Abstract
When 3,972 ground beef enrichments with 6 confirmed to contain a non-O157 Shiga toxin-producing intimin-positive Escherichia coli isolate were tested for Shiga toxin, intimin, and O group (O26, O45, O103, O111, O121, and O145) genes, 183 potential positives and only 2 of the 6 confirmed positives were identified.
TEXT
More than 70 different serotypes of Shiga toxin-producing Escherichia coli (STEC) have been described that cause disease in humans (4). Illnesses range from mild diarrhea to bloody diarrhea to hemorrhagic colitis (HC) and hemolytic-uremic syndrome (HUS). E. coli O157:H7 is the STEC most often associated with the most severe forms of disease (4, 17, 19) and is referred to as an enterohemorrhagic E. coli (EHEC). Numerous non-O157 EHEC types have also been linked to similar illnesses and outbreaks of disease (4, 14). Six serogroups, or O groups (O26, O111, O103, O121, O45, and O145), have been described by the CDC as the cause of 71% of non-O157 EHEC disease (4) and have colloquially become known as the “top (or big) 6 STEC.” It is estimated that non-O157 EHEC may cause diarrhea at frequencies similar to those of other enteric bacterial pathogens, such as Salmonella and Shigella (18), while also producing infections that result in outbreaks and HUS (4). In addition to Shiga toxin, EHEC also contain an additional virulence factor, intimin, that allows EHEC to adhere tightly to the host intestinal lining during infection (15).
Recently, the USDA Food Safety and Inspection Service (FSIS) announced its intention to consider STEC strains of the six most frequent serogroups to be adulterants in ground beef, materials intended for ground beef production, and other nonintact beef products (9). The FSIS plans to test for these adulterants using a PCR protocol that initially targets the detection of Shiga toxin genes, stx1 and/or stx2, and the intimin (eae) gene (20). Sample enrichments identified as containing both stx and eae are then tested for the six O groups (11, 20). A beef sample enrichment that is positive for stx, eae, and an O group is considered a potential positive. Potentially positive enrichments are subjected to immunomagnetic separation (IMS) that specifically targets the suspect O group (20). Once IMS products are grown on selective agar plates and suspect colonies are observed that agglutinate appropriate latex reagents, the sample enrichments are classified as presumptively positive until a such a time that the colonies are confirmed by additional biochemical and PCR testing, at which point they are considered confirmed positive.
The initial screening for stx, eae, and O group to identify sample enrichments that are potentially positive is the basis of many commercial tests entering the market to meet the needs of beef processors and testing laboratories. However, this method of screening may identify a substantial number of potentially positive samples due to the simultaneous presence of multiple non-EHEC E. coli that contain just one or two of the targets. Therefore, to estimate the rate of potential positives, we turned to sample enrichments from a recent study (3). In that study, 4,133 samples of commercial ground beef had been collected throughout the United States over a period of 2 years. An stx gene was identified in 24.3%, and an STEC isolate was obtained from 7.3% of the sample enrichments (3). Further analysis of the STEC isolates found that 0.15% of the initial sample enrichments contained a non-O157 EHEC (3) that would now be considered an adulterant. We describe here the rates of prevalence of the O group targets identified by PCR in those sample enrichments and, in conjunction with stx and eae frequencies, the number of samples identified that would be considered potential positives. Using the previously known culture results, we assess the ability of this screening for stx, eae, and an O group to accurately predict the presence of an adulterant non-O157 EHEC.
Screening for Shiga toxin, intimin, and O group genes was performed on DNA (n = 3,972) that had been previously prepared from ground beef-tryptic soy broth (TSB) enrichments (3) and stored at −70°C. Since some of the earliest DNA samples had been in storage for up to 36 months, we first repeated the eae and stx (stx1 and stx2) PCR screenings (3, 16) for this analysis. The identities and numbers of samples that contained stx and eae were compared to those previously reported (3) by using t tests in Prism 5 statistical analysis software (GraphPad, La Jolla CA), which showed there were no significant differences in the levels of prevalence of stx and eae (P > 0.05) between the two data sets. Next, O groups were identified using PCR conditions (16) and primers as previously described that targeted the wzx and wzy genes in the biosynthetic operons of O26 (7), O45 (6), O103 (13), O121 (12), and O145 (10), and open reading frame (ORF) 3.4 of the E. coli O111 rfb region (8).
The results of this analysis identified 1,684 (42.4%) sample enrichments that contained one or more of the nine targets (Fig. 1). Four hundred fifteen (10.4%) of the enrichments contained both eae and an stx gene. Twenty-five contained eae and stx1, 196 contained eae and stx2, and 194 contained eae and both stx1 and stx2. Although the presence of stx2 in E. coli O157:H7 has been associated with more severe disease outcomes (5) the same cannot be said of the non-O157 EHEC (4); therefore, the type of stx present does not influence the identification of potentially positive sample enrichments. The eae target was present in 400 enrichments which did not contain stx, while stx1 and/or stx2 were present in 544 enrichments which did not contain eae. Similar to our previous study of these samples, where 41.5% of stx-positive sample were eae positive (3), 43.3% were found so here. The difference in the prevalence values is due to the smaller number of sample enrichments in this analysis, 3,972 instead of 4,133.
Fig 1.
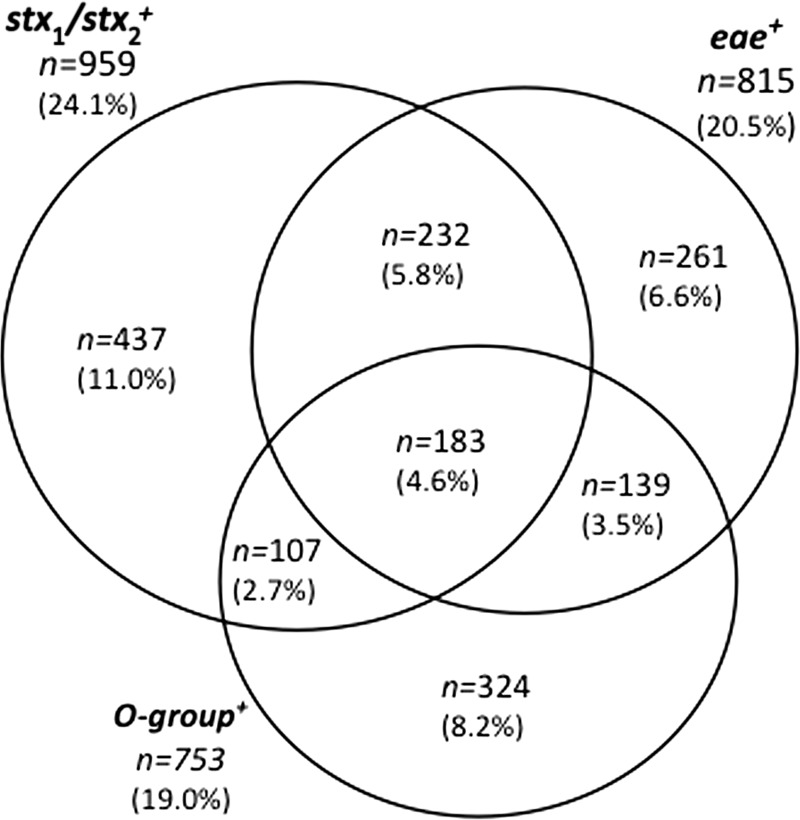
Venn diagram of the relationship of gene targets predicting the presence of the top six adulterant EHEC types in ground beef enrichments (n = 3,972), where 1,684 samples were positive for one or more targets. stx1 and/or stx2 (upper left) was present in 959 enrichments, eae (upper right) was present in 815 enrichments, and one or more O groups (bottom) were present in 753 enrichments. Values represent the number of samples within each region or in overlapping regions, with the percentage of the total number of samples in parentheses.
At least one of the six O group-specific markers was observed in 753 (19.0%) of the enrichments. Three hundred twenty-four (8.2%) of the enrichments contained only O group markers. This is an equal or greater number of enrichments containing the O group than of enrichments containing stx and/or eae. The distribution of the O groups in the enrichments (Table 1) showed that O103 was the most frequent (7.7%), while O111 and O145 were the least frequent (0.05% each). O groups O26, O45, and O121 were present in 5.5, 4.7, and 5.5% of the enrichments, respectively. When samples that were stx positive, eae positive, or stx and eae positive were compared, the rates of prevalence of the O group markers increased to 107 (2.7%), 139 (3.5%), and 183 (4.6%), respectively. The proportion of stx-positive samples also positive for an O group marker was about half that of the samples positive for stx and eae.
Table 1.
Distribution according to presence of stx and eae of non-O157 EHEC O groups based on PCR of commercial ground beef sample enrichments
| Genotypic characterization of sample(s) | Total no. of samples | Total no. (%) O group positivea | No. (%) with O group: |
|||||
|---|---|---|---|---|---|---|---|---|
| O26 | O45 | O103 | O111 | O121 | O145 | |||
| All samples | 3,972 | 753 (19.0) | 218 (5.5) | 186 (4.7) | 306 (7.7) | 2 (0.1) | 220 (5.5) | 2 (0.1) |
| stxb eae negative | 2,613 | 324 (12.4) | 74 (2.8) | 90 (3.4) | 112 (4.3) | 1 (0.03) | 79 (3.0) | 1 (0.0) |
| stx only | 544 | 107 (19.7) | 24 (4.4) | 28 (5.1) | 49 (9.0) | 0 (0.0) | 37 (6.8) | 0 (0.0) |
| eae only | 400 | 139 (34.8) | 46 (11.5) | 34 (8.5) | 61 (15.3) | 1 (0.3) | 33 (8.3) | 0 (0.0) |
| stx eae | 415 | 183 (44.1) | 74 (17.8) | 34 (8.2) | 84 (20.2) | 0 (0.0) | 71 (17.1) | 1 (0.2) |
Number of sample enrichments that had one or more O groups identified as present by PCR.
stx characterization includes stx1 and stx2.
Multiple O groups were identified in 152 (3.9%) of all enrichments tested. Thirty (0.8%) enrichments that did not contain stx or eae were positive for multiple O group markers, while 122 (3.1%) of the enrichments positive for stx and/or eae had multiple O group markers present (data not shown). One hundred twenty-seven enrichments contained two O group markers, 21 enrichments contained three O group markers, and four enrichments contained four O group markers (O26, O45, O103, and O121). Whereas 27 of the 127 enrichments that contained two O group markers lacked both stx and eae, 3 of the 21 enrichments that contained three O group markers lacked both stx and eae. Of the four enrichments that contained 4 O groups, all four also contained stx, while just two contained eae. The only O group not observed in an enrichment that was multiply positive was O145.
A group of 183 enrichments was found to be positive for stx, eae, and an O group. Forty-two STEC isolates were previously obtained from this group (3), as well as 12 enteropathogenic E. coli (EPEC) isolates that contained eae and lacked stx. Three of the EPEC isolates were serogroup O26. In addition, other isolates were found to contain stx, eae, and/or O group genes that would have contributed to a potentially positive screen result for adulterant E. coli types in these enrichments. For example, among STEC isolates of the top six serotypes, 11 were eae negative (3); these STEC isolates were of serotypes O26:Hunt, O103:H2 and H21, and O121:H7, H8, H16, and H19. All of the sample enrichments from which these STEC isolates were obtained were PCR positive for stx, but nine were also PCR positive for eae, presumably from a different source. The presence of these various E. coli isolates is probably the cause for many of the potentially positive interpretations of sample enrichments from which no EHEC isolates were obtained. It cannot be ruled out, however, that a nonisolated EHEC was also present in the samples, because the current PCR testing methods cannot distinguish whether the targets are in one cell or multiple cells.
Only 2 of the 42 STEC isolates present in the 183 samples that screened positive for stx, eae, and an O group were identified as adulterant EHEC (Table 2). The presence of an O103:H2 EHEC isolate and an O145:H28 EHEC isolate was accurately predicted by the PCR screen. However, in the previous study, all stx-positive enrichments were cultured regardless of the presence of eae, and four adulterant EHEC strains were isolated from four enrichments that lacked eae and any of the targeted O groups. Three O103:H2 EHEC isolates were obtained from enrichments that lacked either the eae or O103 screening targets. Furthermore, an O26:H11 EHEC isolate that contained eae and stx1 was obtained from an enrichment that only screened positive for stx2. Therefore, although the screening for stx, eae, and O group markers to identify potentially positive samples appears logical, in practical use, it failed to identify two-thirds of the sample enrichments that contained an adulterant non-O157 EHEC.
Table 2.
Distribution of gene targets (stx and eae) in commercial ground beef sample enrichments that were confirmed by culture to contain an adulterant EHEC
| Serotypic and genotypic characterization of EHEC isolates | Enrichment PCR screen result(s) for: |
Adulterant E. coli present according to screen | |||
|---|---|---|---|---|---|
| O group | stx1 | stx2 | eae | ||
| O26:H11; stx1 eae | None | − | + | − | No |
| O103:H2; stx1 eae | None | + | + | − | No |
| O103:H2; stx1 stx2 eae | None | + | + | + | No |
| O103:H2; stx1 eae | None | + | + | − | No |
| O103:H2; stx1 eae | O45, O103 | + | − | + | Yes |
| O145:H28; stx1 eae | O145 | + | − | + | Yes |
In three other cases, non-O157 EHEC isolates of O groups other than O26, O45, O103, O111, O121, and O145 were isolated. These non-O157 EHEC isolates were serotypes O117:Hunt, Ount:H8, and Ount:H25 and, in addition to stx and eae, also contained other virulence factors that correlate with EHEC disease (3, 5). The screening tests correctly identified two enrichments as containing stx and eae while failing to identify the third, in which eae was not detected. Appropriately, the six O groups were not detected in these three enrichments either. The isolates recovered from these enrichments would not be considered adulterants since they are not of the six most frequent O groups, but their presence suggests that other potentially significant EHEC types may go unrecognized.
There were unavoidable limitations to our PCR screening and culture methods. Standard PCR was performed and the results assessed on agarose gels; this system, while robust, may lack the sensitivity of quantitative PCR methods, such as that in the FSIS Microbiology Laboratory Guidebook (MLG) (20). The primers used in our studies have all been described previously (6, 7, 8, 10, 12, 13, 16), and while they target the same genes as the FSIS MLG primer set (20) and other commercially developed test methods, they are not the same as those used in the other methods. The primers we used may lack specificity or sensitivity to certain subtypes of stx and/or eae compared to those in the most recent revision of the FSIS MLG non-O157 STEC protocol (20) or other, commercial test methods. In addition, the DNA used in our studies was obtained by a boiling lysis method that may not have offered the best template for optimal PCR amplification. It is possible, then, that the rates of prevalence of the targets presented here may underestimate the true values.
Culture confirmation of PCR-positive enrichments is challenging. In our previous study, we noted that the rate at which an STEC isolate was obtained in a ground beef sample enrichment was not different if the enrichment had screened positive for stx1, stx2, or stx1 and stx2 (3). Furthermore, fewer STEC isolates were obtained from samples that had screened positive for the presence of eae in addition to stx (22%) than from samples that were only positive for stx (35%, 3). The isolation method used phenotypic characteristics, such as hemolysis of blood agar, as a means of identifying suspect colonies of STEC. Additional isolation methods that do not rely on colony phenotypes may identify additional isolates, but in comparison studies, methods such as stx oligonucleotide colony hybridization were not able to confirm more enrichments than the phenotypic method used (2). The above-described limitations can explain some of the inconsistencies between the screening and culture results. Since culture isolation had been performed on all sample enrichments that had screened positive for the presence of stx1 and/or stx2, no culture information is available for the enrichments that screened positive for eae only, an O group only, or eae and an O group. It is suspected that a number of EPEC strains would be identified in the eae-positive groups.
In the group of sample enrichments examined here, it appears that the most rapid order for narrowing the set down to the potentially positive group with individual PCR tests would start by targeting the O groups (n = 753), followed by stx (reducing the number to 290) and then eae (reducing the number to 183). However, the FSIS MLG protocol and other commercial test kit methods that detect the presence of non-O157 STEC focus on either initial detection of stx and eae and then by a secondary O group detection or on antigenic O group capture followed by detection of eae and then stx. The results presented here suggest that approaches similar to the latter, which initially target O groups, will identify fewer samples for subsequent analysis than methods that initially target eae and stx. Regardless of the approach taken, our results suggest that not all adulterant STEC isolates may be detected by screening for stx, eae, and O groups. In use, this approach identifies a large number of potentially positive enrichments that cannot be culturally confirmed. In our opinion, the current method casts a wide net to capture few EHEC isolates, and alternative targets, such as virulence-associated genes (5) or EHEC-specific single-nucleotide polymorphisms (1), may provide increased predictive ability and decrease the number of potentially positive enrichments that require confirmatory testing.
ACKNOWLEDGMENT
Names are necessary to report factually on available data; however, the USDA neither guarantees nor warrants the standard of the product, and the use of the name by USDA implies no approval of the product to the exclusion of others that may also be suitable.
Footnotes
Published ahead of print 27 July 2012
REFERENCES
- 1. Bono JL, et al. 2007. Association of Escherichia coli O157:H7 tir polymorphisms with human infection. BMC Infect. Dis. 7:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bosilevac JM, Guerini MN, Koohmaraie M. 2009. Comparison of colony hybridization to phenotype screening on washed sheep's blood agar for the isolation of Shiga toxin producing Escherichia coli from complex matrixes, P02.5.3. Abstr. 7th Int. Symp. Shiga Toxin (Verocytotoxin)-Producing Escherichia coli Infect. Buenos Aires, Argentina, 10 to 13 May 2009 [Google Scholar]
- 3. Bosilevac JM, Koohmaraie M. 2011. Prevalence and characterization of non-O157 Shiga toxin-producing Escherichia coli isolates from commercial ground beef in the United States. Appl. Environ. Microbiol. 77:2103–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brooks JT, et al. 2005. Non-O157 Shiga toxin-producing Escherichia coli infections in the United States, 1983-2002. J. Infect. Dis. 192:1422–1429 [DOI] [PubMed] [Google Scholar]
- 5. Coombes BK, et al. 2008. Molecular analysis as an aid to assess the public health risk of non-O157 Shiga toxin-producing Escherichia coli strains. Appl. Environ. Microbiol. 74:2153–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. DebRoy C, Fratamico PM, Roberts E, Davis MA, Liu Y. 2005. Development of PCR assays targeting genes in O-antigen gene clusters for detection and identification of Escherichia coli O45 and O55 serogroups. Appl. Environ. Microbiol. 71:4919–4924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. DebRoy C, et al. 2004. Detection of Escherichia coli serogroups O26 and O113 by PCR amplification of the wzx and wzy genes. Appl. Environ. Microbiol. 70:1830–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Durso LM, Bono JL, Keen JE. 2007. Molecular serotyping of Escherichia coli O111:H8. J. Microbiol. Methods 69:381–383 [DOI] [PubMed] [Google Scholar]
- 9.Federal Register 2011. Shiga toxin-producing Escherichia coli in certain raw beef products. Fed. Regist. 76:58157–58165 [Google Scholar]
- 10. Feng L, et al. 2005. Structural and genetic characterization of enterohemorrhagic Escherichia coli O145 O antigen and development of an O145 serogroup-specific PCR assay. J. Bacteriol. 187:758–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fratamico PM, et al. 2011. Detection by multiplex real-time polymerase chain reaction assays and isolation of Shiga toxin-producing Escherichia coli serogroups O26, O45, O103, O111, O121, and O145 in ground beef. Foodborne Pathog. Dis. 8:601–607 [DOI] [PubMed] [Google Scholar]
- 12. Fratamico PM, Briggs CE, Needle D, Chen C-Y, DebRoy C. 2003. Sequence of the Escherichia coli O121 O-antigen gene cluster and detection of enterohemorrhagic E. coli O121 by PCR amplification of the wzx and wzy genes. J. Clin. Microbiol. 41:3379–3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fratamico PM, DebRoy C, Strobaugh TP, Jr, Chen C-Y. 2005. DNA sequence of the Escherichia coli O103 O antigen gene cluster and detection of enterohemorrhagic E. coli O103 by PCR amplification of the wzx and wzy genes. Can. J. Microbiol. 51:515–522 [DOI] [PubMed] [Google Scholar]
- 14. Gould H. 2009. Update on the epidemiology of STEC in the United States. Centers for Disease Control and Prevention. 2009 Annual Meeting of the Capital Area Food Protection Association, Non-O157 STEC: Waiting for the Other Shoe To Drop. Washington, DC, 15 September 2009 [Google Scholar]
- 15. Nataro JP, Kaper JB. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Paton AW, Paton JC. 1998. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J. Clin. Microbiol. 36:598–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rivero MA, Passucci JA, Rodriguez EM, Parma AE. 2010. Role and clinical course of verotoxigenic Escherichia coli infections in childhood acute diarrhoea in Argentina. J. Med. Microbiol. 59:345–352 [DOI] [PubMed] [Google Scholar]
- 18. Slutsker L, et al. 1997. Escherichia coli O157:H7 diarrhea in the United States: clinical and epidemiologic features. Ann. Intern. Med. 126:505–513 [DOI] [PubMed] [Google Scholar]
- 19. Tozzi AE, et al. 2003. Shiga toxin-producing Escherichia coli infections associated with hemolytic uremic syndrome, Italy, 1988-2000. Emerg. Infect. Dis. 9:106–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.USDA 2011. Detection and isolation of non-O157 Shiga-toxin producing Escherichia coli (STEC) from meat products. In Microbiology laboratory guidebook. Food Safety and Inspection Service, Office of Public Health Science, USDA, Athens, GA: www.fsis.usda.gov/PDF/Mlg_5B_02.pdf. Accessed 1 February 2012 [Google Scholar]


