimic other fungal agents, such as Cryptococcus neoformans, Candida glabrata, endospores of Coccidioides immitis, or the protozoan Leishmania species, in tissue. Culture therefore is the gold standard to confirm Histoplasma capsulatum (6, 12).
Reported molecular methods to detect H. capsulatum in fresh tissues include nested PCR with nucleotide sequencing (2), real-time PCR (1, 11), conventional PCR with nucleotide sequencing (4), and digoxigenin-labeled PCR (9). In situ hybridization using formalin-fixed, paraffin-embedded (FFPE) tissue has also been done to confirm the presence of H. capsulatum (5). Like all assays, each method has limitations and advantages. For utility in an anatomic pathology service, a real-time PCR assay to test FFPE biopsy material for the presence of H. capsulatum would be useful. This study evaluates a novel PCR assay for the direct identification of H. capsulatum using FFPE human tissue samples.
Real-time PCRs were performed on a LightCycler 1.5 using software version 3.5.3 (Roche, Indianapolis, IN). Primers specific for a 99-bp portion of the H. capsulatum-specific 100-kDa protein gene were generated as previously described (9). A fluorescent probe specific for the same H. capsulatum-specific 100-kDa protein gene amplicon using a 3′-black hole quencher (BHQ) and a 5′-6-carboxyfluorescein (FAM) fluorophore with the sequence 5′-CCAAGCCACCGATACAGTT-3′ was purchased (Eurofins, Hunstville, AL). The reaction volume was 20 μl, with a final concentration of 2 mM MgCl2, 1 μM each primer, 0.2 μM probe, and 0.5 unit of Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA). The real-time PCR assay was performed with these parameters: 95°C for 2 min followed by 40 cycles of 60°C for 20 s and 95°C for 1 s. DNA was isolated from FFPE tissue using the QIAamp DNA FFPE tissue kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. Briefly, a 10-micrometer-thick section of the paraffin block was cut, deparaffinized with xylene, digested with proteinase K, heated to reverse formalin-induced cross-linking of the nucleic acids, and purified using a silica-based column. DNA quantitation was performed using a Nanodrop 1000 spectrophotometer (Thermo Scientific, Wilmington, DE).
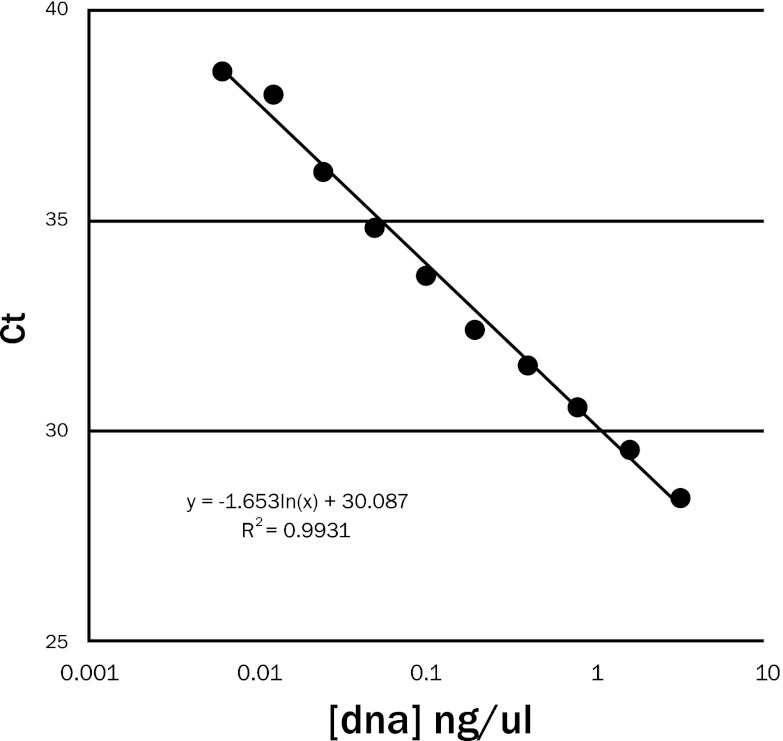
To test analytical sensitivity, a positive DNA control was prepared from a pure culture of H. capsulatum after 10 days of growth at 25°C that was formalin fixed, paraffin embedded, and processed in the same manner as the clinical samples. Serial 10-fold dilutions of DNA extracted from spiked FFPE tissue were tested. The PCR assay became positive at 6 pg/μl of fungal DNA after 39 cycles. The positive threshold cycle (CT) in relation to the logarithmic concentrations of fungal DNA showed a linear response with a high coefficient of determination (R2) at 0.9931 (Fig. 1).
Fig 1.
Results showing the analytical sensitivity of the Histoplasma capsulatum real-time PCR assay. The cycle threshold (CT) for assay positivity plotted with the logarithmic concentration of the fungal DNA showed a linear response with a coefficient of determination (R2) of 0.9931.
To evaluate clinical sensitivity, nine FFPE specimens positive for Histoplasma-like yeast by histology with a concordant positive culture for H. capsulatum from fresh tissue (four lymph node specimens and one each of lung, mediastinal mass, pericardium, small bowel, and spleen) were evaluated using the new real-time PCR assay. Eight of the specimens were PCR positive, for a clinical sensitivity of 88.9%. The PCR-negative specimen did have amplifiable genomic DNA present, as evidenced by a positive PCR for the beta-heme gene product. The inability to amplify the DNA from this culture-positive sample could be due to a number of issues and might include the amount of time the original specimen was in formalin before processing, the handling and storage of the paraffin block since the time of the original staining, and variations in processing the samples prior to DNA extraction.
To evaluate the analytical specificity, the assay was performed using DNA extracted from culture isolates that had previously been amplified and sequenced to confirm identification to include Coccidioides immitis, Blastomyces dermatitidis, Paracoccidioides brasiliensis, Cryptococcus neoformans, Candida glabrata, Candida lusitaniae, Candida krusei, Candida parapsilosis, Rhizopus oryzae, and Geotrichum capitatum. The PCR assay remained negative after 40 cycles using these DNA samples for a 100% specificity, similar to what has been reported with an in situ hybridization probe testing FFPE tissue (5).
An additional 20 clinical specimens with histopathologic evidence of fungal elements resembling Histoplasma species but without concurrent cultures were evaluated using the new PCR assay (Table 1). Eleven of the samples (52.4%) yielded a positive real-time PCR result for H. capsulatum DNA, which confirmed the anatomic pathologist's morphological identification. The discrepancy between the pathology report and PCR result for nine of the samples could be due to issues as described earlier for the inability to detect H. capsulatum in a culture-positive sample but could also represent an incorrect interpretation of the yeast forms by the pathologist.
Table 1.
Real-time PCR testing of formalin-fixed paraffin-embedded tissues where the histological report suggested histoplasmosis without culture confirmation
| Code | Specimen source | Histopathology reporta | PCR resultb |
|---|---|---|---|
| 1 | Lung | Numerous yeast forms compatible with Histoplasma | + |
| 2 | Lung | Numerous budding yeast forms compatible with Histoplasma species | + |
| 3 | Lung | Budding yeast forms morphologically consistent with Histoplasma capsulatum | + |
| 4 | Lung | Fungal organisms morphologically consistent with histoplasmosis identified | + |
| 5 | Lung | Necrotizing granulomatous inflammation with associated abscess formation suggesting Histoplasma | + |
| 6 | Lung | Caseating granulomatous inflammation with numerous fungal yeast forms morphologically consistent with Histoplasma species | + |
| 7 | Lung | Fungal yeast forms consistent with Histoplasma | + |
| 8 | Lung | Necrotizing granulomatous inflammation consistent with histoplasmosis | + |
| 9 | Lung | Necrotizing granulomatous inflammation with abundant fungal organisms consistent with Histoplasma species | + |
| 10 | Lung | Small budding yeasts morphologically consistent with Histoplasma capsulatum | − |
| 11 | Lung | Small budding fungal yeast forms morphologically consistent with Histoplasma capsulatum | − |
| 12 | Lung | Focal necrotizing granuloma formation containing fungal organisms consistent with histoplasmosis | − |
| 13 | Lung | Necrotizing granuloma with small budding yeast forms suggestive of Histoplasma | − |
| 14 | Lung | Rare yeast forms compatible with Histoplasma species | − |
| 15 | Lung | Bilateral multiple calcified granulomata with yeast forms consistent with histoplasmosis | − |
| 16 | Lymph node | Rare yeast-shaped fungal organisms suggesting Histoplasma | + |
| 17 | Lymph node | Necrotizing granuloma containing budding yeast consistent with Histoplasma | + |
| 18 | Lymph node | Yeast forms morphologically consistent with Histoplasma | − |
| 19 | Lymph node | Caseating granulomas, rare yeast morphologically suggestive of Histoplasma | − |
| 20 | Spleen | Foci of hylanized centrally necrotic granulomatous inflammation with small yeast forms consistent with Histoplasma capsulatum | − |
| 21 | Liver | Necrotizing granuloma with small budding yeast forms morphologically consistent with Histoplasma capsulatum | − |
Using Gomori's methenamine silver stain method.
+, positive; −, negative.
Regarding test performance, Simon et al. reported on the ability to detect Histoplasma DNA at a concentration to 10 fg/μl while testing fresh tissue using a real-time PCR assay (11). In comparing the lower analytical sensitivity of our real-time PCR assay to these results, the difference would not be unexpected since FFPE tissue was used in our study and the PCR target and primers were different between the studies. Additionally, differences in the DNA extraction process have shown variations in sensitivity of the assays (8).
In comparing the sensitivity to other studies to detect H. capsulatum in tissue, Simon et al. found a slightly higher sensitivity, at 95.4%; however, they extracted DNA from fresh tissue (11). Comparing the sensitivity of our real-time PCR assay with results reported by Hayden et al. when testing FFPE tissues for the presence of H. capsulatum DNA, our assay showed a higher sensitivity (88.9%) than the sensitivity following the evaluation of GMS-stained slides (75.1%) or in situ hybridization testing (50.0%) (5). Interestingly, the same kit used in our study to extract DNA from FFPE tissue was also previously reported to have a similar PCR efficiency of 90.3% when analyzing fungal DNA as a template for PCR (8).
In summary, this proof-of-concept study showed the utility of a real-time PCR assay that provides an analytical sensitivity to at least 6 pg/μl of H. capsulatum DNA extracted from FFPE tissue, with 88.9% clinical sensitivity and 100% specificity. The application of a molecular test such as this can be advantageous to reduce the turnaround time for the diagnosis of histoplasmosis and provide more accuracy compared to histopathology alone. Additional testing is ongoing to evaluate this assay in a larger sampling of tissues, with further testing utilized to analyze discrepancies between histology and PCR results.
ACKNOWLEDGMENTS
The authors would like to graciously thank Amanda Bartling for her assistance with the real-time PCR assay.
Footnotes
Published ahead of print 1 August 2012
REFERENCES
- 1. Babady NE, et al. 2011. Detection of Blastomyces dermatitidis and Histoplasma capsulatum from culture isolates and clinical specimens by use of real-time PCR. J. Clin. Microbiol. 49:3204–3208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bialek R, et al. 2002. Evaluation of two nested PCR assays for detection of Histoplasma capsulatum DNA in human tissue. J. Clin. Microbiol. 40:1644–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. De Pauw B, et al. 2008. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin. Infect. Dis. 46:1813–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guedes HL, et al. 2003. PCR assay for identification of Histoplasma capsulatum based on the nucleotide sequence of the M antigen. J. Clin. Microbiol. 41:535–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hayden RT, Qian X, Roberts GD, Lloyd RV. 2001. In situ hybridization for the identification of yeastlike organisms in tissue section. Diagn. Mol. Pathol. 10:15–23 [DOI] [PubMed] [Google Scholar]
- 6. Kauffman CA. 2007. Histoplasmosis: a clinical and laboratory update. Clin. Microbiol. Rev. 20:115–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mukhopadhyay S. 2011. Role of histology in the diagnosis of infectious causes of granulomatous lung disease. Curr. Opin. Pulm. Med. 17:189–196 [DOI] [PubMed] [Google Scholar]
- 8. Muñoz-Cadavid C, et al. 2010. Improving molecular detection of fungal DNA in formalin-fixed paraffin-embedded tissues: comparison of five tissue DNA extraction methods using panfungal PCR. J. Clin. Microbiol. 48:2147–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Qualtieri J, Stratton CW, Head DR, Tang YW. 2009. PCR detection of Histoplasma capsulatum var. capsulatum in whole blood of a renal transplant patient with disseminated histoplasmosis. Ann. Clin. Lab. Sci. 39:409–412 [PubMed] [Google Scholar]
- 10. Schwarz J, Silverman FN, Adriano SM, Straub M, Levine S. 1955. The relation of splenic calcification to histoplasmosis. N. Engl. J. Med. 252:887–891 [DOI] [PubMed] [Google Scholar]
- 11. Simon S, Veron V, Boukhari R, Blanchet D, Aznar C. 2010. Detection of Histoplasma capsulatum DNA in human samples by real-time polymerase chain reaction. Diagn. Microbiol. Infect. Dis. 66:268–273 [DOI] [PubMed] [Google Scholar]
- 12. Weydert JA, Van Natta TL, DeYoung BR. 2007. Comparison of fungal culture versus surgical pathology examination in the detection of Histoplasma in surgically excised pulmonary granulomas. Arch. Pathol. Lab. Med. 131:780–783 [DOI] [PubMed] [Google Scholar]