Abstract
The high genetic variation of hepatitis C virus (HCV) results in rapid selection of drug resistance mutations (DRMs) during monotherapy with direct-acting antivirals (DAAs). It has been proposed that each possible single mutant preexists in infected individuals; however, the levels of preexisting DRMs are too low to be directly quantified in most patients using current techniques. In this study, we evaluated the presence of DRMs in HCV-infected patients treated with the HCV protease inhibitors GS-9256 or GS-9451 as monotherapy using deep sequencing in 137 longitudinal samples from 45 patients. Software was developed to analyze deep-sequencing results with an assay cutoff of 0.25%. No NS3 DRMs that confer resistance to GS-9256 and GS-9451 (R155K, A156T, and D168V/E) were observed in 33 baseline samples at >0.25%. In contrast, these and other substitutions at NS3 positions 155, 156, and 168 were detected in 19/27 patients at day 2 (24 h) and 21/21 at day 4 (84 h) of monotherapy but not in placebo-treated patients. Based on the DRM growth kinetics during drug treatment, pretreated NS3 mutations at amino acids 155, 156, and 168 were estimated on average at 0.025% and 0.015% per genotype 1a and 1b HCV-infected patients, respectively. Relative fitness of the DRM viruses was shown to be significantly lower than the wild type. Deep-sequencing analyses of NS3 protease inhibitor-treated HCV-infected patients suggest a limit of HCV viral load suppression of 3.6 to 3.8 log10 with NS3 protease inhibitor monotherapy that does not suppress the identified preexisting NS3 DRMs and thus a need for a combination therapy.
INTRODUCTION
In the United States, about 4 million people are estimated to be living with hepatitis C virus (HCV) (1). Treatment of HCV infection with pegylated interferon combined with ribavirin has substantial limitations, especially for patients infected with HCV genotype 1 (5). A single-nucleotide polymorphism near the IL-28B gene strongly predicts a limited virological response to pegylated interferon plus ribavirin treatment for chronic HCV infection (8). To improve the success of HCV treatment with the current standard of care, new direct-acting antivirals (DAAs) that target HCV NS3 protease, NS5A, and NS5B RNA polymerase are undergoing intensive investigation. Two NS3 protease inhibitors, telaprevir and boceprevir, have been recently approved by the FDA to be administered with pegylated interferon-ribavirin as part of a three-drug combination therapy. While the development of DAA inhibitors looks very promising, the rapid selection of drug-resistant viral variants still presents a challenge for the effectiveness of DAAs (10). In prior phase 1 monotherapy studies with NS3 and NS5B inhibitors, it was demonstrated that drug-resistant variants can be detected by standard population sequencing within a few days after initiating anti-HCV treatment, resulting in treatment failure and a limited HCV viral load decrease.
HCV is a positive-strand RNA virus with a viral genome of 9,600 bases that replicates via a minus-strand RNA intermediate using a viral RNA-dependent RNA polymerase (NS5B). The mutation rate of HCV NS5B is estimated to be 10−3 to 10−5 per copied base pair (2). In an infected, untreated individual, plasma HCV RNA levels can reach up to 107 to 108 IU/ml, potentially resulting in the presence of every possible mutant, including mutants that can confer resistance to DAAs (4). The numerous viral variants exist in infected individuals as a pool of closely related but distinct variants. With initiation of antiviral pressure with DAA monotherapy, replication of sensitive wild-type (WT) viral variants are inhibited, resulting in selective amplification of drug-resistant variants (4). Upon termination of DAA drug pressure, better fit wild-type viruses outgrow the presumably less fit drug-resistant viruses. Since HCV does not archive the viral RNA, theoretically, the less fit viruses should return to the pretreatment equilibrium frequencies. However, it has yet to be determined how long it takes to reach this equilibrium and what other factors affect the prevalence of drug-resistant mutants (DRMs) in DAA-experienced patients.
Pretreatment viral quasispecies may include drug-resistant variants existing within a predominantly wild-type virus population. The presence of drug-resistant variants at different low-level frequencies in HCV-infected patients subsequently results in various degrees of viral response and mutant enrichment upon treatment with HCV inhibitors. Thus, the determination of natural levels of low-frequency resistant variants at baseline offers the potential for the interpretation and prediction of the viral response to HCV inhibitors.
The most common method of detecting drug-resistant variants in HCV-infected patients is population-based DNA sequencing. The method provides a composite of the major sequences present but is limited in the detection of minor viral mutant subpopulations that present at <20% to 25% (7). With the development of allele-specific PCR, single-genome sequencing, and deep-sequencing technologies, the detection of drug-resistant variants became more sensitive with the current assay cutoff at about 0.1% to 1% (3, 6, 9). Further improvement of the assay sensitivity is constrained by uniform amplification of viral quasispecies and background noise from mutations generated during PCR amplification and sequencing. Several recent studies clearly demonstrated the utility of deep sequencing and other sensitive technologies to detect HCV quasispecies (3, 6, 9). The deep-sequencing method was first used to detect the emergence of NS3 mutants in an elegant infectious model of HCV using human hepatocyte chimeric mice. Deep sequencing demonstrated de novo selection of telaprevir-resistant NS3 mutants in mice injected with wild-type HCV about 2 weeks after the start of treatment (6). Another recent study described the development of a new mismatch amplification mutation assay PCR for the specific detection of naturally occurring drug-resistant HCV mutants, which were then confirmed by deep sequencing. This deep amplicon sequencing allowed a detailed analysis of the structure of the viral population in four tested patient samples (3).
GS-9451 and GS-9256 are novel potent NS3 protease inhibitors. Single dose (SD) or multiple ascending dose (MAD) monotherapy phase 1 clinical trials with GS-9451 and GS-9256 resulted in potent antiviral activity in HCV genotype 1 patients (K. A. Wong, A. Bae, K. Ku, A. Worth, J. Waters, S. Sun, M. D. Miller, and H. Mo, presented at the 61st Annual Meeting of the American Association for the Study of Liver Diseases, Boston, MA, 29 October to 2 November 2010; K. A. Wong, H. Dvory-Sobol, A. Bae, K. Ku, J. Waters, M. D. Miller, and H. Mo, presented at the 46th Annual Meeting of the European Association for the Study of the Liver, Berlin, Germany, 30 March to 3 April 2011). As detected by standard population sequencing, resistance mutations to GS-9451 and GS-9256 included NS3 R155K, A156T, and D168E/G/V (K. A. Wong et al., presented at the 61st Annual Meeting of the American Association for the Study of Liver Diseases, Boston, MA, 29 October to 2 November 2010; K. A. Wong et al., presented at the 46th Annual Meeting of the European Association for the Study of the Liver, Berlin, Germany, 30 March to 3 April 2011). In the current study, the HCV NS3 protease region was analyzed using deep sequencing from 137 longitudinal plasma samples from 45 patients enrolled in 3 clinical trials investigating monotherapy with single or multiple doses of the novel NS3 HCV protease inhibitors (PIs) GS-9451 and GS-9256. The pretreatment DRM levels were estimated. Additionally, in vivo relative fitness of the DRM viruses was evaluated during the off-drug follow-up period. Results presented here provide a comprehensive view of the viral population dynamics during monotherapy with NS3 protease inhibitors.
MATERIALS AND METHODS
Clinical trial designs and sample selection for deep sequencing analyses.
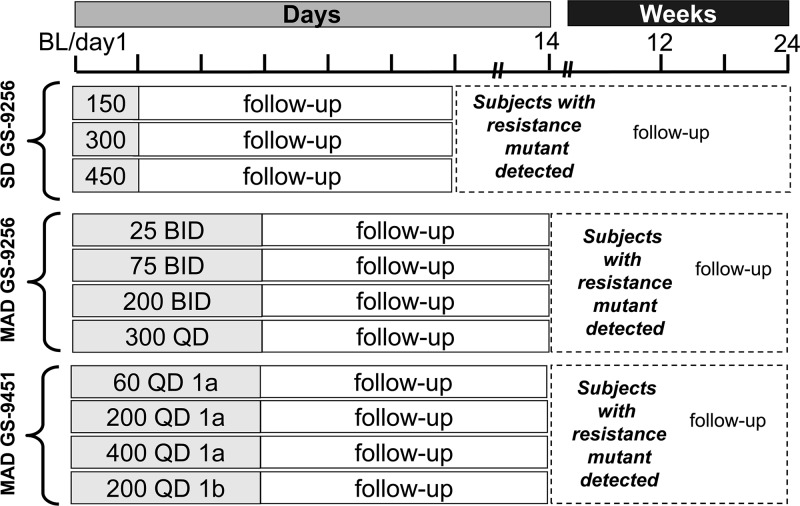
The patient population and clinical trial designs for the SD and MAD studies of GS-9256 and GS-9451 were previously presented and described elsewhere (R. Goldwater, M. DeMicco, J. Zong, G. Chittick, G. Yuen, S. West, J. Kagel, A. Bae, H. Mo, D. Oldach, W. Delaney, and J. Findlay, presented at the 61st Annual Meeting of the American Association for the Study of Liver Diseases [AASLD], Boston, MA, 29 October to 2 November 2010; E. Lawitz, J. B. Hill, T. Marbury, M. Rodriguez-Torres, M. DeMicco, J. Quesada, P. Shaw, S. Gordon, M. Shelton, D. Coombs, J. Zong, A. Bae, K. Wong, H. Mo, E. Mondou, K. Hirsch, and W. Delaney, presented at the 61st Annual Meeting of the American Association for the Study of Liver Diseases [AASLD], Boston, MA, 29 October to 2 November 2010; E. Lawitz, T. Marbury, B. Vince, N. Grunenberg, M. Rodriguez-Torres, M. DeMicco, J. Tarro, M. Shelton, S. West, J. Zong, A. Bae, K. Wong, H. Mo, D. Oldach, W. Delaney, and F. Rousseau, presented at the 45th Annual Meeting of the European Association for the Study of the Liver, Vienna, Austria, 14 to 18 April 2010). Briefly, the randomized, double-blind, placebo-controlled, multiple-dose, dose-escalation studies GS-US-208-0103, GS-US-208-0104, and GS-US-169-0103 investigated the anti-HCV activities of single-dose GS-9256, multiple-dose GS-9256, and multiple-dose GS-9451, respectively, in genotype 1 HCV-infected patients. All cohorts of these studies are outlined in Fig. 1. A total of 138 patient samples from these studies were selected to represent the pretreatment, on-treatment, and follow-up off-treatment time points. Tables 1 and 2 show the sample distribution between time points and studies. Three placebo control patients were included in the analysis. The 1a and 1b replicon plasmid controls were also tested in parallel with patient samples to determine the assay background.
Fig 1.
Single and multiple ascending dose GS-9256 and GS-9451 clinical trial designs.
Table 1.
Cohort distribution of the samples analyzed by deep sequencing from the GS-9256 SD study
| Study | Cohort | No. of samples at: |
|||
|---|---|---|---|---|---|
| BL | Day 2 | Day 3 | Day 8 | ||
| SD GS-9256 | 150 mg SD GS-9256 | 1 | 1 | ||
| 300 mg SD GS-9256 | 2 | 2 | |||
| 450 mg SD GS-9256 | 3 | 3 | 3 | 3 | |
Table 2.
Cohort distribution of the samples analyzed by deep sequencing from GS-9451 and GS-9256 MAD studies
| Study | Cohort | No. of samples at: |
|||||
|---|---|---|---|---|---|---|---|
| BL | Day 2 | Day 4 | Day 6/7 | Day 14 | Week 12 | ||
| MAD GS-9451 | 200 mg QD GS-9451 | 4 | 4 | 2 | 2 | 4 | 2 |
| 400 mg QD GS-9451 | 3 | 3 | 2 | 1 | 2 | 4 | |
| MAD GS-9256 | 200 mg BID GS-9256 | 4 | 4 | 2 | 3 | 2 | 2 |
| 300 mg QD GS-9256 | 1 | 1 | 4 | 1 | 3 | 1 | |
| 75 mg BID GS-9256 | 12 | 9 | 11 | 9 | 11 | ||
| Placebo | 3 | 2 | 2 | ||||
NS3 amplification and deep sequencing.
All RNA isolations, reverse transcription followed by PCR (RT-PCR) amplifications, and deep sequencing were performed at Virco DBA (Virco, Belgium) (11). Up to 1 ml of patient plasma sample was processed to isolate RNA. The NS3 protease region was amplified as two overlapping amplicons in a nested PCR using genotype-specific bar-coded primers. To maximize the number of input templates and to minimize variation due to PCR drift, each patient RNA sample was divided into 7 aliquots, and 7 parallel RT-PCRs were performed. The addition of 12 distinct 10-nucleotide (nt)-long barcode sequences to the primers allowed the simultaneous processing of amplicons originating from multiple patient samples (11). Each 454 deep-sequencing run included 48 samples. Bar-coded amplicons were pooled equimolarly and sequenced on the GS-FLX instrument according to the manufacturer's amplicon sequencing protocol (454 Life Sciences, Branford, CT). A total of 738,535 sequences and accompanying quality score files were obtained for 137 samples from Virco (Belgium).
Deep sequencing data analysis.
To analyze sequencing data generated using the 454/Roche sequencing technology, a database-driven software package called DB454 was developed. The DB454 software was built using the Perl programming language, an Oracle 10g database, and interfaced with the PyroMap sequence alignment tool (http://hivdb.stanford.edu/pages/resources.html). To eliminate the errors produced by the 454/Roche sequencing technology, the following algorithms were employed to exclude sequence reads with low quality scores, homopolymer-related errors, or characteristics of primer-dimer formation. Sequence reads containing an ambiguous base call (N), any base with a quality score of <10, an average quality score of <25 across all bases, or a sequence length of <150 bp were excluded from the analysis. Sequence reads for HCV genotype 1a and genotype 1b plasmid controls were aligned to H77 (AF009606) and Con1 (AJ238799), respectively. For clinical samples, all sequence reads were aligned to the sample's baseline pretreatment sequence generated using standard population-based sequencing methods. After alignment, sequence positions with coverage of >100 reads were considered for mutation analysis. Any amino acid change present in clinical samples in >2 reads were included in the results.
Viral kinetics of HCV mutants, back-calculation of pretreatment DRM levels, and relative replication fitness.
To evaluate the viral kinetics of HCV mutants, the percentage of mutant virus at each time point was converted to the IU per ml using a total HCV RNA viral load. To quantify wild-type viruses, a linkage analysis was performed by querying the number of reads that contained all three amino acid coding positions. Viruses that contained all three WT amino acids (R155, A156, and D168) were considered triple wild type (tWT). The replication kinetics of all detectable mutations with available day 2 and day 4 time points was used to estimate the viral growth of DRMs during the 3-day monotherapy with PIs.
The DRM frequencies at day 2 and day 4 were used to back-calculate the baseline frequencies of preexisting DRMs. The pretreatment levels of drug-resistant mutants were calculated from the percentage of mutant virus detected at day 2 (24 h) and the decrease in the HCV viral load as follows: XBL = Yday2 × VLday2/VLBL, where XBL is the percentage of mutant virus at baseline, Yday2 is the percentage of the same mutant virus at day 2 (24 h) as measured by deep sequencing, VLday2 is HCV viral load at day 2, and VLBL is HCV viral load at baseline. This calculation was based on the assumption that the DRM copy number did not change significantly within the first 24 h of drug treatment and the HCV viral load was declining due to the suppression of wild-type virus. If a mutant was not detected at day 2, the pretreatment levels of DRMs were calculated from the percentage of mutant virus detected at day 4 with an adjustment of 4-fold for the average change in DRM copy number/ml from day 2 to day 4.
Changes in the viral load of the wild type and mutants between day 4 and day 6/7 were used to evaluate replication fitness of DRMs. The relative fitness of DRMs was evaluated only in patients where data were available at both the day 4 and day 6/7 time points and a mutant was observed at those time points. To evaluate the relative fitness of DRM, an average change of viral load from day 4 to day 6/7 was determined and compared to the change in viral load of the wild-type viruses. The viral load of the tWT viruses was calculated based on the linkage analysis between amino acid positions 155, 156, and 168 as described above.
RESULTS
NS3 deep-sequencing data analysis and assay cutoff.
Deep-sequencing results of the NS3 protease-encoding region were obtained for 137 longitudinal samples derived from 45 patients and genotype 1a and 1b plasmid controls. A total of 738,535 sequences were obtained and processed using DB454 software that applied the quality checks as described in Materials and Methods. The resultant 441,070 sequences were used to generate a list of mutations present in the tested samples. On average, 1,129 reads were obtained per each nucleotide position of the first 181 amino acids of NS3, with an average length of 207 nt. To establish the background noise derived from PCR amplification and 454 pyrosequencing, the genotype 1a and 1b replicon plasmid controls were sequenced, with an average coverage of 4,453 reads per nucleotide position. On average, the background noise for plasmid controls was 0.06% amino acid substitutions per NS3 amino acid position, with the most frequent amino acid substitution observed at 0.21% in the 1a plasmid (S37P) and at 0.06% in the 1b plasmid (R117H and L143Q). For the purpose of this study, the analysis assay cutoff was set at 0.25% for both genotypes 1a and 1b. The selected background cutoff was at least four standard deviations above the average detected background noise for plasmid controls and was outside the upper 95% confidence interval of 0.08%.
NS3 DRM detection by deep sequencing.
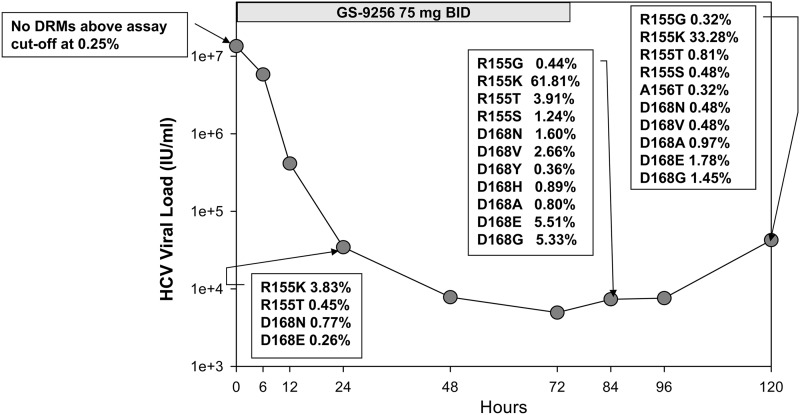
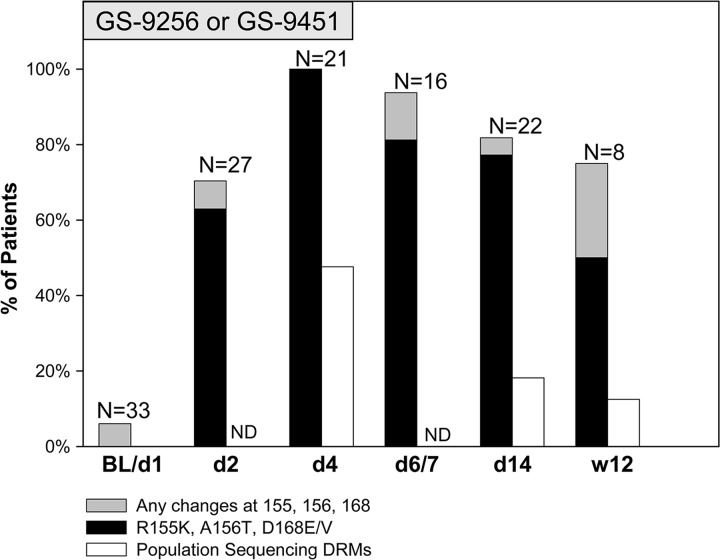
The numbers of samples selected for deep sequencing analysis from different studies, cohorts, and time points are shown in Tables 1 and 2. A total of 33/138 samples were pretreatment samples, 48/138 were on-treatment samples, and 53/138 were follow-up off-treatment samples, with 1/53 follow-up samples failing deep sequencing. In addition, 4/138 samples from follow-up points for placebo controls were also deep sequenced. Table 3 shows the baseline characteristics of patients analyzed in this study. The average HCV RNA levels at baseline were 4.3 × 106 to 6.3 × 106 IU/ml across three studies with 59% to 75% of genotype 1a patients. Deep-sequencing analysis showed that, at baseline, only 2/33 patients contained detectable variations at DRM positions: R155M at 0.44% and R155G at 0.93%. In addition, a T54S mutation was detected as a full mutation by population and deep sequencing in one placebo patient and an I170T mutation was also observed in 1/33 patients at baseline with a frequency of 0.27%. No variations at NS3 amino acid positions 155, 156, and 168 were found in placebo-treated patient samples at baseline or subsequent time points. In contrast, deep-sequencing analysis of samples from patients treated with NS3 protease inhibitors revealed multiple DRMs as early as 24 h after initiation of monotherapy treatment. Figure 2 illustrates an example of HCV RNA from one representative patient over time with mutations detected by deep sequencing. This patient received GS-9256 for 3 days with a twice-a-day (BID) dose of 75 mg and developed an R155K drug-resistant NS3 mutation at the end of treatment as detected by population sequencing. For this genotype 1a patient, no DRMs were detected at baseline (day 1); however, 4, 11, and 10 substitutions at DRM positions were detected on day 2 (24 h after first dose), day 4 (12 h after last dose), and day 6 (2 days off-drug follow-up), respectively. The frequencies of DRM variants increased from day 2 to day 4 with the suppression of wild-type viruses, with only R155K reaching the detection limit of population sequencing (>20%). The frequencies of DRM variants decreased at day 6 compared to day 4 upon termination of antiviral treatment. Similar results were observed in the majority of the patients. Figure 3 summarizes the number of patients with detectable DRMs by population and deep sequencing at all time points. At baseline, only 2/33 patients had detectable changes at DRM positions, whereas 17/19 (70%) and 21/21 (100%) patients had detectable mutations at DRM positions at day 2 and day 4, respectively. The average number of substitutions observed at DRM positions among patients receiving GS-9256 or GS-9451 was 4.7 (ranging from 0 to 12). The DRMs were observed at lower frequencies during the follow-up period after the termination of treatment but were still detected in 15/16 and 18/22 patients on day 6/7 and day 14, respectively. R155K was detected in 15/21, 5/16, and 11/22 patients at day 4, 6/7, and 14, respectively; A156T was detected in 8/21 4/16 1/22 patients at day 4, 6/7, and 14, respectively; D168E was detected in 13/21, 9/16, and 12/22 patients at day 4, 6/7, and 14, respectively; and D168V was detected in 10/21, 10/16, and 5/22 patients at day 4, 6/7, and 14, respectively. Only eight selected patients who had DRMs at earlier time points were tested on week 12, with 6/8 patients still harboring detectable DRMs above the 0.25% assay cutoff: R155K (1.0%, 1.5%, and 4.1%), R155W (0.3%), D168E (15.4% and 39.6%), D168G (0.3%), and D168N (1.1%).
Table 3.
Baseline characteristics of patients analyzed by deep sequencing
| Study | Average baseline HCV RNA (IU/ml) | % of patients with genotype 1a and 1b |
|---|---|---|
| SD GS-9256 | 6.3 × 106 | 75 (1a), 25 (1b) |
| MAD GS-9451 | 4.7 × 106 | 67 (1a), 33 (1b) |
| MAD GS-9256 | 4.3 × 106 | 59 (1a), 41 (1b) |
Fig 2.
Deep sequencing analysis of one patient from the MAD GS-9256 study. Only mutants detected at DRM positions are shown with the percentage of mutant virus indicated for each time point.
Fig 3.
Substitutions at NS3 amino acid positions 155, 156, and 168, as detected by population and deep sequencing. Percentage of patients with detectable mutations at R155, A156, and D168 by deep sequencing is shown as a gray bar with a black bar, indicating the percentage of patients that had detectable R155K, A156T, or D168E/V mutations. In addition, the white bar shows the percentage of patients that had detectable mutations at R155, A156, and D168 by population sequencing at baseline, day 4, day 14, and week 12. ND (no data) indicates that population sequencing was not performed at day 2 and day 6/7.
A genetic linkage analysis was performed to evaluate the presence of double and triple mutants at NS3 positions 155, 156, and 168. In most of the patients tested, double and triple mutants were not detected or detected at much lower levels than those of the single mutants. For example, in one genotype 1a patient, R155K was present at 32%, D168E was present at 34%, and R155K and D168E were present at 1.2% at the end of treatment with GS-9456. In another genotype 1a patient, R155K was observed at 54%, A156T was observed at 0.5%, and D168E was observed at 13% at the end of treatment with GS-9256. In this patient, the R155K D168E double mutation was observed at 1% and no triple mutations at positions 155, 156, and 168 were detected above the assay cutoff. On average, across all baseline samples, silent changes were about 4 times more frequent than nonsilent coding changes.
The levels of the mutants observed at the end of treatment were higher for the higher drug dose, suggesting that the wild-type virus was suppressed to higher levels. The analysis of the linkage between NS3 amino acid positions 155, 156, and 168 also allowed us to evaluate the presence of the NS3 wild-type variants by quantifying the percentage of reads that contain all three wild-type amino acids at these positions. We evaluated the correlation of the residual wild-type virus at the end of the treatment with the drug dose. As expected, the residual NS3 wild-type levels were inversely proportional to the drug dose with only about 20% of viral population remaining wild-type at the end of 3 days of treatment, with the highest 200-mg BID dose of GS-9256 or the 400-mg QD dose of GS-9451.
Back-calculation of pretreatment frequencies of NS3 DRMs.
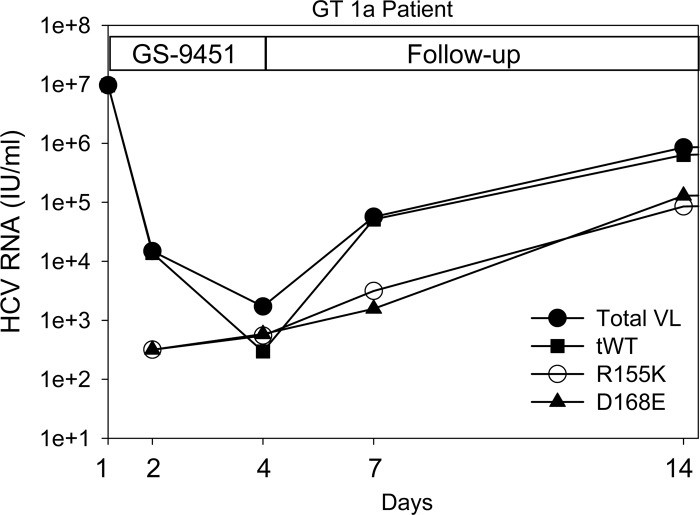
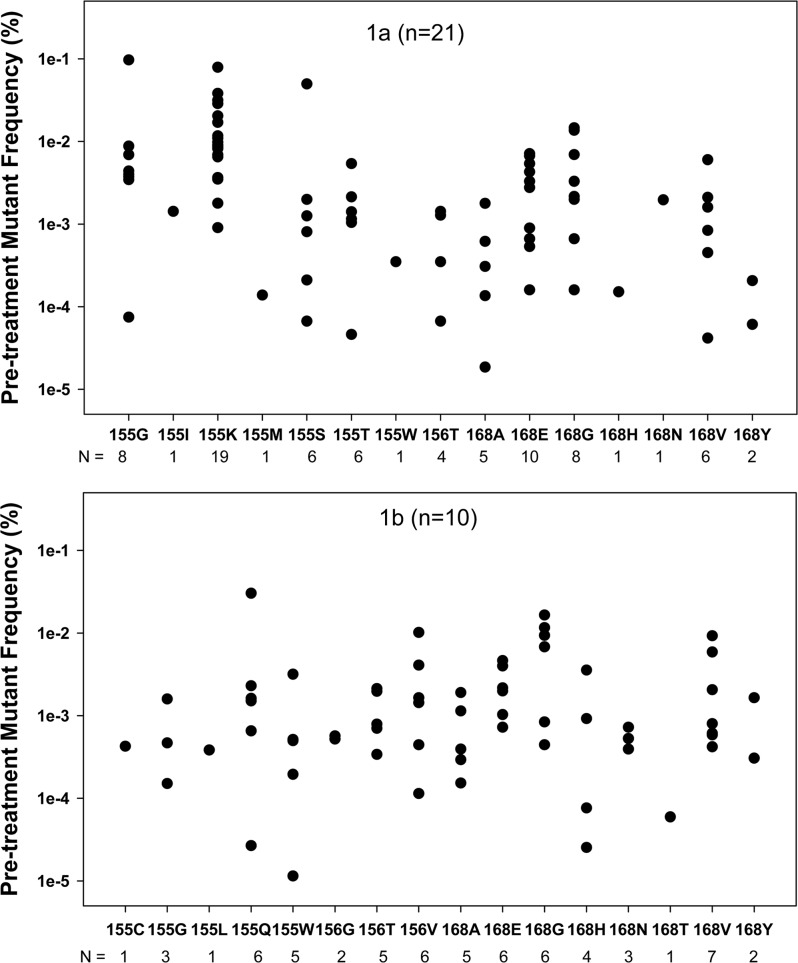
About 70% of the patients who experienced a strong reduction of HCV viral load within 24 h of treatment with GS-9256 or GS-9451 had shown the presence of minor subpopulations of DRMs. It appeared that the DRMs became detectable due to the efficient clearance of wild-type viruses. To evaluate the viral kinetics of wild-type and DRM variants during drug treatment and the follow-up period, the viral load for mutants was calculated based on the observed percentage of a mutant virus as detected by deep sequencing for each time point. The viral load of the tWT viruses was calculated based on the percentage of reads that contained a wild-type amino acid at all three DRM positions by deep sequencing. Viruses that contained all three wild-type amino acids (R155, A156, and D168) were considered tWT. The proportion of wild-type virus was evaluated based on the reads that contained all three amino acid positions. Viral load kinetics of the wild-type virus were plotted based on the total HCV viral load and the determined frequency of the NS3 wild-type virus variants at each time point. Viral load kinetics of the mutant virus were also plotted based on the total HCV viral load and the determined frequency of each NS3 mutant variant at each time point. Figure 4 shows the viral kinetics of R155K, D168E, WT, and total viral load in one representative patient. The result indicates that, on a log scale, the R155K and D168E mutants exhibit near-linear growth, suggesting that the quantity of these mutants did not change significantly during the first 24 h. Similar mutant viral kinetics were observed in the other analyzed patients. Based on the observation that the DRM viral growth between day 2 and day 4 was on average only 4-fold and that the replication cycle of HCV is 6 to 12 h, it is reasonable to assume that the copy number per ml of NS3 DRMs did not change more than 2-fold during the first 24 h of drug treatment. Based on this assumption, a DRM copy number per ml at day 2 for each sample was divided by the HCV total viral load at baseline, deriving the estimated pretreatment levels of DRMs. For example, if R155K was detected at 1% on day 2 with a viral load at day 2 of 105 IU/ml and a baseline viral load of 107 IU/ml, the pretreatment R155K would be estimated at a frequency of 0.01% or 10−4. For the samples where DRMs were only detectable at day 4, the calculations were adjusted to account for the mutant growth between day 2 and day 4. The back-calculated pretreatment levels of all observed DRMs are summarized in Fig. 5. R155K was the most frequent DRM among the patients infected with HCV genotype 1a, with an average baseline frequency of 0.016% ± 0.018%. R155K was detected during drug treatment in 19/21 genotype 1a patients. Other mutations that were observed in genotype 1a patients included D168E (10/21), R155G (8/21), and D168G (8/21). The following substitutions were also observed in 1 to 6 of 21 1a patients: R155I, R155M, R155S, R155T, R155W, A156T, D168A, D168H, D168N, D168V, and D168Y (Fig. 5, first panel). In genotype 1b patients, D168V was the most frequent DRM observed in 7/10 patients during treatment, with an average baseline frequency of 0.003% ± 0.003%. Other mutations that were observed in genotype 1b patients included R155Q (6/10), D168E (6/10), A156V (6/10), and D168G (6/10). The following substitutions were also observed in 1 to 5 of 10 1b patients: R155C, R155G, R155L, R155W, R155G, R156T, D168A, D168H, D168N, D168T, and D168Y (Fig. 5, second panel). Distributions of baseline frequencies of these genotype 1a and 1b mutants are shown in Fig. 5. On average, pretreatment frequencies of mutations observed in genotype 1a patients were as follows: R155K > R155G > R155S > D168G > D168E > D168N > R155T > D168V > R155I > A156T > D168A > R155W > D168H > R155M. For genotype 1b patients, the observed mutations preexisted at average frequencies in the following order: D168G > R155Q > A156V > D168V > D168E > A156T > D168H > D168Y > R155W > D168A > R155G > D168N > R156G > R155C > R155L > D168T.
Fig 4.
Viral kinetics of HCV in one representative genotype 1a patient. The viral load for the R155K and D168E mutants was calculated based on the observed percentage as detected by deep sequencing for each time point. The viral load of the triple wild-type viruses (tWT) was calculated based on the linkage analysis between amino acid positions 155, 156, and 168 as described in Materials and Methods.
Fig 5.
Pretreatment levels of NS3 DRMs as determined by back-calculating from day 2 and day 4 deep-sequencing results. The first panel represents results from 21 genotype 1a patients; the second panel represents results from 10 genotype 1b patients. The number under each mutant indicates the number of patients in which this mutant was detected within the first 3 days of therapy. The pretreatment levels of drug-resistant mutants were calculated from the percentage of mutant detected at day 2 (24 h) and the decrease in HCV viral load was as follows: XBL = Yday2 × VLday2/VLBL, where XBL is the percentage of a mutant at baseline and Yday2 is the percentage of the same mutant at day 2 (24 h) as measured by deep sequencing.
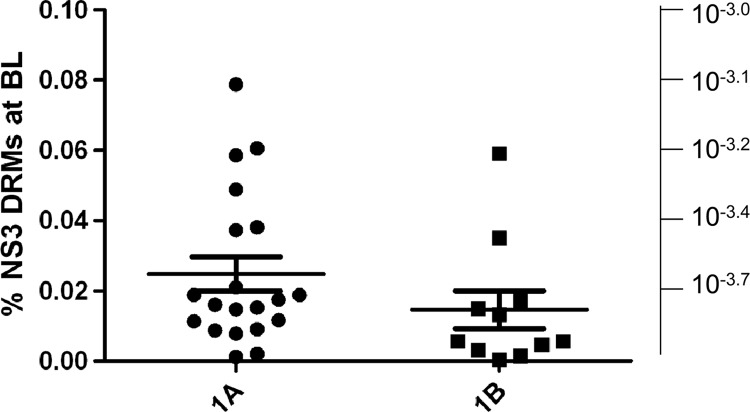
To evaluate the potential limit for HCV viral load suppression in patients treated with optimal concentrations of PI due to the presence of NS3 DRMs, the frequencies of detected DRMs were added for each patient to calculate the cumulative pretreatment NS3 DRM level per patient (Fig. 6). Mean pretreatment levels of resistant variants at NS3 amino acid positions 155, 156, and 168 per patient were 0.025% and 0.015% in genotype 1a and 1b patients, respectively. In genotype 1a patients, the median pretreatment level of NS3 DRMs was 0.017%, ranging from 0.001% to 0.079% (25% and 75% of 0.010% and 0.034%, respectively). In genotype 1b patients, the median pretreatment level of NS3 DRMs was 0.006%, ranging from 0.0004% to 0.059% (25% and 75% of 0.004% and 0.017%, respectively). These levels of preexisting DRMs would theoretically limit the average HCV viral load suppression in patients treated with an optimal dose of current PI at the drug dose that is insufficient to suppress the described DRMs to 3.6 and 3.8 log10 in genotype 1a and 1b HCV-infected patients, respectively. In contrast, in the presence of PIs that are capable to suppress the described DRMs, a higher HCV viral load suppression would be possible.
Fig 6.
Cumulative baseline pretreatment DRM frequencies per patient. Cumulative per patient pretreatment levels of DRMs derived as a sum of all back-calculated DRMs for each patient. The left scale shows cumulative per patient DRM frequency represented as a percentage of the total viral population; the right scale shows cumulative per patient DRM frequency represented by log10 frequency of the total viral population to be directly comparable with the expected viral load decrease in patients optimally treated with NS3 inhibitors.
In vivo relative fitness of NS3 DRMs.
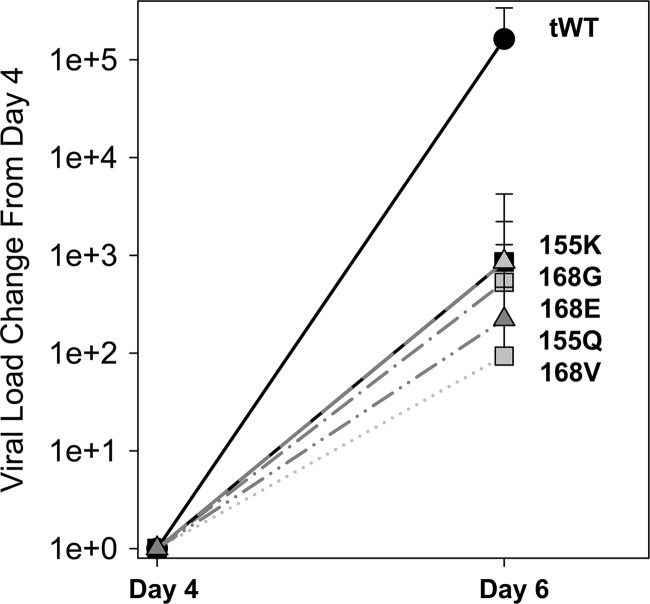
The relative fitness of observed DRMs was evaluated based on the early kinetics of wild-type and mutant viruses during the off-drug follow-up period. We found that the replication kinetics between day 4 and day 6/7 were the most representative of the fitness differences of the mutants and were the least affected by target cell availability. The relative fitness of DRMs was evaluated only in patients where data were available at both the day 4 and day 6/7 time points and a mutant was observed at those time points. An average change of viral load of each DRM was plotted and compared to the change in viral load of the wild-type viruses (Fig. 7). The viral load of the tWT viruses was calculated based on the linkage analysis between amino acid positions 155, 156, and 168 as described in Materials and Methods. The relative fitness of all DRMs was found to be significantly lower than the fitness of the wild-type viruses. There was a small difference in relative fitness between the mutations observed with the order of the viral fitness as follows: WT > R155K ≈ D168G ≈ D168E ≈ R155Q ≈ D168V. Despite the significantly lower replication fitness of the DRMs, we found that in 6/8 selected patients the following DRMs persisted through week 12: R155K (1.0%, 1.5%, and 4.1%), R155W (0.3%), D168E (15.4% and 39.6%), D168G (0.3%), and D168N (1.1%).
Fig 7.
Replication fitness of DRM viruses in patients during the off-treatment follow-up period. The relative fitness of each DRM and WT, shown as an average change of viral load from day 4 to day 6/7, was determined and compared to the change in viral load of the wild-type viruses. The mean change in viral load from day 4 is shown. Error bars represent standard deviations.
DISCUSSION
A deep-sequencing technology that allows the detection of minor viral subpopulations and a developed data analysis tool were utilized in this study to detect NS3 protease mutations at amino acid positions R155, A156, and D168 down to a detection limit of 0.25% in patients undergoing monotherapy treatment with NS3 protease inhibitors. The results showed that the variety of PI-resistant mutations that arise early during treatment are often missed by standard population sequencing. In fact, all patients tested (21/21) at the end of the 3-day monotherapy treatment with GS-9256 or GS-9451 (two novel NS3 protease inhibitors) had detectable DRMs by deep sequencing, whereas only about half of these patients had detectable DRMs by standard population sequencing. Lack of detection of DRMs by population-based sequencing was always consistent with the presence of the DRMs below the limit of detection for mutant subpopulations which is ∼20% using standard sequencing.
At baseline, only 2/33 patients had detectable changes at DRM position 155, 156, or 168, whereas 17/19 (70%) and 21/21 (100%) patients had detectable mutations at these DRM positions at day 2 and day 4, respectively. An average of five substitutions was observed at DRM positions among patients receiving GS-9256 or GS-9451. R155K was the most frequently observed DRM among genotype 1a patients. Other mutations that were observed in genotype 1a patients included D168E, R155G, D168G, R155I, R155M, R155S, R155T, R155W, A156T, D168A, D168H, D168N, D168V, and D168Y. In genotype 1b patients, D168V was the most frequent DRM observed during treatment. Other mutations that were observed in genotype 1b patients included R155Q, D168E, A156V, D168G, R155C, R155G, R155L, R155W, R155G, R156T, D168A, D168H, D168N, D168T, and D168Y. Interestingly, 7/7 (K, G, I, S, T, W, and M) variants observed at position 155 of genotype 1a samples only require a single nucleotide change from AGR codons that are predominant in genotype 1a HCV. R155 in genotype 1b patients is mostly encoded by a CGN codon with a requirement of 2 nucleotide changes for the R155K mutant, and this was not observed in any genotype 1b patient at day 2 or day 4 above the 0.25% cutoff. All 5 substitutions (Q, G, L, C, and W) observed at position R155 among genotype 1b patients are possible via a single nucleotide change from the CGN genotype 1b codon at R155. A recent presentation has shown reduced susceptibility of the observed NS3 mutants to GS-9256 and GS-9451 (H. Dvory-Sobol, T. Schwartz, B. Han, A. Worth, S. Rajyaguru, E. Svarovskaia, M. D. Miller, and H. Mo, presented at the 47th Annual Meeting of the European Association for the Study of the Liver, Barcelona, Spain).
The early detection of DRMs correlated with the HCV viral load decline, suggesting that clearance of wild-type viruses revealed the preexisting DRM variants. Based on the early viral load kinetics, we back-calculated the pretreatment frequencies of NS3 mutants that were not sensitive to and therefore not suppressed by GS-9256 or GS-9451. Even though the pretreatment DRM levels are back-calculated estimates and not directly measured baseline levels, we find these estimates to be useful to define the viral load reduction “floor” during monotherapy drug treatment. Average pretreatment frequencies of NS3 DRMs per patient were estimated at 0.025% and 0.015% in genotype 1a and 1b patients, respectively. These levels of preexisting DRMs would predict a limit to the HCV viral load suppression in patients treated with optimal doses of protease inhibitors of 3.6 and 3.8 log10 in genotype 1a and 1b HCV-infected patients, respectively, if these DRMs are not suppressed. This prediction is consistent with observed viral load reductions by PIs of about 3.5 log10 with genotype 1b and showing greater viral load reductions compared to genotype 1a (K. A. Wong et al., presented at the 61st Annual Meeting of the American Association for the Study of Liver Diseases, Boston, MA, 29 October to 2 November 2010; K. A. Wong et al., presented at the 46th Annual Meeting of the European Association for the Study of the Liver, Berlin, Germany, 30 March to 3 April 2011). Similar analyses of pretreatment levels of DRMs at other HCV drug targets, such as NS5B and NS5A, need to be performed to provide useful insights into the development of the most potent drug combinations. The accuracy of viral load reduction estimates might be affected by the limited sensitivity of deep sequencing analysis in on-treatment samples with lower viral loads, as only patients with detectable DRMs can be included in the calculations.
Even though the results presented in this work provide valuable evidence supporting the preexistence and persistence of DRMs, it is important to emphasize the limitations of deep sequencing. The depth of the sequencing of samples is not only dependent on the number of reads obtained but is also strongly limited by the amount of viral template input into the RT-PCR amplification. Therefore, plasma samples with low viral loads can provide only limited results for mutant subpopulations. Another limitation of deep-sequencing technologies is related to errors generated during PCR amplification and the deep-sequencing procedure. It is critical to evaluate the errors occurring during sample manipulation using controls such as plasmids or in vitro-transcribed RNA from a clonal sequence. Development of software that can recognize the 454 pyrosequencing-related errors, such as insertions and deletions at the homopolymeric regions as described in this analysis, improves the accuracy of the data analysis.
The analysis described was also limited only to the viral variants observed at NS3 positions R155, A156, and D168. The other investigational and approved NS3 protease inhibitors were reported to select for mutations at NS3 positions 36, 54, 80, and 170, in addition to 155, 156, and 168. Mutants at NS3 positions 36, 54, and 170 confer resistance to telaprevir and boceprevir. The viral mutants at these positions confer no resistance to GS-9256 or GS-9451 and were not selected by these compounds. Based on population sequencing, the Q80K mutant is present at high frequencies in about 50% of genotype 1a HCV-infected patients. In this study, in one of the placebo patients at baseline, the T54S mutant was detected as a full mutation by population and deep sequencing. The I170T mutant was also observed in 1/33 patients at baseline with a frequency of 0.27%; in 6/33 patients a wild-type mixture of V/I170 was also observed. Further studies are needed to determine the preexisting levels of DRMs at positions 36, 54, and 170 of NS3. In a recent study that used deep sequencing for the detection of HCV drug-resistant variants, the presence of HCV PI-resistant variants was found in 12/146 samples from treatment-naive individuals (S. Margeridon-Thermet, S. Le Pogam, T. Liu, B. Hanczaruk, T. Arnold, B. Simen, N. Shulman, R. Shafer, and I. Najera, presented at the HIV and Hepatitis Resistance Workshop, Los Cabos, Mexico, 8 to 10 June 2011). Drug-resistant variants were detected at NS3 amino acid positions V36, R155, A156, D168, and V/I170 at frequencies ranging from 0.5% to 3.9%. This observation is in close agreement with results presented here, showing the overall presence of pretreatment mutants at positions 36, 54, 155, 156, 168, and 170 in 4/33 (12%) samples as measured directly by deep sequencing with an assay cutoff of 0.25%. Interestingly, using linkage analysis, we were able to establish that the residual NS3 wild-type levels at the end of treatment were inversely proportional to the drug dose. This analysis may provide a useful tool to determine an optimal drug dose that is capable of suppressing the majority of the wild-type virus.
In addition to the evaluation of pretreatment levels for DRMs, persistence of DRMs was assessed during the off-drug follow-up period. The observation that the number of patients with detectable DRMs decline with time is consistent with previous reports.
The greatest difference in the replication kinetics between DRMs and wild-type viruses was observed in the first few days posttreatment (day 4 to day 6/7). In several patients where DRMs were observed after day 6/7, it appeared that the replication kinetics of mutants and wild-type viruses were somewhat similar with a less pronounced defect of the DRM replication. It is possible that strong suppression of the wild-type virus during the drug treatment resulted in an increased availability of target cells for new infections. Once the drug was removed, the wild-type and mutant viruses were competing for the available target cells. Within a few days, the availability of the target cells possibly became a limiting factor in the infection of new cells and the replication of mutants and wild-type viruses is mainly driven by the replication of infected cells. Because the samples for follow-up analysis at week 12 were selected based on the presence of results at the earlier time points, it remains to be established, in the larger set of long-term follow-up samples, how long and at what frequency DRMs persist in patients treated with DAA monotherapy. Further studies are needed to test this hypothesis and elucidate the persistence of DRMs during off-drug follow-up periods in patients who were exposed to DAAs.
In summary, the deep-sequencing analyses of NS3 PI-treated HCV-infected patients revealed the quantity and diversity of minority drug-resistant NS3 variants as early as 24 h after initiating antiviral treatment. Estimates of the pretreatment levels of NS3 DRMs strongly suggests a limit of HCV viral load suppression with NS3 protease inhibitor monotherapy due to preexistence of DRMs and a need for a combination therapy to durably treat HCV infection.
ACKNOWLEDGMENTS
We are employees and stockholders of Gilead Sciences, Inc.
This study was sponsored by Gilead Sciences, Inc.
Footnotes
Published ahead of print 25 July 2012
REFERENCES
- 1. Colvin HM, Mitchell AE. 2010. Hepatitis and liver cancer: a national strategy for prevention and control of hepatitis B and C. The National Academies Press, Washington, DC: [PubMed] [Google Scholar]
- 2. Duarte EA, et al. 1994. RNA virus quasispecies: significance for viral disease and epidemiology. Infect. Agents Dis. 3:201–214 [PubMed] [Google Scholar]
- 3. Fonseca-Coronado S, et al. 2012. Specific detection of naturally occurring hepatitis C virus mutants with resistance to telaprevir and boceprevir (protease inhibitors) among treatment-naive infected individuals. J. Clin. Microbiol. 50:281–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guedj J, Rong L, Dahari H, Perelson AS. 2010. A perspective on modelling hepatitis C virus infection. J. Viral Hepat. 17:825–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hadziyannis SJ, et al. 2004. Peginterferon-alpha2a and ribavirin combination therapy in chronic hepatitis C: a randomized study of treatment duration and ribavirin dose. Ann. Intern. Med. 140:346–355 [DOI] [PubMed] [Google Scholar]
- 6. Hiraga N, et al. 2011. Impact of viral amino acid substitutions and host interleukin-28b polymorphism on replication and susceptibility to interferon of hepatitis C virus. Hepatology 54:764–771 [DOI] [PubMed] [Google Scholar]
- 7. Leitner T, et al. 1993. Analysis of heterogeneous viral populations by direct DNA sequencing. Biotechniques 15:120–127 [PubMed] [Google Scholar]
- 8. McHutchison JG. 2011. The role of genetic markers in hepatitis C virus therapy: a major step for individualized care. Liver Int. 31(Suppl 1):29–35 [DOI] [PubMed] [Google Scholar]
- 9. Ninomiya M, et al. 2012. Use of Illumina deep sequencing technology to differentiate hepatitis C virus variants. J. Clin. Microbiol. 50:857–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pawlotsky JM. 2011. Treatment failure and resistance with direct-acting antiviral drugs against hepatitis C virus. Hepatology 53:1742–1751 [DOI] [PubMed] [Google Scholar]
- 11. Vandenbroucke I, et al. 2011. Minor variant detection in amplicons using 454 massive parallel pyrosequencing: experiences and considerations for successful applications. Biotechniques 51:167–177 [DOI] [PubMed] [Google Scholar]