Abstract
Spores of thermophilic Geobacillus species are a common contaminant of milk powder worldwide due to their ability to form biofilms within processing plants. Genotyping methods can provide information regarding the source and monitoring of contamination. A new genotyping method was developed based on multilocus variable-number tandem-repeat (VNTR) analysis (MLVA) in conjunction with high-resolution melt analysis (MLV-HRMA) and compared to the currently used method, randomized amplified polymorphic DNA PCR (RAPD-PCR). Four VNTR loci were identified and used to genotype 46 Geobacillus isolates obtained from retailed powder and samples from 2 different milk powder processing plants. These 46 isolates were differentiated into 16 different groups using MLV-HRMA (D = 0.89). In contrast, only 13 RAPD-PCR genotypes were identified among the 46 isolates (D = 0.79). This new method was then used to analyze 35 isolates obtained from powders with high spore counts (>104 spores · g−1) from a single processing plant together with 27 historical isolates obtained from powder samples processed in the same region of Australia 17 years ago. Results showed that three genotypes can coexist in a single processing run, while the same genotypes observed 17 years ago are present today. While certain genotypes could be responsible for powders with high spore counts, there was no correlation to specific genotypes being present in powder plants and retailed samples. In conclusion, the MLV-HRMA method is useful for genotyping Geobacillus spp. to provide insight into the prevalence and persistence of certain genotypes within milk powder processing plants.
INTRODUCTION
The spores of thermophilic bacilli Geobacillus spp. are common contaminants of milk powder (24). Spores present in the raw milk can adhere to surfaces, germinate, and grow as biofilms, which results in the contamination of the milk product (10). While not pathogenic, the presence of these heat-stable spores can result in spoilage of reconstituted milk. Recent studies have focused on identifying the predominant spore-forming species within dairy plants (11) and understanding spore adhesion to surfaces (28, 36) and subsequent biofilm development within a dairy plant (27, 35). However, to date, there has been a lack of a discriminatory and reproducible genotyping assay to track individual strains of Geobacillus spp. through a milk processing plant and its products.
Random amplification of polymorphic DNA PCR (RAPD-PCR) profile analysis is a method that has previously been used to genotype isolates of Geobacillus spp. (42). This technique uses short arbitrary primers that bind to nonspecific sections of the genome. The amplification of these sequences using PCR results in different banding patterns between strains, creating a fingerprint. This technique has been used to characterize strains from Geobacillus spp., Anoxybacillus flavithermus, and Bacillus licheniformis, isolated from milk powders from New Zealand (31) and around the world (33). These studies found a predominant genotype for Geobacillus stearothermophilus, which was designated strain A, along with 2 other genotypes. RAPD-PCR profiling requires no genome sequence information and is quick and easy. However, this technique is known to have poor reproducibility between laboratories and the interpretation of the banding patterns can also be difficult due to weak bands in an isolate's profile, resulting from various efficiencies of the PCR and mismatches between the primer and the DNA template. Another method for genotyping aerobic dairy sporeformers involves sequencing variations found within the housekeeping gene rpoB (7). However, this may not be a suitable genotyping method for Geobacillus spp. since the rpoB gene is highly conserved in this genus compared with other bacilli. Previous studies have shown that sequencing of the variable regions within the rpoB gene could replace 16S rRNA gene sequencing in Geobacillus spp. as a species identification method (22, 41).
Multilocus variable-number tandem-repeat (VNTR) analysis (MLVA) has been used to discriminate between different genotypes within species by analyzing length polymorphisms found within several VNTR loci. Length polymorphisms arise in VNTRs due to the variability of the copy number of tandem repeats found within genes or the noncoding regions of a genome. Copy number differences occur when one or more tandem repeats are added or removed during replication of the genome by slipped-strand mispairing events, giving rise to a genotype different to that of the mother cell. These changes in VNTR length can be analyzed using gel electrophoresis and can be an important source of genetic variation within an organism because of their rapid evolution rates (17, 30, 40). Recently, MLVA has been used to genotype a number of different pathogens such as Salmonella enterica (3), Clostridium difficile (4), Mycobacterium tuberculosis (19), Listeria monocytogenes (23), and Bacillus anthracis (16). In most cases, the MLVA method developed for each of these species provided discrimination values equal to, or greater than, those of the “gold” standard genotyping methods such as pulsed-field gel electrophoresis (PFGE), restriction fragment length polymorphism (RFLP), and multilocus sequence typing (MLST). While a previous study examined the presence of different strains of Listeria monocytogenes directly in food samples (5), there have been no applications of MLVA to genotype food spoilage organisms.
High-resolution melt analysis (HRMA) is a rapid, closed-tube, post-PCR method used for analyzing genetic variations in PCR products using DNA-binding fluorescent dyes and a PCR machine with a highly precise temperature control. Amplicons with different DNA sequences, G+C contents, and lengths are differentiated on the basis of their melt curves. Previous studies have employed HRMA in conjunction with MLVA (MLV-HRMA) for genotyping the pathogens B. anthracis (12) and Pseudomonas aeruginosa (25). These studies found MLV-HRMA to be reproducible and the results to be consistent with those of the original MLVA technique.
This study aimed to develop an MLVA-based genotyping method in conjunction with HRMA to genotype Geobacillus isolates obtained from retailed milk powder and milk powder processing plants. The discriminatory power of this technique was also compared to that found using the RAPD-PCR typing technique. Finally, the MLVA method was applied to investigate genotypes found in milk powders with high spore counts from specific processing plants and used to compare these types to historical isolates obtained from different plants in the same region, isolated up to 17 years ago.
MATERIALS AND METHODS
Milk powder samples and bacterial isolates.
Geobacillus isolates were obtained from dairy samples from (i) milk powder processing plants in 2010, (ii) milk powder processing plants from 1995 to 2000, (iii) specific high-spore-count powders from processing plants in 2010, and (iv) retailed milk powder from samples sourced from Brisbane, Queensland, Australia, and Christchurch, New Zealand, in 2010. These samples were maintained at −20°C or below until used for microbiological analysis.
To obtain isolates, 1 g of each reconstituted powder was treated at 100°C for 30 min before being cultured on tryptic soy agar (TSA) at 55°C overnight. Twenty-seven historical isolates were heat treated at 100°C for 30 min before being subcultured on plate count agar and incubated at 65°C for 48 h. Individual colonies were then isolated and restreaked on TSA and grown under the same conditions to ensure the purity of isolates. Finally, a single colony was picked and grown in tryptic soy broth (TSB) under the same conditions described above. Isolates were stored in 30% (wt/vol) glycerol at −80°C.
In order to obtain isolates from specific high-spore-count milk powders (>104 spores · g−1), seven separate high-spore-count powders were obtained from a single processing plant over a 1-month period in 2010. Isolates from these powders were obtained as described above; however, 5 colonies were purified from each powder sample. This provided a total of 35 isolates from the 7 powder samples.
DNA extraction and identification.
One milliliter of culture from each isolate grown in TSB was centrifuged at 14,000 × g (Centrifuge 5418; Eppendorf). Genomic DNA was then extracted from cells using a DNeasy blood and tissue lysis kit (Qiagen, Australia) according to the manufacturer's instructions for Gram-positive bacteria. Genomic DNA was finally eluted using two individual centrifugation steps (14,000 × g) of 50 μl of elution buffer providing a final volume of 100 μl and was stored at −20°C until required. Isolates were identified as Geobacillus spp. using 16S rRNA gene sequencing (primers are listed in Table 1) and V6 16S rRNA gene HRMA analysis (K. Chauhan et al., unpublished results). Several isolates were also characterized by partial rpoB sequencing based on a previous study by Weng et al. (41). Primers were generated to amplify two variable regions designated A and B within the rpoB gene (Table 1). PCRs were performed in a PTC-200 Peltier thermal cycler (MJ Research) using the following protocol: 94°C for 2 min, followed by 34 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 2 min, followed by 72°C for 5 min. DNA sequencing was performed at the Australian Genome Research Facility (AGRF), Brisbane, Queensland, Australia, or at Macrogen, Seoul, South Korea. The genera were identified by comparing 700 bp of 16S rRNA gene sequences in the 16S Ribosomal Database Project (RDP) (http://rdp.cme.msu.edu/), while rpoB sequences were compared to the sequences with accession numbers listed in a previous study (41).
Table 1.
Geobacillus VNTR loci with primer designations and sequences
| Locus | Primer name | Primer sequence | Predicted annealing temp (°C) |
|---|---|---|---|
| rpoB | rpoB_A_F | 5′-GATGATCGACCGCCGAACGCT-3′ | 59 |
| Region A | rpoB_A_R | 5′-ACGGCGGTTGCCTAAATGGTCG-3′ | 59 |
| rpoB | rpoB_B_F | 5′-GAATCGCGCGACACGAAGCTC-3′ | 59 |
| Region B | rpoB_B_R | 5′-AGCGCCCATACTTCCATCTCGC-3′ | 59 |
| 16S | 16S_F | 5′-AGAGTTTGATCCTGGCTC-3′ | 53 |
| 16S_R | 5′-CGGGAACGTATTCACCG-3′ | 53 | |
| GV4 | GV4_F | 5′-GCCTTCAAGGAGAGCTTGCTGGA-3′ | 57 |
| GV4_R | 5′-CGTCGAAAGCCACAAACGGCTG-3′ | 57 | |
| GV10 | GV10_F | 5′-GTTGCAGCCGATTCATGAACGC-3′ | 57 |
| GV10_R | 5′ATGGCTGCGCGTCTCGATGATG-3′ | 57 | |
| GV12 | GV12_F | 5′-TGCGTTTCGACCCGATGAAGCG-3′ | 53 |
| GV12_R | 5′-ACAGGAAATGGGCGCGGTGAA-3′ | 53 | |
| GV18 | GV18_F | 5′-AGCACTGCTTGTTGGACGAAAACAA-3′ | 57 |
| GV18_R | 5′-AGCCAACATGGCAACGAAACGAG-3′ | 57 |
Primer development for VNTR loci.
The program Tandem Repeats Finder (TRF) (2) was used to locate repeat regions within the genome sequences of Geobacillus kaustophilus HTA426 (NC_006510) and Geobacillus sp. Y4.1MC1 (NC_014650). One hundred potential VNTR loci were identified, and 35 loci were chosen based on repeat size (>20 bp) and copy number (>2). Primers were designed for the flanking regions within 100 bp of either side of the repeat region using PrimerBLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). Primer sets were designated Geobacillus VNTR 1 (GV1) to GV35. Initially, 15 isolates were used to test the primers to determine the amplification and length variation of each locus. Real-time PCRs were performed in a Rotor-Gene Q (Qiagen, Valencia, CA) using Platinum SYBR green quantitative PCR (qPCR) SuperMix-UDG (Invitrogen, Carlsbad, CA). Thermal cycling consisted of 95° for 1 min, followed by 28 cycles of 95°C for 1 min, 55°C for 30 s, and 72°C for 40 min, followed by 72°C for 2 min. HRMA conditions were from 75 to 90°C at 0.1°C · s−1. The MLV-HRMA results were interpreted using the Rotor-Gene Q series software (version 1.7; Qiagen) difference graphs with a single isolate as a control. Melt curves were considered different if the apex position along the x axis differed by more than 0.1°C and if normalized fluorescence differed by ±8 units of the mean curve. For the traditional MLVA method, size differences of PCR products were visualized after 2% agarose gel electrophoresis and staining with Serva DNA stain G (Serva, Heidlberg, Germany).
VNTR loci stability.
In order to determine the stability of the copy number within each locus, 5 isolates of different genotypes were chosen and subcultured on TSA every 24 h for a period of 12 days. DNA was extracted using the DNeasy blood and tissue kit, as described above, after days 1 and 12. The loci in the 5 isolates were analyzed using MLV-HRMA and traditional MLVA as described above.
RAPD-PCR analysis.
RAPD-PCR profiles were used to differentiate strains and were generated using the primer OPR 13 (5′-GGACGACCAAG-3′) (31). The reaction mixture contained GoTAQ green (Promega, Madison, WI) according to the manufacturer's instructions. The total reaction size was 25 μl, which contained 2 μl of template DNA and 47.5 μM OPR 13 primer. PCRs were performed in a PTC-200 Peltier thermal cycler (MJ Research) using the following protocol: 94°C for 2 min, followed by 36 cycles of 94°C for 1 min, 35°C for 1 min, and 72°C for 3 min, followed by 72°C for 5 min. PCR products were visualized after 2% agarose gel electrophoresis and staining with Serva DNA stain G. In order to check for reproducibility of RAPD-PCR analysis between two different DNA extracts from the same 14 isolates, the RAPD-PCR was performed on two separate DNA extractions from the same isolate.
The RAPD-PCR profiles were analyzed using a previous method (1). The molecular size (bp) of each potential band position was determined across all RAPD-PCR profiles. At each band position, two possible alleles were considered either present (a score of 1) or absent (a score of 0). Different RAPD profiles were designated by different scores and classified as different genotypes.
Data analysis and statistics.
The MLV-HRMA genotypes were clustered using PHYLOViZ 1.0 software (downloadable at http://www.phyloviz.net/) (13). This software uses goeBURST algorithm to generate a minimum spanning tree and group the genotypes on various locus variant levels. A genotype was created for each isolate by assigning an MLV-HRMA genotype (MHT) or MLVA genotype (MT). The goeBURST algorithm is essentially the global optimal eBURST algorithm, and therefore, the same approach can be used to interpret the result generated by both algorithms as described previously (8). To compare the discriminatory powers of MLVA, MLV-HRMA, and RAPD-PCR, the Simpson's diversity coefficient (D) was calculated for the 46 isolates genotyped by the 3 different typing methods (14, 15).
RESULTS
Initial screening of Geobacillus VNTR loci.
One hundred VNTR loci were identified using the TRF; however, only 35 loci had a suitable copy number and repeat length. In order to find suitable VNTR loci for genotyping isolates of Geobacillus spp., primers were developed to amplify 35 different VNTR loci and were tested on 15 different Geobacillus isolates. Of these 35 loci, only 7 primer sets could produce single amplicons for all 15 isolates. Of these 7 loci, 4 produced suitable length variations between the 15 different isolates and were chosen for further testing. The characteristics of these VNTR loci, designated GV4, GV10, GV12, and GV18, can be seen in Table 2. These loci were selected as markers for MLVA and MLV-HRMA genotyping of Geobacillus isolates. Six isolates of B. licheniformis and A. flavithermus were also included in the initial screening and did not produce any PCR products using the primers described above (data not shown).
Table 2.
Geobacillus VNTR locus characteristics
| Locus | Genome location (bp) | Repeat size (bp) | Copy no. | Coding region | MLVA D value | MLV-HRMA D value |
|---|---|---|---|---|---|---|
| GV4 | 1,445,932 | 42 | 6.9 | Hypothetical protein | 0.63 | 0.66 |
| GV10 | 2,444,419 | 21 | 5.6 | Hypothetical protein | 0.56 | 0.59 |
| GV12 | 3,131,493 | 42 | 6.5 | Hypothetical protein (preprotein translocase) | 0.67 | 0.73 |
| GV18 | 784,978 | 39 | 14.1 | Hypothetical protein | 0.52 | 0.56 |
| Total | 0.88 | 0.89 |
MLVA and MLV-HRMA of GV loci.
The 4 VNTR loci described above were chosen to screen 46 Geobacillus isolates obtained from retailed and processing plant milk powder samples. MLVA results differentiated the 46 isolates into 15 MTs (D = 0.88). An example of a gel showing length polymorphisms for GV18 between isolates can be seen in Fig. 1. The MLV-HRMA differentiated the 46 isolates into 16 different MHTs (D = 0.89). HRMA difference curves for each locus can be seen in Fig. 2.
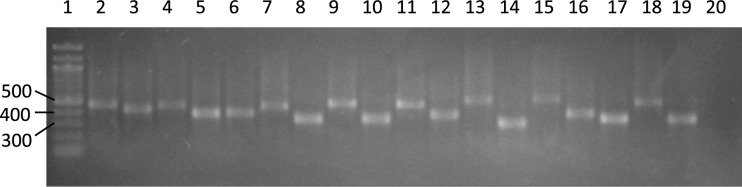
Fig 1.
Examples of GV18 locus amplicons from 18 Geobacillus isolates separated in a 2% agarose gel. Lane 1, DNA ladder; lanes 2 to 19, Geobacillus isolates; lane 20, negative control.
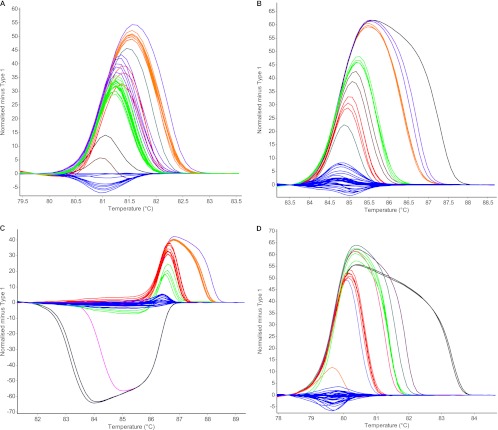
Fig 2.
HRMA difference curves for the four separate VNTR loci GV4 (A), GV10 (B), GV12 (C), and GV18 (D). Different colors in each series of curves represent different MHTs. The same five isolates were used as a reference control for every locus.
Analysis of individual loci by MLVA revealed that GV4, GV10, GV12, and GV18 differentiated all 46 isolates into 4, 5, 5, and 4 MTs, respectively. Analysis of MLV-HRMA curves could differentiate more genotypes for all four loci than MLVA, with GV4, GV10, GV12, and GV18 revealing 9, 8, 7, and 8 MHTs, respectively. The individual MLVA locus GV12 had the greatest D value (0.67), while GV18 had the lowest (0.52). Individual D values were higher for MLV-HRMA, with GV4 being the highest (0.76), while GV18 was the lowest (0.56) (Table 2).
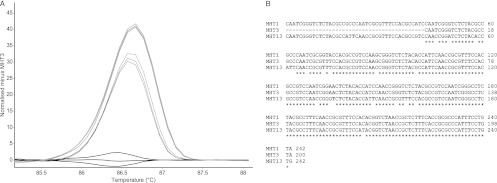
In order to determine the cause of the MLV-HRMA technique differentiating more groups than MLVA, several amplicons from each GV locus were sequenced. In particular, MHTs which did not fall within major groups in difference melt curves were chosen and compared. In all cases, these amplicons had the same copy number but had various G+C contents. An example of this can be seen in Fig. 3. MHTs 1 and 13 for the GV12 locus had the same number of repeat units, while MHT 3 lacked 1 repeat unit (42 bp). MHT 1 and 13 PCR amplicons had different G+C contents of 60% and 59%, respectively.
Fig 3.
(A) HRMA difference curve for the locus GV12, displaying the difference between MHTs 1 (solid gray line), 3 (solid black line), and 13 (dashed line). Each line of the same shading represents a replicate. (B) The nucleotide sequence alignment of these three genotypes. Note that MHT 3 lacks one repeat unit (42 nucleotides) compared to MHTs 1 and 13.
VNTR stability.
In order to determine the stability of the VNTR loci within Geobacillus, 5 isolates were chosen and subcultured daily on TSA for 12 days. DNA was extracted from isolates after 1 and 12 days subculture and their HRMA profiles of 4 loci were compared to their original profiles before subculturing. No changes were observed in either MLVA or MLV-HRMA profiles for any of the isolates or loci, suggesting that these loci are stable over the course of short-term culturing (data not shown).
Minimum-spanning-tree analysis.
Minimum-spanning trees were generated using the goeBURST PHYLOViZ program in order to obtain insight into the evolutionary descent of MHTs (Fig. 4). Clonal groups are distinguished by MHTs at a single locus variant (SLV) level (8). Three clonal groups were revealed among the 46 isolates. Clonal group 1 contained 63% of the isolates tested and all of the isolates obtained from powders with high spore counts. Two other clonal groups, consisting of only two MHTs each, were located. The remaining MHTs differed by two or more loci.
Fig 4.
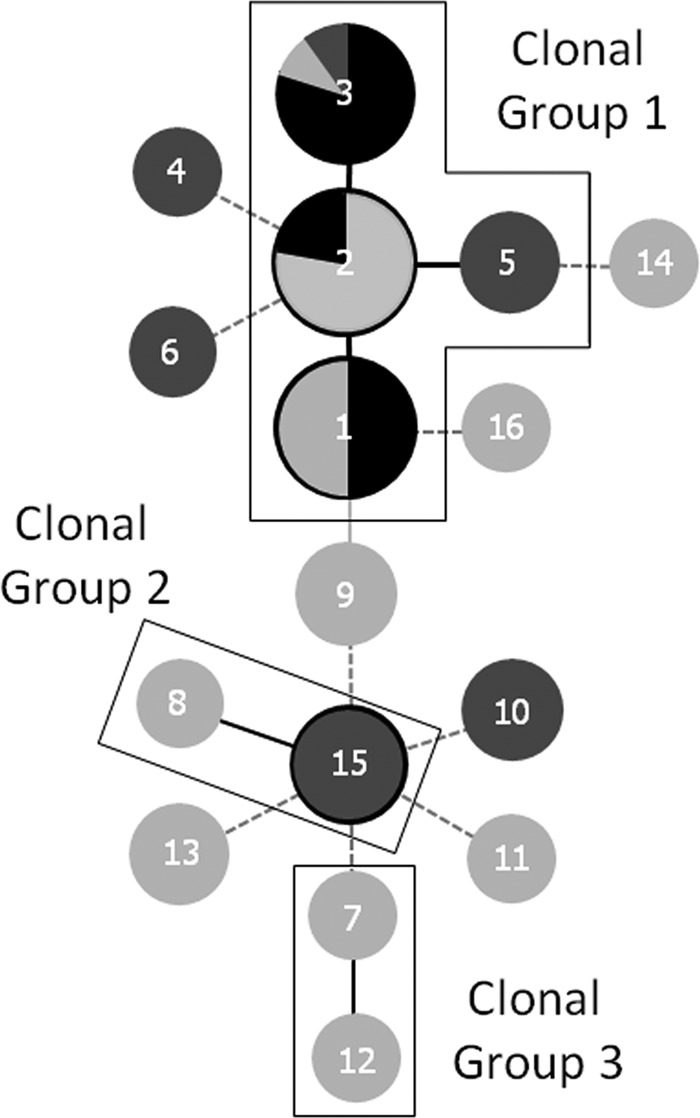
Minimum spanning tree of 16 MLV-HRMA genotypes obtained from 46 isolates of Geobacillus spp. Each node in a tree represents a different MHT, and the genotype number is indicated by the number in the node itself. Clonal groups are encased in a thin black line. Single-locus-variant genotypes are linked by a solid black line, while double- and triple-locus-variant genotypes are linked by solid gray and dashed lines, respectively. Node size represents the number of isolates associated with each genotype. Black shading indicates powders with high spore counts (>104 CFU ml−1), dark gray shading indicates powders with medium spore counts (102 to 104 CFU ml−1), and light gray shading indicates powders with low spore counts (<102 CFU ml−1).
Information such as the spore concentration of powders and the isolation source could be overlaid on the minimum spanning tree. Isolates from powders with high spore counts were genotypes present only in clonal group 1, while isolates found in powders with medium to low counts were dispersed throughout the tree (Fig. 4). However, genotypes were ubiquitous and certain milk powder processing plants did not contain their own unique genotypes.
RAPD-PCR genotyping and comparison to MLV-HRMA.
The same 46 isolates analyzed by MLV-HRMA were analyzed by RAPD-PCR in order to compare the discriminatory powers of both methods. An example of the profiles for the isolates can be seen in Fig. 5A. Two major banding patterns were observed for the 46 isolates. The first can be seen in lanes 2 to 6 and contained a cluster of three bands between 1,000 and 1,300 bp. Examples of the second major banding pattern can be seen in lanes 7 to 15. These contained bands at 3,000, 1,600, 1,300, 1,000, 800, 600, and 300 bp. This second banding pattern closely resembles that of G. stearothermophilus strain A, found in a previous study (33). Slight variations of these two major banding patterns were also observed between isolates. For example, the isolate in lane 11 had stronger intensity bands at ∼3,000 and 1,300 bp but a diminished band at 1,600 bp compared with other isolates consisting of a similar banding pattern. Isolates in lanes 9, 10, and 12 also had an intense band at 900 bp compared to other isolates.
Fig 5.
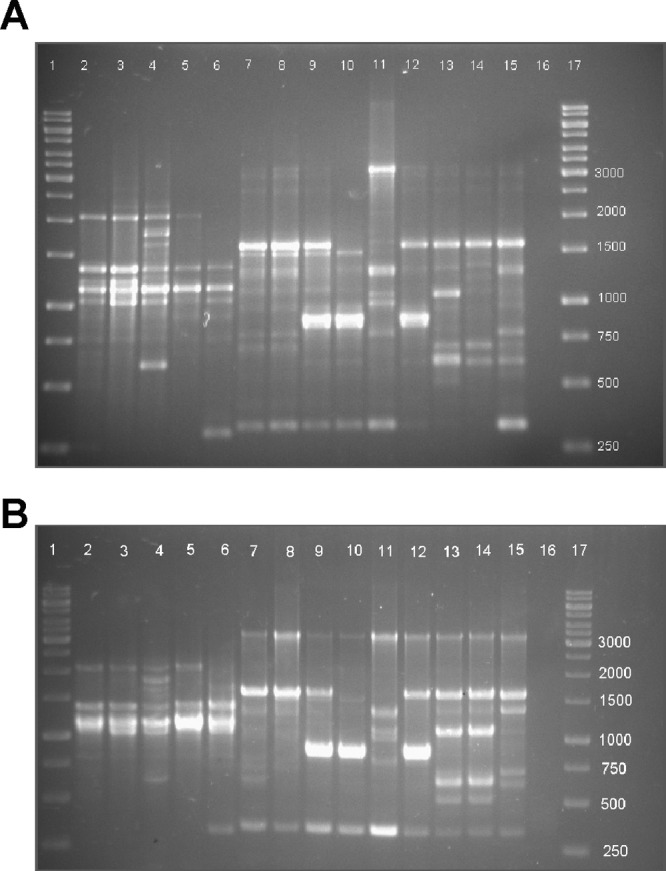
(A and B) Examples of RAPD-PCR banding profiles of 15 isolates from two separate DNA extractions. Lanes 1 and 17, DNA ladder; lanes 2 to 15, isolates; lane 16, negative control. Two major banding patterns can be seen, with the first pattern in lanes 2 to 6 and the second pattern in lanes 7 to 15.
In order to examine the reproducibility of RAPD-PCR profiles, PCRs were performed on different DNA extracts from the same isolates using the same PCR machine. Generally, the profiles from the two extracts were similar; however, minor differences were observed. For example, in lane 14, a minor band was visible at 1,200 bp (Fig. 5A) but faint in the second DNA extract (Fig. 5B). There were also 3 bands present between 500 and 800 bp in lane 15 from the first DNA extract, while there were only 2 from the second extract. Only profiles that were consistently different in both the RAPD-PCR analyses were called different genotypes. Out of 46 isolates analyzed, 13 different genotypes could be identified using RAPD-PCR (D = 0.79).
The discriminatory abilities of RAPD-PCR, MLVA, and MLV-HRMA were compared and produced D values of 0.79, 0.88, and 0.89, respectively. Interestingly, a majority of the isolates grouped together in both the RAPD-PCR and MLV-HRMA techniques (data not shown). The MLV-HRMA technique could distinguish between the four genotypes in clonal group 1, while the RAPD-PCR could not.
rpoB sequencing.
For species identification, 6 isolates (from MHTs 1, 2, 3, 7, 10, and 13) were chosen and 2 variable regions (A and B) within the rpoB gene were sequenced. The highest sequence identities for all 6 isolates using GenBank were to those of Geobacillus thermoleovorans or Geobacillus stearothermophilus (39). However, it was found that, for the same isolate, the two fragments gave conflicting results, whereby one fragment would share greater sequence identity with G. thermoleovorans while the other fragment had greater identity with G. stearothermophilus. This result further confirms the close relatedness of Geobacillus species.
MLV-HRMA of isolates obtained from powders with high spore counts and historical isolates.
Thirty-five isolates (different from those analyzed above) were obtained from 7 different skim milk powders with high spore counts (>1 × 104 spores · g−1), taken from individual processing runs over a 1-month period from a single processing plant and genotyped using the MLV-HRMA technique. All these isolates were found to fall into clonal complex 1 (Fig. 4). The most common genotypes observed matched MHTs 1 and 3, along with a third genotype that matched MHT 2 (data not shown). The GV12 locus was able to differentiate between MHTs 1 and 3, while the GV4 locus could differentiate between MHTs 1, 2, and 3. The two other loci GV10 and GV18 were unable to differentiate between the isolates. Interestingly, up to three different genotypes (MHTs 1, 2, and 3) were detected in a single gram of milk powder obtained from a single processing run, indicating that multiple genotypes can exist in a processing line at one time. However, several powder samples had only one MHT, which was either MHT 1 or 3.
Twenty-seven historical isolates obtained from milk powder processing plants other than the ones tested above but from the same region of Australia between 11 and 17 years ago were also genotyped using the MLV-HRMA technique. Of the 27 isolates, 23 matched MHT 1 and 1 matched MHT 13, while the 3 remaining isolates generated 3 different MHTs not observed before in the present study.
DISCUSSION
The goal of this study was to develop an easy, reproducible, and highly discriminatory genotyping method to provide insight into Geobacillus isolates from milk powder production plants and retailed powders. This new technique (MLV-HRMA) was then compared with MLVA gel-based interpretations and the previously used typing method RAPD-PCR. Finally, the technique was applied to genotype potentially problematic isolates obtained from powders with high spore counts and historical isolates obtained up to 17 years ago from processing plants in the same region of Australia.
The number of VNTR loci tested in this study is similar to what has been developed in previous studies. Most studies have used 5 to 15 loci (20, 23, 29, 34, 37, 39); however, other studies have used either a single locus (25) or as many as 42 loci (18). Increasing and optimizing the number of loci would likely improve the discriminatory power of the current method; however, as mentioned above, primer sets for 35 loci were initially screened. To achieve this, more VNTR loci would likely need to be located within the genome sequences of more closely related Geobacillus spp. isolated from dairy plants rather than deep-sea thermal vents, which is where the genome-sequenced G. kaustophilus strain was isolated. This MLV-HRMA typing method could be employed to genotype environmental Geobacillus spp., since the VNTRs were first located in genomes of environmental isolates. Furthermore, the number of loci that could be amplified in all strains may be low since the technique genotypes strains of Geobacillus at a genus level rather than at a single species level. The genus Geobacillus has been known to be highly conserved, and 16S rRNA gene sequences have failed to accurately identify different species (22, 26, 41, 43). Identification solely on the basis of 16S rRNA genes for Geobacillus spp. has resulted in misnaming a number of isolates (6). Previous studies have chosen other housekeeping genes such as recA, recN, or rpoB as targets for species identification of Geobacillus spp. (41, 43). In the current study, we sequenced two variable regions of the rpoB gene of 6 isolates and found >99% identity with both G. thermoleovorans and G. stearothermophilus. This is in agreement with previous research, which suggested that these two species are predominant in milk powder manufacturing plants (11).
The comparison of the Simpson's index of diversity between the different typing techniques revealed that MLV-HRMA provided a greater level of discrimination than RAPD-PCR while only a small improvement compared to the gel-based analysis of the same amplicons in MLVA. Similar groupings of isolates were observed in both MLV-HRMA and RAPD-PCR techniques. Most of the isolates that gave the second banding pattern (Fig. 5, lanes 7 to 15) similar to that of G. stearothermophilus strain A were the same MHTs in clonal complex group 1 from the MLV-HRMA. While the RAPD-PCR technique grouped all the isolates together in clonal group 1 (lanes 7 and 8 in RAPD-PCR gels), the MLV-HRMA method could differentiate these isolates and divide them into MHTs 1, 2, and 3, giving rise to a higher discriminatory power for the method. Isolates displaying the first banding pattern (lanes 2 to 6) in the RAPD-PCR profiles were found in clonal complex groups 2 and 3, as well as all related double-locus variants. Here we found that the reproducibility of RAPD-PCR profiles was reasonable, but there were still clear banding profile differences between replicate experiments, making it difficult to ascertain true genotypic variations.
The MLV-HRMA technique could differentiate a larger number of genotypes for each individual locus than the traditional MLVA method. This suggests that MLV-HRMA can differentiate amplicons which have the same or similar lengths. This could be due to two possible mechanisms. First, the basis of HRMA depends on an amplicon's size and G+C content. Tandem-repeat regions could be more prone to base mutations, resulting in the presence of single nucleotide polymorphisms (SNPs) within the VNTR loci of isolates with the same number of repeats. While Keim et al. have already discussed the discriminatory power of MLVA and SNP analysis separately, along with their respective mutation frequency (17), there has been no mention of the presence of SNPs occurring within tandem-repeat regions. A second possible explanation for the presence of these SNPs may be the result of convergent evolution, where two unrelated strains of Geobacillus with slightly different G+C contents have the same number of repeats within a single VNTR locus. A similar study looked at the number of repeat units in an MLVA study looking at the genus Brucella; however, the sequence of these repeats was not examined across species (20). Further research would need to be done to confirm either of these possibilities.
The VNTR loci used in this study were found to be stable over a short period of time (12 days). This is similar to what other studies have found previously when checking VNTR stability (38). Keim et al. discussed the relationship of VNTR stability and growth rates in B. anthracis (17). For example, B. anthracis can replicate only in a host and not in soil, and therefore, the evolution rate of VNTR loci may be very low. A similar conclusion may be applicable for Geobacillus spp., since they have a higher death rate than growth rate in soil below 40°C (21); however, they have a very high growth rate in dairy processing plants due to the abundance of nutrients and thermophilic conditions (9). The milk powder production process is also a single-pass system, meaning spores that end up in the final product are not reintroduced at the start of a processing line. However, some spores and cells associated with biofilms may remain in the plant after a cleaning-in-place (CIP) regime. This may be the reason for the relatively low number of genotypes observed compared to the number of isolates obtained from samples and that the same genotypes are present today, as they were up to 17 years ago, in processing plants in the same region of Australia. This is not surprising, since the same genotypes of B. licheniformis found in present-day milk powder were also discovered in 90-year-old milk powder from Shackleton's hut in Antarctica (32).
From the initial screening of 46 isolates, specific plants did not contain unique genotypes, indicating that genotypes were ubiquitous across all plants in Victoria, Australia, and even retailed powders from New Zealand. However, isolates obtained from powders with high spore counts in the initial screening were found in clonal group 1. To confirm this, we screened additional spore count powders. Two different MHTs were predominant in powders with high spore counts from a single processing plant within Australia. One of these MHTs was also present in historical isolates obtained from different plants 17 years ago. These results indicate that these two MHTs could potentially be “problem” strains with unique attributes that allow them to proliferate in a milk powder processing environment. Rueckert et al. found that one RAPD-PCR genotype of Geobacillus spp. was ubiquitous in different milk powder samples from around the world and that A. flavithermus, rather than Geobacillus spp., was present in powders with high spore counts (33). While the present study failed to isolate A. flavithermus from Australian milk powders with high spore counts, the development of an MLVA-based typing method for A. flavithermus using the same approach described here could also prove useful, as a genome sequence for this organism is available.
In conclusion, the MLV-HRMA typing method developed in the current study is easy and reproducible and had a greater discriminatory power and reproducibility than RAPD-PCR. This method could be enhanced if more VNTR loci are located as more suitable Geobacillus genomes from a dairy environment become available. This new typing method could aid plant operators in identifying and monitoring persistent and potentially problematic isolates that could be causing high spore levels in milk powder and could trace the origin of contamination in dairy processing plants.
ACKNOWLEDGMENTS
This study was funded by The Geoffrey Gardiner Dairy Foundation and Dairy Innovation Australia Limited. Kanika Chauhan and Rajat Dhakal are in receipt of postgraduate scholarships awarded by The University of Queensland and Endeavor Australia, respectively.
We thank the dairy processing plants that supplied milk samples for this study.
Footnotes
Published ahead of print 3 August 2012
REFERENCES
- 1. Adzitey F, Rusul G, Huda N, Cogan T, Corry J. 2012. Prevalence, antibiotic resistance and RAPD typing of Campylobacter species isolated from ducks, their rearing and processing environments in Penang, Malaysia. Int. J. Food Microbiol. 154:197–205 [DOI] [PubMed] [Google Scholar]
- 2. Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boxrud D, et al. 2007. Comparison of multiple-locus variable-number tandem repeat analysis, pulsed-field gel electrophoresis, and phage typing for subtype analysis of Salmonella enterica serotype enteritidis. J. Clin. Microbiol. 45:536–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Broukhanski G, Low DE, Pillai DR. 2011. Modified (M) MLVA for rapid identification and typing of Clostridium difficile during institutional outbreaks. J. Clin. Microbiol. [Epub ahead of print.] doi:10.1128/JCM.02359-02310. [DOI] [PMC free article] [PubMed]
- 5. Chen S, Li J, Saleh-Lakha S, Allen V, Odumeru J. 2011. Multiple-locus variable number of tandem repeat analysis (MLVA) of Listeria monocytogenes directly in food samples. Int. J. Food Microbiol. 148:8–14 [DOI] [PubMed] [Google Scholar]
- 6. Dinsdale AE, et al. 2011. Emended descriptions of Geobacillus thermoleovorans and Geobacillus thermocatenulatus. Int. J. Syst. Evol. Microbiol. 61:1802–1810 [DOI] [PubMed] [Google Scholar]
- 7. Durak MZ, Fromm HI, Huck JR, Zadoks RN, Boor KJ. 2006. Development of molecular typing methods for Bacillus spp. and Paenibacillus spp. isolated from fluid milk products. J. Food Sci. 71:M50–M56 [Google Scholar]
- 8. Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Flint S, Bremer P, Brooks J. 1997. Biofilms in dairy manufacturing plant—description, current concerns and methods of control. Biofouling 11:81–97 [Google Scholar]
- 10. Flint S, Palmer J, Bloemen K, Brooks J, Crawford R. 2001. The growth of Bacillus stearothermophilus on stainless steel. J. Appl. Microbiol. 90:151–157 [DOI] [PubMed] [Google Scholar]
- 11. Flint S, Ward LJH, Walker K. 2001. Functional grouping of thermophilic Bacillus strains using amplification profiles of the 16S-23S internal spacer region. Syst. Appl. Microbiol. 24:539–548 [DOI] [PubMed] [Google Scholar]
- 12. Fortini D, et al. 2007. Optimization of high-resolution melting analysis for low-cost and rapid screening of allelic variants of Bacillus anthracis by multiple-locus variable-number tandem repeat analysis. Clin. Chem. 53:1377–1380 [DOI] [PubMed] [Google Scholar]
- 13. Francisco AP, et al. 2012. PHYLOViZ: phylogenetic inference and data visualization for sequence based typing methods. BMC Bioinformatics 13:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grundmann H, Hori S, Tanner G. 2001. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J. Clin. Microbiol. 39:4190–4192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hunter P, Gaston M. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Keim P, et al. 2000. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 182:2928–2936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Keim P, et al. 2004. Anthrax molecular epidemiology and forensics: using the appropriate marker for different evolutionary scales. Infect. Genet. Evol. 4:205–213 [DOI] [PubMed] [Google Scholar]
- 18. Klevytska AM, et al. 2001. Identification and characterization of variable-number tandem repeats in the Yersinia pestis genome. J. Clin. Microbiol. 39:3179–3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Le Flèche P, Fabre M, Denoeud F, Koeck J, Vergnaud G. 2002. High resolution, on-line identification of strains from the Mycobacterium tuberculosis complex based on tandem repeat typing. BMC Microbiol. 2:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Le Flèche P, et al. 2006. Evaluation and selection of tandem repeat loci for a Brucella MLVA typing assay. BMC Microbiol. 6:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Marchant R, Bannat IM, Franzetti A. 2011. Thermophilic bacteria in cool soils: metabolic activity and mechanisms of dispersal. In Fontaneto D. (ed), Biogeography of microscopic organisms: is everything small everywhere? Cambridge University Press, New York, NY [Google Scholar]
- 22. Meintanis C, et al. 2008. Application of rpoB sequence similarity analysis, REP-PCR and BOX-PCR for the differentiation of species within the genus Geobacillus. Lett. Appl. Microbiol. 46:395–401 [DOI] [PubMed] [Google Scholar]
- 23. Murphy M, et al. 2007. Development and application of multiple-locus variable number of tandem repeat analysis (MLVA) to subtype a collection of Listeria monocytogenes. Int. J. Food Microbiol. 115:187–194 [DOI] [PubMed] [Google Scholar]
- 24. Murphy P, Lynch D, Kelly P. 1999. Growth of thermophilic spore forming bacilli in milk during the manufacture of low heat powders. Int. J. Dairy Technol. 52:45–50 [Google Scholar]
- 25. Naze F, et al. 2010. Pseudomonas aeruginosa outbreak linked to mineral water bottles in a neonatal intensive care unit: fast typing by use of high-resolution melting analysis of a variable-number tandem-repeat locus. J. Clin. Microbiol. 48:3146–3152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nazina TN, et al. 2001. Taxonomic study of aerobic thermophilic bacilli: descriptions of Geobacillus subterraneus gen. nov., sp. nov. and Geobacillus uzenensis sp. nov. from petroleum reservoirs and transfer of Bacillus stearothermophilus, Bacillus thermocatenulatus, Bacillus thermoleovorans, Bacillus kaustophilus, Bacillus thermoglucosidasius and Bacillus thermodenitrificans to Geobacillus as the new combinations G. stearothermophilus, G. thermocatenulatus, G. thermoleovorans, G. kaustophilus, G. thermoglucosidasius and G. thermodenitrificans. Int. J. Syst. Evol. Microbiol. 51:433–446 [DOI] [PubMed] [Google Scholar]
- 27. Parkar SG, Flint SH, Brooks JD. 2003. Physiology of biofilms of thermophilic bacilli—potential consequences for cleaning. J. Ind. Microbiol. Biotechnol. 30:553–560 [DOI] [PubMed] [Google Scholar]
- 28. Parkar SG, Flint SH, Palmer JS, Brooks JD. 2001. Factors influencing attachment of thermophilic bacilli to stainless steel. J. Appl. Microbiol. 90:901–908 [DOI] [PubMed] [Google Scholar]
- 29. Ramisse V, et al. 2004. Variable number of tandem repeats in Salmonella enterica subsp. enterica for typing purposes. J. Clin. Microbiol. 42:5722–5730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reyes JF, Tanaka MM. 2010. Mutation rates of spoligotypes and variable numbers of tandem repeat loci in Mycobacterium tuberculosis. Infect. Genet. Evol. 10:1046–1051 [DOI] [PubMed] [Google Scholar]
- 31. Ronimus RS, et al. 2003. A RAPD-based comparison of thermophilic bacilli from milk powders. Int. J. Food Microbiol. 85:45–61 [DOI] [PubMed] [Google Scholar]
- 32. Ronimus RS, Rueckert A, Morgan HW. 2006. Survival of thermophilic spore-forming bacteria in a 90+ year old milk powder from Ernest Shackleton's Cape Royds hut in Antarctica. J. Dairy Res. 73:235–243 [DOI] [PubMed] [Google Scholar]
- 33. Rueckert A, Ronimus RS, Morgan HW. 2004. A RAPD-based survey of thermophilic bacilli in milk powders from different countries. Int. J. Food Microbiol. 96:263–272 [DOI] [PubMed] [Google Scholar]
- 34. Schouls LM, van der Heide HGJ, Vauterin L, Vauterin P, Mooi FR. 2004. Multiple-locus variable-number tandem repeat analysis of Dutch Bordetella pertussis strains reveals rapid genetic changes with clonal expansion during the late 1990s. J. Bacteriol. 186:5496–5505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Scott SA, Brooks JD, Rakonjac J, Walker KMR, Flint SH. 2007. The formation of thermophilic spores during the manufacture of whole milk powder. Int. J. Dairy Technol. 60:109–117 [Google Scholar]
- 36. Seale RB, Flint SH, McQuillan AJ, Bremer PJ. 2008. Recovery of spores from thermophilic dairy bacilli and effects of their surface characteristics on attachment to different surfaces. Appl. Environ. Microbiol. 74:731–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Titze-de-Almeida R, et al. 2004. Multilocus variable-number tandem-repeat polymorphism among Brazilian Enterococcus faecalis strains. J. Clin. Microbiol. 42:4879–4881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Top J, Schouls LM, Bonten MJM, Willems RJL. 2004. Multiple-locus variable-number tandem repeat analysis, a novel typing scheme to study the genetic relatedness and epidemiology of Enterococcus faecium isolates. J. Clin. Microbiol. 42:4503–4511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Van Ert M, et al. 2007. Global genetic population structure of Bacillus anthracis. PLoS One 2:e461 doi:10.1371/journal.pone.0000461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vogler AJ, et al. 2007. Mutations, mutation rates, and evolution at the hypervariable VNTR loci of Yersinia pestis. Mutat. Res. 616:145–158 [DOI] [PubMed] [Google Scholar]
- 41. Weng FY, Chiou CS, Lin PHP, Yang SS. 2009. Application of recA and rpoB sequence analysis on phylogeny and molecular identification of Geobacillus species. J. Appl. Microbiol. 107:452–464 [DOI] [PubMed] [Google Scholar]
- 42. Williams JGK, Kubelik AR, Livak KJ, Rafallski JA, Tingey SV. 1990. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 18:6531–6535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zeigler DR. 2005. Application of a recN sequence similarity analysis to the identification of species within the bacterial genus Geobacillus. Int. J. Syst. Evol. Microbiol. 55:1171–1179 [DOI] [PubMed] [Google Scholar]