Abstract
Respiratory virus infections cause significant morbidity and mortality in immunocompromised patients. Timely diagnosis is needed to provide optimal clinical care. Diagnostic tests routinely available at most institutions are limited by poor sensitivity and a slow turnaround time. We collected 90 respiratory samples from 87 immunocompromised patients (56 bronchoalveolar lavage and 34 nasopharyngeal aspirate samples) in order to compare the performance of routine respiratory virus testing available at our institution to the FilmArray respiratory panel assay, a novel diagnostic tool which utilizes multiplex PCR to test for 21 respiratory pathogens with a 1-h turnaround time. Samples with discordant results and 13 samples with concordant results underwent further verification testing by laboratory-developed real-time PCR. The FilmArray assay identified viral pathogens in more samples than did clinical testing (30/90 versus 16/90; McNemar P = 0.001). Most of the additional viral pathogens identified by the FilmArray respiratory panel assay that were confirmed by verification testing were pathogens not assessed by routine clinical tests, including rhinovirus/enterovirus, human metapneumovirus, and coronavirus. The FilmArray respiratory panel assay allowed for increased identification of respiratory viral pathogens in this cohort of immunocompromised patients.
INTRODUCTION
Patients with hematologic malignancy and recipients of stem cell and solid organ transplants are at significant risk for severe illness due to viral respiratory tract infection (1, 5). While infection with an upper respiratory tract virus, such as rhinovirus or parainfluenza virus, typically results in a self-limited illness in a normal host, this type of infection can result in significant morbidity and mortality in an immunocompromised host. The severity of illness in this population is typically attributed to the frequent development of secondary infection with bacteria, fungi, or other viruses and also to the spread of the virus to involve the lower respiratory tract (1, 3–6).
In order to provide optimal patient care, rapid and accurate diagnosis of viral respiratory pathogens is needed for immunocompromised patients. Though there are several respiratory viruses that can cause significant illness in this population, the symptoms of different viral respiratory tract infections are similar and do not help in distinguishing the specific pathogen, and thus patients in whom viral respiratory tract infection is suspected need to be tested for a battery of pathogens (1, 5). Rapid and accurate identification of the specific viral pathogen(s) causing illness allows for targeted therapy where treatments exist, timely institution of appropriate infection control measures, appropriate monitoring for secondary infections, and minimization of empirical treatment for possible concerning alternative conditions. Furthermore, while there are no FDA-approved treatments for many respiratory viruses, such as parainfluenza virus or rhinovirus, accurate identification of these viruses will allow a better understanding of the need for and development of investigational treatment options (2).
Until recently, the primary diagnostic tools for respiratory viruses included direct fluorescent antibody (DFA) assays, enzyme immunoassays, and viral culture. While DFA assays and enzyme immunoassays have a rapid turnaround time, sensitivity is limited. Viral culture is more sensitive but requires several days of incubation before results are available. More recently, PCR-based tests and specifically multiplex PCR assays for respiratory viruses have greatly improved respiratory viral diagnostics, particularly in the immunocompromised population (7, 9, 10, 12, 14). However, the technical complexity of PCR-based testing has limited its usefulness. The FilmArray respiratory panel (RP) is a multiplexed, fully automated PCR assay, which is capable of detecting 18 viral respiratory pathogens and three atypical bacterial pathogens with a turnaround time of approximately 1 h (12). The performance of this assay in the general adult and pediatric populations in comparison to that of DFA, other multiplex PCR-based assays, and laboratory-based PCR assays has been described (8, 11–13). The goals of the present study are to characterize the performance of the FilmArray RP assay on bronchoalveolar lavage (BAL) and nasopharyngeal aspirate (NPA) samples in the immunocompromised host population in comparison to standard clinical testing for respiratory viruses.
MATERIALS AND METHODS
Patients and samples.
The study population included 87 adult patients with hematologic malignancy or recipients of hematopoietic stem-cell transplant (HSCT) or solid organ transplant (SOT) who underwent testing for viral respiratory pathogens for any clinical indication at Dana-Farber Cancer Institute/Brigham and Women's Hospital (DFCI/BWH) between November 2009 and September 2010. The clinical indications for testing included symptoms of an upper respiratory tract infection (URI) or lower respiratory tract infection (LRI), or testing was done for surveillance of other infectious or noninfectious conditions.
Study samples were collected consecutively Monday through Friday for those BAL or NPA samples (collected from transplant recipients or patients with hematologic malignancy) from which there was fluid remaining after all aliquots necessary for clinically indicated tests were obtained. Three of the eighty-seven patients each contributed two samples that were collected at least 1 month apart for new clinical indications, for a total of 90 samples. Fluid samples were diluted 3:1 with M4 viral transport medium, aliquoted, and stored at −80°C until study testing with a research version of the FilmArray RP assay (Idaho Technology, Inc., Salt Lake City, UT) or individual PCR testing for verification was carried out. Both FilmArray and verification PCR testing were performed retrospectively such that the results had no impact on clinical decision making.
Electronic medical records were reviewed for clinical details of patients who contributed respiratory samples, including gender, age, underlying malignancy, type of transplant, and the reason for the respiratory virus testing. This study was approved by the Office of Human Research Services at DFCI/BWH.
Clinical testing.
All study BAL and NPA samples were tested for one or more respiratory viruses based on clinical indications determined by the patient's clinical providers. During the study period, the following DFA respiratory virus tests were available at DFCI/BWH: influenza A and B viruses (Millipore, Billerica, MA), adenovirus (Millipore, Billerica, MA), respiratory syncytial virus (RSV) (Millipore, Billerica, MA, and Trinity Biotech USA Inc., Jamestown, NY), and parainfluenza viruses 1, 2, and 3 (PIV1, -2, and -3) (Diagnostic Hybrids, Athens, OH). Additionally, for BAL samples only, culture for adenovirus and multiplex PCR for influenza A virus, influenza B virus, and RSV (Prodesse, Gen-Probe Incorporated, San Diego, CA) were also available. Twelve BAL samples were also sent to a reference lab for human metapneumovirus (HMPV) DFA (Focus Diagnostics, Cypress, CA) based on clinical provider orders.
FilmArray testing.
Patient samples were retrospectively tested at DFCI/BWH for respiratory pathogens with a premarket version of the FilmArray RP panel which included testing for the following pathogens: influenza A virus (H1N1, H1N1 2009, and H3N2), influenza B virus, RSV, parainfluenza viruses 1 to 4, adenovirus, rhinovirus/enterovirus (the assay does not distinguish between these two pathogens), HMPV, coronavirus (229E, HKU1, OC43, and NL63), bocavirus, Mycoplasma pneumoniae, Chlamydophila pneumoniae, and Bordetella pertussis. The FilmArray instrument and pouch system have been described in detail elsewhere (11–13). The research use only version of the FilmArray RP system reported a cycle threshold for each positive PCR assay.
Verification PCR testing.
Study samples for which clinical respiratory virus testing and FilmArray RP assay results were discordant, as well as one sample for which FilmArray and clinical testing were concordant, identified as parainfluenza virus 3, and 12 samples which were negative by both methods, underwent further verification testing at Idaho Technology using validated real-time singleplex PCR assays. The 12 samples which were negative by both methods were randomly selected from all samples which were negative by FilmArray and clinical testing and that had more than one remaining sample aliquot.
Three separate sample preparation methods were used for verification testing, including a DNA preparation to assess for bocavirus, B. pertussis, C. pneumoniae, and M. pneumoniae, a standard RNA preparation to assess for multiple pathogens, including coronaviruses (229E, HKU1, NL63, and OC43), enterovirus, HMPV, influenza A virus (H1, H1N1 2009, and H3), PIV4, and rhinovirus, and a separate standard RNA preparation to assess for RSV. Three samples underwent verification testing using the DNA preparation (including two that were negative for pathogens by both clinical testing and the FilmArray RP assay and one that was positive for bocavirus only by the FilmArray RP assay). Twenty-four samples underwent verification testing using the standard RNA preparation for multiple pathogens (including 7 samples that were negative for pathogens by both clinical testing and the FilmArray RP assay, 16 samples that had discordant results by the two testing methods, and 1 sample that was positive for parainfluenza virus 3 by both methods). Six samples underwent verification testing using the standard RNA preparation for RSV (including three samples that were negative for pathogens by both clinical testing and the FilmArray RP assay, two samples that had discordant results by the two testing methods for RSV, and one sample that tested positive for RSV by clinical testing and for RSV and coronavirus by the FilmArray RP assay).
The assays used a chemistry (real-time, singleplex PCR with hydrolysis probes) and targeted sequences for each virus and bacterium different from those in the assay(s) in the FilmArray (12). The targets for each organism, their primer and probe sequences, and the limit of detection 95 (LoD95) (concentrations of organism or nucleic acid at which 95% of the samples are positive) are shown in Table S1 in the supplemental material. Inclusivity and exclusivity testing used essentially the same organisms as were tested on the FilmArray RP (FilmArray respiratory panel instruction booklet, available upon request). The comparator assays distinguish between enterovirus and human rhinovirus, but this information was not used in the comparison with the FilmArray RP assay results which report only a combined result.
The QIAcube (Qiagen, Valencia, CA) was used to purify nucleic acid for the PCR or reverse transcription-PCRs. For the DNA purification, 500 μl of sample was loaded into the instrument and 200 μl was recovered. For the RNA purifications, 140 μl was loaded and 100 μl was recovered. Ten microliters of purified nucleic acid was used in each 20-μl singleplex PCR. Verification testing at Idaho Technology was performed on coded samples without knowledge of either the clinical testing or the FilmArray RP assay results.
Resolution of concordant and discordant results.
Patients were considered to be infected with a specific respiratory virus if results from clinical testing were positive and matched the FilmArray RP results. Patients were considered not to be infected if the clinical testing and FilmArray RP testing both yielded negative results. In cases where clinical test results did not match FilmArray RP results, if verification testing was concordant with the positive clinical test result or FilmArray RP result, then the patient was also considered to be infected with the respiratory virus. In cases where the clinical testing result and the FilmArray RP result were discordant and confirmatory testing was negative, the patient was considered not to be infected with a respiratory virus. If no verification testing was available for a particular pathogen (parainfluenza viruses 1, 2, and 3), then discordant results could not be resolved.
Because verification testing was used primarily in cases where the clinical testing and FilmArray RP testing results were discordant, because both the FilmArray RP and verification panel tested for more pathogens than did clinical testing, and because not all pathogens tested have a “gold standard” test, true positive and negative predictive values could not be estimated. However, patients were designated as having a respiratory viral disease or not (as described above) in order to tabulate a calculated positive predictive value (cPPV) and calculated negative predictive value (cNPV) for the standard clinical testing available at DFCI/BWH and the FilmArray RP assay. The samples that had concordant positive results for clinical testing and FilmArray RP did not have verification assays and may have had false-positive results in both assays, so each of the cPPVs may be optimistic. Among the samples that had concordant negative results for clinical testing and FilmArray RP which did not have verification testing, there may have been some samples that had false-negative results, so each of the cNPVs may be optimistic.
Statistical analysis.
Clinical testing and the FilmArray RP assay were compared using the exact two-sided McNemar's test. The cPPV and cNPV (as defined above) and their corresponding exact 95% binomial confidence intervals were calculated separately for clinical testing and FilmArray RP testing. All statistical analyses were performed using the software program SAS version 9.2 (SAS Institute, Cary, NC).
RESULTS
Ninety samples were obtained from 87 immunocompromised patients who were undergoing respiratory viral testing for clinical indications. Patient characteristics are shown in Table 1. Nearly half (48%) of the patients were HSCT recipients, and one-third (34%) were SOT recipients; the remainder had hematologic malignancy but had not undergone HSCT. Sample characteristics are shown in Table 2. The majority of samples were obtained for URI or LRI symptoms and a minority for surveillance. The majority of NPA samples were collected for URI or LRI symptoms (only 1 of 34 was collected for surveillance), while the majority of BAL samples were collected for LRI symptoms but with a substantial number (19 of 56) collected for surveillance.
Table 1.
Baseline characteristics of 87 patients from whom respiratory samples were collected
| Characteristic | Value for patients |
|---|---|
| Median age, yrs (range) | 55 (19, 80) |
| No. (%) male | 53 (61) |
| No. (%) with underlying condition | |
| SOTa | 30 (34) |
| HSCT | 42 (48) |
| Hematologic malignancyb | 56 (64) |
| No. (%) with type of hematologic malignancyb | |
| Acute leukemia or myelodysplastic syndrome | 24 (43) |
| Chronic leukemia | 10 (18) |
| Lymphoma | 18 (32) |
| Multiple myeloma | 4 (7) |
Includes 28 lung transplant recipients, 1 kidney transplant recipient, and 1 combined heart and kidney transplant recipient.
Includes 15 patients with hematologic malignancy alone, 40 HSCT recipients who underwent transplantation for hematologic malignancy, and one SOT recipient with hematologic malignancy.
Table 2.
Clinical characteristics of the 90 respiratory samples collected for clinical indicationsa
| Sample characteristic | No. (%) of samples |
|---|---|
| Type of sample | |
| NPA | 34 (38) |
| BAL | 56 (62) |
| Clinical indication for test | |
| URI | 28 (31) |
| LRI | 42 (47) |
| Surveillance | 20 (22) |
Samples obtained from 87 patients, of whom 3 had two respiratory samples taken at least 1 month apart for different clinical indications.
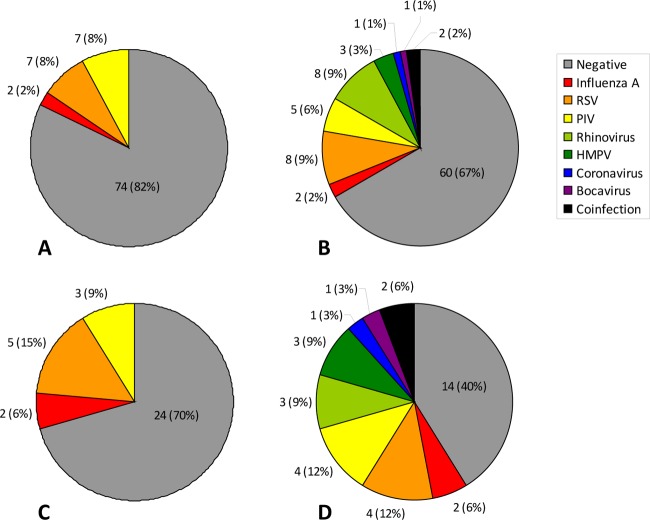
The FilmArray RP assay was significantly more likely to detect a respiratory virus than routine clinical testing at DFCI/BWH. Among 90 samples, the FilmArray RP assay identified 30 with viral pathogens (including 2 samples in which 2 pathogens were detected). In contrast, routine testing at DFCI/BWH identified 16 samples with one viral pathogen each among the 90 samples. Detailed results are shown graphically in Fig. 1 and in Tables 3 and 4. Among the 18 samples on which the two assays disagreed, the FilmArray RP assay identified a viral pathogen in 16 when the clinical testing was negative and the clinical testing identified a viral pathogen in 2 when the FilmArray RP assay was negative. (McNemar P = 0.001). If only verified positive results were counted, there were 13 samples with discordant results, and all had verification of the FilmArray RP result (McNemar P = 0.0002). When FilmArray RP assay results for viruses that could not be detected by clinical testing (coronavirus, rhinovirus/enterovirus, parainfluenza virus 4, and bocavirus) were excluded from analysis, there was no significant difference in the performance of the FilmArray RP assay in comparison to clinical testing (P = 0.51). No bacterial infections with B. pertussis, C. pneumoniae, or M. pneumoniae were identified by either routine clinical testing (in cases where specific testing was pursued) or the FilmArray RP assay in any of the samples.
Fig 1.
Respiratory virus testing results for tests routinely available at DFCI/BWH and the FilmArray RP panel, including the numbers of pathogens and percentages of the total. (A) Results for routine testing at DFCI/BWH for all samples (n = 90). (B) Results for FilmArray RP assay for all samples (n = 90). “Coinfection” includes one sample with RSV and coronavirus and another with HMPV and rhinovirus. (C) Results for routine testing at DFCI/BWH for NPA samples (n = 34). (D) Results for FilmArray RP assay for NPA samples (n = 34). “Coinfection” includes one sample with RSV and coronavirus and another with HMPV and rhinovirus.
Table 3.
Results from clinically indicated testing,a FilmArray RP testing, and verification testing for samples where a respiratory viral disease was considered present based on concordance between two or more testing methods
| No. of samplesb | Clinical result | FilmArray RP result | Verification result |
|---|---|---|---|
| 2 | Influenza virus A | Influenza virus A H1-09 | |
| 6 | RSV | RSV | |
| 2 | Negative | RSV | RSV |
| 1 | RSV | RSV, Coronavirus OC43c | RSVc |
| 1 | PIV1 | PIV1 | |
| 1 | PIV2 | PIV2 | |
| 2 | PIV3 | PIV3d | |
| 1 | Negative | PIV4 | PIV4 |
| 1 | PIV3 | Rhinovirus/enterovirus | Rhinovirus/enterovirus |
| 5 | Negative | Rhinovirus/enterovirus | Rhinovirus/enterovirus |
| 3 | Negative | HMPV | HMPV |
| 1 | Negative | Coronavirus NL63 | Coronavirus NL63 |
Including samples collected for clinical symptoms and for surveillance.
n = 26.
Verification testing for coronavirus OC43 was not performed on this sample, so only RSV infection was confirmed.
Verification testing for other viruses, including coronaviruses (229E, HKU1, NL63, and OC43), enterovirus, HMPV, influenza A virus (H1, H1N1 2009, and H3), PIV4, and rhinovirus was performed on one of these two samples and was negative.
Table 4.
Results from clinically indicated testing,a FilmArray RP testing, and verification testing for samples where respiratory viral disease was not confirmed
| No. of samplesb | Clinical result | FilmArray RP result | Verification result |
|---|---|---|---|
| 58 | Negative | Negative | —c |
| 1 | PIV2 | Negative | —d |
| 1 | PIV3 | Negative | —d |
| 2 | Negative | Rhinovirus/enterovirus | Negative |
| 1 | Negative | HMPV; rhinovirus/enterovirus | Negative |
| 1 | Negative | Bocavirus | Negative |
Including samples collected for clinical symptoms and for surveillance.
n = 64.
—, 12 samples which were negative for respiratory pathogens by DFCI/BWH clinical testing and the FilmArray RP assay underwent verification testing. All 12 samples were negative for viral pathogens by verification testing.
—, verification testing for PIV2 or PIV3 was not performed, but the sample did undergo verification testing for other viruses (coronaviruses [229E, HKU1, NL63, and OC43], enterovirus, HMPV, influenza A virus [H1, H1N1 2009, and H3], PIV4, and rhinovirus), the result of which was negative.
Three patients had two separate samples collected for different clinical indications (different respiratory tract infection symptoms in each case). Two of these patients each had one NPA and one BAL sample each collected more than a month apart, with negative clinical testing and negative FilmArray RP assay results for both samples for both patients. The third patient had two NPA samples collected 3 months apart for different episodes of illness. The first sample tested positive for parainfluenza virus 1 by both clinical testing and the FilmArray RP assay, and the second sample was negative for respiratory virus infection by clinical testing but was positive by FilmArray RP assay for bocavirus. When the second samples were excluded from analysis, the FilmArray RP assay still identified a viral pathogen significantly more often than clinical testing (McNemar P = 0.002).
Verification testing was performed where results between the FilmArray RP and DFCI/BWH clinical testing were discordant (20 samples, including 1 sample with RSV identified by both assays and coronavirus identified only by FilmArray RP), on one sample identified by both assays as parainfluenza virus 3, and on 12 concordant negative samples. Altogether, 33 samples (37%) underwent verification testing. Results of the verification testing are displayed in Tables 3 and 4. Based on these results, viral disease was considered to be present in 26 samples (29%) (Table 3) and absent in 64 samples (71%) (Table 4). Results of verification testing of the 12 samples which were negative in both FilmArray RP and DFCI/BWH clinical testing were all negative.
Among the 26 samples in which viral infection was considered to be present, 13 (50%) had a positive concordant result by both the FilmArray RP assay and clinically indicated testing (including the sample in which RSV and coronavirus were detected by FilmArray but only the RSV was present by clinical testing). The remaining 13 (50%) had a positive result on the FilmArray RP assay that was concordant with the result of verification testing only (for one of them, PIV3 was identified by clinical testing and rhinovirus/enterovirus was identified by FilmArray RP and verification assays). Other than 2 samples in which RSV was identified, the pathogens detected by FilmArray RP assay and verification testing included pathogens not routinely assessed for by clinically indicated testing, including parainfluenza virus 4, rhinovirus/enterovirus, HMPV, and coronavirus.
Among the 64 samples in which viral infection was not considered to be present, 4 samples tested positive by the FilmArray RP assay for at least one virus but had negative results in clinical testing and verification testing. These four samples included two that tested positive for rhinovirus/enterovirus, one that tested positive for rhinovirus/enterovirus and HMPV, and one that tested positive for bocavirus. The median cycle threshold for the viruses detected by FilmArray RP in these four samples was higher than the median cycle threshold for the other 27 viruses detected by FilmArray RP assay and confirmed by validation testing (26.5 versus 12.6). Two samples tested positive by clinical testing for parainfluenza virus 2 and parainfluenza virus 3 but had negative FilmArray RP assay results, and one sample tested positive by clinical testing for parainfluenza virus 3 but tested positive for rhinovirus/enterovirus by FilmArray and verification testing. Because the verification testing panels did not include parainfluenza virus 2 and parainfluenza virus 3, these discrepancies could not be resolved.
The cPPV and cNPV of the FilmArray RP assay and clinical testing to detect a respiratory viral infection are displayed in Table 5. The two samples that tested positive for parainfluenza virus 2 or 3 by clinical testing and negative by the FilmArray RP assay and the third sample that tested positive for parainfluenza virus 3 by clinical testing but positive for rhinovirus by FilmArray RP assay and verification testing did not undergo verification testing for parainfluenza virus 2 or 3 and were therefore excluded from this analysis since the discrepancies could not be resolved. In this context, the overall cPPV of DFCI/BWH clinical testing (1.00) was greater than that of the FilmArray RP (0.87), while the cNPV of the FilmArray (1.00) was greater than that of the DFCI/BWH clinical testing (0.84). Because the indication for collection of BAL samples differed from that for NPA in that some BAL samples were collected for surveillance while NPA samples were collected mostly for symptoms, the cPPV and cNPV were also calculated for BAL and NPA samples individually. Among both BAL and NPA samples, the cPPV for clinical testing was greater than that for the FilmArray (BAL fluid, 1.00 versus 0.89; NPA, 1.00 versus 0.85), while the cNPV for the FilmArray remained greater than that for clinical testing (BAL fluid, 1.00 versus 0.90; NPA, 1.00 versus 0.71). The low cPPV of the FilmArray RP assay can be attributed to the four samples described above that tested positive by the FilmArray RP assay but were not confirmed by validation testing (including two samples that tested positive for rhinovirus/enterovirus, one sample that tested positive for rhinovirus/enterovirus and HMPV, and one sample that tested positive for bocavirus). The cPPV and cNPV of the FilmArray RP assay and clinical testing overall and for NPA and BAL samples specifically did not change much when only the first samples obtained from each patient in the cohort were considered.
Table 5.
Calculated positive and negative predictive values for FilmArray RP and DFCI/BWH clinically indicated testing
| Sample typea | Test | Calculated positive predictive value (CI)b | Calculated negative predictive value (CI)b |
|---|---|---|---|
| Allc | DFCI/BWH | 1.00 (0.79, 1.00) | 0.84 (0.74, 0.91) |
| FilmArray RP | 0.86 (0.68, 0.96) | 1.00 (0.95, 1.00) | |
| BALc | DFCI/BWH | 1.00 (0.37, 1.00) | 0.90 (0.78, 0.97) |
| FilmArray RP | 0.89 (0.52, 1.00) | 1.00 (0.93, 1.00) | |
| NPA | DFCI/BWH | 1.00 (0.74, 1.00) | 0.71 (0.49, 0.87) |
| FilmArray RP | 0.85 (0.62, 0.97) | 1.00 (0.81, 1.00) |
BAL, bronchoalveolar lavage; NPA, nasopharyngeal aspirate.
The estimated positive and negative predictive values did not change much when the second sample obtained from the three patients who underwent testing twice was excluded. CI, confidence interval.
These estimates excluded the three samples in which parainfluenza virus 2 and parainfluenza virus 3 were detected by clinical testing but not by FilmArray RP assay, since verification testing did not test for either of these pathogens.
DISCUSSION
These data demonstrate that in immunocompromised patients, the FilmArray RP assay identified significantly more viral pathogens in BAL and NPA samples than the standard clinical testing available during the study period at our institution. Predictably, the majority of additional pathogens identified by the FilmArray RP assay included those not available by routine testing at DFCI/BWH (rhinovirus/enterovirus, coronavirus, and bocavirus) and those that were available only through reference lab testing (HMPV), which is seldom utilized due to slow turnaround time. The performance of the FilmArray RP assay for this patient population was similar to that reported previously for a general adult and pediatric patient population, in which approximately 50% more viral pathogens were identified by FilmArray than by traditional clinical methods, the majority of which were due to viral pathogens not typically detected by traditional methods (13). Both the wider array of pathogens tested for and the rapid turnaround time of the FilmArray RP assay in comparison to routine testing at DFCI/BWH would fill the need for rapid diagnoses for immunocompromised patients such as those included in this cohort.
This study assessed the performance of the FilmArray RP with both NPA and BAL samples and included the largest number of BAL samples for which the performance of the FilmArray RP has been studied to date (13). The majority of BAL samples in the present study were obtained for symptoms of LRI, though some samples were obtained for surveillance of other conditions, such as rejection in lung transplant recipients. In this context, in which the overall number of BAL samples with any respiratory viruses detected was relatively low (9/56 [16%]), the cNPVs of the FilmArray RP panel on BAL samples were higher than those of routinely available clinical testing at DFCI/BWH, while the cPPV was lower. In contrast, all NPA samples in the present study except one were collected from a symptomatic immunocompromised host, and thus the overall number of samples with respiratory viruses present was relatively high (17/34 [50%]). In this context, the cNPV of the FilmArray RP assay was also higher than that of clinical testing, while the cPPV was lower than that for clinical testing. This low cPPV for the FilmArray RP assay for BAL and NPA samples was likely due to the detection of viral pathogens in one BAL sample and three NPA samples by the FilmArray RP assay that were not confirmed by validation testing.
Clinical testing identified parainfluenza virus in three samples whose results were not confirmed by FilmArray RP assay, including one sample with parainfluenza virus 2 and two samples with parainfluenza virus 3. All three samples were obtained by BAL in patients with symptoms of lower respiratory tract infection. Because only parainfluenza virus 4 was included in the verification testing panel utilized for the study, it is not clear if these results reflect false-positive clinical testing results or false-negative FilmArray RP assay results, and thus these samples were excluded from the cPPV and cNPV calculations.
The FilmArray RP assay identified viral pathogens in four samples (three NPA samples and one BAL sample) that were not confirmed either by clinically indicated testing or by verification testing, including two samples with rhinovirus/enterovirus, one sample with rhinovirus/enterovirus and HMPV, and one sample with bocavirus. Though these results may be false-positive FilmArray RP assay results, it is also possible that these viruses were indeed present in the samples but in a low enough quantity that they were not detected by verification testing. The median cycle threshold for these viruses in the FilmArray RP assay was much higher than that for viruses detected by the FilmArray RP assay in other samples and confirmed by validation testing. This difference in median cycle threshold suggests that there may have been very small amounts of virus present in these samples, leading to false-negative validation testing. This issue also highlights the difficulty of studying the performance of novel respiratory virus diagnostics where there is no gold standard test for many viral pathogens, such as bocavirus.
In addition to the challenge presented by studying viral diagnostics for pathogens in which there is no diagnostic gold standard, this exploratory study was also limited by the relatively small number of samples and the lack of verification testing on all samples. The latter specifically limited our ability to estimate a true positive or negative predictive value, and thus the cPPV and cNPV calculated with the available results are optimistic estimates.
In summary, in comparison to routine clinical testing for respiratory viruses, the FilmArray RP assay detected more viral pathogens among samples obtained from an immunocompromised population. In addition, this assay system performed well on BAL samples. This study provides a practical real-world assessment of the performance of the FilmArray RP assay in a population for whom rapid and accurate diagnosis of viral pathogens is crucial for appropriate clinical management and development of novel therapeutics for respiratory viruses.
Supplementary Material
ACKNOWLEDGMENTS
M.A.P. is employed by Idaho Technology, Inc. The other authors declare no competing financial interests.
This work was supported by a Small Business Innovation Research grant, 1 R43 AI 082843-01, from the NIH/NIAID, by the Harvard Clinical and Translational Science Center, grant no. 1 UL1 RR025758-01 from the NIH/NCRR, and also by the Harvard Center for AIDS Research, grant P30 AI060354-08 from the NIH/NIAID.
We acknowledge Alex McAdam and Richard Rossi for assistance with sample collection, Kody Nilsson for technical assistance, and Sam Richards for directing the verification PCR work.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIAID, NCRR, or the NIH. This article contains information on assays and/or samples types that have not been approved by the FDA for in vitro diagnostic use. Idaho Technology does not promote these products for in vitro diagnostic use.
Footnotes
Published ahead of print 18 July 2012
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Boeckh M. 2008. The challenge of respiratory virus infections in hematopoietic cell transplant recipients. Br. J. Haematol. 143:455–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen YB, et al. 2011. Treatment of parainfluenza 3 infection with DAS181 in a patient after allogeneic stem cell transplantation. Clin. Infect. Dis. 53:e77–e80 doi:10.1093/cid/cir501 [DOI] [PubMed] [Google Scholar]
- 3. Englund JA, et al. 2006. Brief communication: fatal human metapneumovirus infection in stem-cell transplant recipients. Ann. Intern. Med. 144:344–349 [DOI] [PubMed] [Google Scholar]
- 4. Gutman JA, Peck AJ, Kuypers J, Boeckh M. 2007. Rhinovirus as a cause of fatal lower respiratory tract infection in adult stem cell transplantation patients: a report of two cases. Bone Marrow Transplant. 40:809–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ison MG, Michaels MG. 2009. RNA respiratory viral infections in solid organ transplant recipients. Am. J. Transplant 9(Suppl. 4):S166–S172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kamboj M, et al. 2008. Clinical characterization of human metapneumovirus infection among patients with cancer. J. Infect. 57:464–471 [DOI] [PubMed] [Google Scholar]
- 7. Kuypers J, Campbell AP, Cent A, Corey L, Boeckh M. 2009. Comparison of conventional and molecular detection of respiratory viruses in hematopoietic cell transplant recipients. Transpl. Infect. Dis. 11:298–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Loeffelholz MJ, et al. 2011. Comparison of the FilmArray Respiratory Panel and Prodesse real-time PCR assays for detection of respiratory pathogens. J. Clin. Microbiol. 49:4083–4088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Murali S, et al. 2009. Detection of respiratory viruses with a multiplex polymerase chain reaction assay (MultiCode-PLx Respiratory Virus Panel) in patients with hematologic malignancies. Leuk. Lymphoma 50:619–624 [DOI] [PubMed] [Google Scholar]
- 10. Peck AJ, et al. 2007. Respiratory virus infection among hematopoietic cell transplant recipients: evidence for asymptomatic parainfluenza virus infection. Blood 110:1681–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pierce VM, Elkan M, Leet M, McGowan KL, Hodinka RL. 2012. Comparison of the Idaho Technology FilmArray system to real-time PCR for detection of respiratory pathogens in children. J. Clin. Microbiol. 50:364–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Poritz MA, et al. 2011. FilmArray, an automated nested multiplex PCR system for multi-pathogen detection: development and application to respiratory tract infection. PLoS One 6:e26047 doi:10.1371/journal.pone.0026047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rand KH, Rampersaud H, Houck HJ. 2011. Comparison of two multiplex methods for detection of respiratory viruses: FilmArray RP and xTAG RVP. J. Clin. Microbiol. 49:2449–2453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Weinberg A, Zamora MR, Li S, Torres F, Hodges TN. 2002. The value of polymerase chain reaction for the diagnosis of viral respiratory tract infections in lung transplant recipients. J. Clin. Virol. 25:171–175 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.