Abstract
Obacunone belongs to a class of unique triterpenoids called limonoids, present in Citrus species. Previous studies from our laboratory suggested that obacunone possesses antivirulence activity and demonstrates inhibition of cell-cell signaling in Vibrio harveyi and Escherichia coli O157:H7. The present work sought to determine the effect of obacunone on the food-borne pathogen Salmonella enterica serovar Typhimurium LT2 by using a cDNA microarray. Transcriptomic studies indicated that obacunone represses Salmonella pathogenicity island 1 (SPI1), the maltose transporter, and the hydrogenase operon. Furthermore, phenotypic data for the Caco-2 infection assay and maltose utilization were in agreement with microarray data suggesting repression of SPI1 and maltose transport. Further studies demonstrated that repression of SPI1 was plausibly mediated through hilA. Additionally, obacunone seems to repress SPI2 under SPI2-inducing conditions as well as in Caco-2 infection models. Furthermore, obacunone seems to repress hilA in an EnvZ-dependent fashion. Altogether, the results of the study seems to suggest that obacunone exerts an antivirulence effect on S. Typhimurium and may serve as a lead compound for development of antivirulence strategies for S. Typhimurium.
INTRODUCTION
Salmonella species continue to be an important health problem. According to the FoodNet surveillance network 2009 preliminary report on infectious diseases, Salmonella species were among the leading causes of food-borne illnesses in the United States. A total of 7,039 cases of Salmonella infection were reported in 2009 (8). Salmonella enterica serovar Enteritidis and Salmonella enterica serovar Typhimurium were among the 10 major serotypes identified as causative agents. An estimated 1.4 million cases of nontyphoidal Salmonella infections occur in the United States annually, resulting in 15,000 hospitalizations and 400 deaths (57). This grim situation may worsen further, especially as the spread of antibiotic-resistant strains of Salmonella from various sources is being reported (61). In order to counter Salmonella infections, it is imperative to identify and develop strategies toward nonconventional targets. The antivirulence approach targets the functions essential for infection rather than viability (9). Bacterial virulence systems such as the secretion system, quorum sensing systems, pilus, and adhesins are some of the actively pursued nonconventional targets for development of antivirulence drugs (9, 20, 51).
S. Typhimurium contains several virulence determinants, such as fimbrial/nonfimbrial adhesins, flagella, the virulence plasmid, the spv gene cluster, and Salmonella pathogenicity islands (SPIs) (46). In particular, SPI1 is required for initial attachment and subsequent internalization of the pathogen to the intestinal cells (2). SPI2, on the other hand, plays an important role in intracellular survival and systemic infection (38, 40, 53). In addition, several SPI1-encoded effectors contribute to the pathogen's successful intracellular existence (5). Numerous genetic and environmental factors regulate expression of SPI1- and SPI2-encoded type III secretion systems (TTSSs) and other virulence determinants. Furthermore, motility in Salmonella Typhimurium LT2 is regarded as an important virulence factor (46). Identification of an antivirulence agent which influences all or any of these virulence factors may have preventive and therapeutic potential.
Limonoids are a unique class of secondary metabolites present in Citrus species. Obacunone, a limonoid (Fig. 1), is present in significant quantities as glucoside and aglycone in citrus juices and seeds (17, 18, 39). Commonly consumed citrus fruits and juices such as grapefruit and orange juice may contain up to 11 ppm obacunone (17, 18, 39), while lemon seeds are a rich source of obacunone (as much as 29 ppm) (15) and may serve as a good source of raw material for purification purposes. Chemically, limonoids are triterpenoids, characterized by the presence of a furan ring and a high degree of oxygenation. In vitro and animal studies suggests that obacunone and a few other limonoids may have potential anticarcinogenic activity against certain types of cancers (34, 41, 42, 52). Furthermore, experiments on normal cell lines and animal models (41, 42) as well as with human volunteers suggest that limonoids may have low toxicity (30). A 500-mg/kg (of body weight) dose of obacunone per day was well tolerated and did not have adverse effects in rats (52), indicating a low toxicity.
Fig 1.
Structure of obacunone.
Recent data from our laboratory suggest that certain limonoids may act as inhibitors of bacterial quorum sensing and associated phenotypes such as TTSS, biofilm, and motility (55, 56). Specifically, obacunone appears to interfere with autoinducer-mediated cell-cell signaling and to repress the locus of enterocyte effacement, which encodes TTSS in Escherichia coli O157:H7 (56), and may potentially serve as a lead compound to develop novel antivirulence agents. We wondered if obacunone has an antivirulence effect on Salmonella. The present report demonstrates that obacunone exerts an antivirulence effect on S. Typhimurium LT2 by repressing SPI1 and SPI2.
MATERIALS AND METHODS
Chemicals.
Previously purified obacunone (56) was used in the current study. Briefly, defatted powder of grapefruit seeds was extracted with acetone, concentrated under vacuum, and chromatographed on silica gel column. Obacunone was eluted with dichloromethane-ethyl acetate (95:5). A stock solution of obacunone was prepared by dissolving 20 mg obacunone in 1 ml dimethyl sulfoxide (DMSO).
Bacterial strains, plasmid, and growth conditions.
Bacterial strains used in this study are listed in Table 1. Unless otherwise specified, bacterial cultures were grown in Luria-Bertani (LB) medium at 37°C with shaking at 200 rpm. When appropriate, the medium was supplemented with 10 μg of chloramphenicol or 10 μg of tetracycline per ml medium.
Table 1.
Bacterial strains, plasmids, and primers used in the current study
| Strain, plasmid, or primer | Description or sequence (5′–3′) | Catalog no. or reference |
|---|---|---|
| Strains | ||
| Salmonella Typhimurium LT2 | Wild type | ATCC 15277 |
| Salmonella Typhimurium SL1344 EE658 | hilA080::Tn5lacZY (Tetr) | 26 |
| Salmonella Typhimurium SL1344 RL829 | ΔaraBAD22 envZ182::cam hilA080::Tn5lacZY (Camr Tetr) | 26 |
| Plasmid pBAD33 | Camr, arabinose-inducible expression vector | 14 |
| Primers | ||
| Gene | ||
| hilA | (F) GTCCGGTCGTAGTGGTGTCT | 54 |
| (R) CGCATACTGCGATAATCCCT | ||
| prgK | (F) GGGTGGAAATAGCGCAGATG | 54 |
| (R) TCAGCTCGCGGAGACGATA | ||
| invB | (F) CTGGGCGCAATTGGGTGCTG | 54 |
| (R) GCTCCCCATTCTGCTCCCCC | ||
| sopE2 | (F) ATACCGCCCTACCCTCAGAAG | This study |
| (R) GCCTGCATCAACAAACAGACA | ||
| hyaA | (F) GCCTGCTCCTCCACACGCTG | This study |
| (R) CCCCACCGTATCGGCGGTTG | ||
| malF | (F) ATTGACCAACGGCGGGCCAG | This study |
| (R) ATTGCCAGCGCGCCTACCAG | ||
| malK | (F) AATCGTGGTGCTGGACGCCG | This study |
| (R) CAATCGCGGTGGCGGTCACT | ||
| rpoA | (F) GCGCTCATCTTCTTCCGAAT | This study |
| (R) CGCGGTCGTGGTTATGTG | ||
| ssrA | (F) ACGGTTTGGCCAGTGAGCGA | This study |
| (R) CGCACAACCTTGCATGGCCC | ||
| ssrB | (F) ACGCTCGCGCAAAGTAAGCAGT | This study |
| (R) AGCCGCAGGTGCTAATGGCT | ||
| spiC | (F) TGGGCATCCTGCCAGAGGAGA | This study |
| (R) AACCATCCCCCATCCGCTGTGA | ||
| envZ cloning | (F) GGCTGGAATGGTACCGGCA | This study |
| (R) CAGGCGCGCAAGCTTATCTG |
A 1.96-kb fragment containing the envZ gene was amplified from S. Typhimurium LT2 genomic DNA using Deep Vent DNA polymerase (New England BioLabs, Ipswich, MA) and a primer pair presented in Table 1. The PCR was performed under the following conditions: initial denaturation at 95°C for 5 min followed by 30 cycles with denaturation at 95°C for 15 s, annealing at 55°C for 30 s, and extension at 72°C for 2 min. The last cycle was followed by a final extension for 5 min at 72°C. Primers were designed to create restriction sites by altering one base in the sequence. The amplified fragment (≈1 μg) was double digested with KpnI and HindIII in NEB 2 buffer for 1 h at room temperature and cloned into pBAD33, generating plasmid pBADenvZ. The generated plasmid was electroporated into S. Typhimurium SL1344 strain RL829 (37).
Growth and metabolic activity.
Diluted (100-fold in LB medium) overnight cultures of S. Typhimurium LT2 (200 μl) were incubated in 96-well plates with 100 μg/ml obacunone or an equivalent amount of DMSO. Optical density at 600 nm (OD600) was monitored for 16 h at 15-min intervals in a Synergy HT MultiMode microplate reader (BioTek Instruments, Winooski, VT). The instrument was set to maintain 37°C with shaking at medium speed. The viability of S. Typhimurium was measured using alamarBlue as described before (55).
Total RNA extraction and array hybridization.
Overnight cultures of S. Typhimurium LT2 were diluted 100-fold in LB medium and grown to an OD600 of ≈1.0 (approximately 11 generation cycles) at 37°C in the presence of 100 μg/ml obacunone or DMSO. The RNA was stabilized by addition of 5 volumes of RNAProtect bacterial reagent (Qiagen Inc., Valencia, CA) and isolated with an RNeasy minikit (Qiagen Inc.). To remove all DNA, samples were treated with DNase I (30 units) for 15 min on column, followed by elution of RNA with elution buffer. RNA quality was analyzed with a Bioanalyser 2100 (Agilent Technologies Inc., Santa Clara, CA), and quantity was determined with an ND-1000 spectrophotometer (NanoDrop Technologies; Fisher Scientific).
S. Typhimurium LT2 genome microarrays, version 4, containing 5,462 open reading frames (ORFs) were obtained from the Pathogen Functional Genome Resource Center (PFGRC), National Institute of Allergy and Infectious Diseases. The arrays were hybridized as described earlier (54). Briefly, 10 μg total RNA from each treatment was converted to cDNA using a random primer (Invitrogen Life Technologies, Carlsbad, CA) and labeled with Cy3 and Cy5 monoreactive dyes (GE Healthcare, Piscataway, NJ). The labeled cDNA was column purified (Qiaquick PCR purification kit; Qiagen Inc.), and equal quantities of labeled cDNA from obacunone- and DMSO-treated samples were used to hybridize microarrays in dye-swap fashion. A total of 6 dye-swapped hybridizations, corresponding to three biological replicates, were conducted. Hybridizations were carried out at 42°C for 18 h. The arrays were washed and scanned on a GenePix 4100A scanner (Molecular Devices Corporation, Sunnyvale, CA) at 532 and 635 nm.
Microarray data collection and analysis.
Signal intensities from the images were extracted using GenePix 6.0 software (Molecular Devices Corporation). The median signal intensities were normalized using LOWESS (43) by Acuity 4.0 software. Genes differentially expressed between obacunone- and DMSO-treated samples were identified by Student's t test. P values were adjusted for multiple comparisons using the Benjamini-Hochberg method (3). Genes with a P value of <0.01 were considered significantly differentially expressed and reported.
Quantitative reverse transcription-PCR (qRT-PCR).
For validation of the microarray study, three independent total RNA samples were collected from S. Typhimurium cultures grown in LB medium to an OD600 of ≈1.0 in the presence of obacunone or DMSO. RNA was extracted and processed under conditions similar to those for the microarray study. Gene-specific primers (Table 1), designed with Primer 3 (48), were used to amplify selected genes using 25 ng cDNA. For measurement of ssrAB and spiC expression, S. Typhimurium cultures, grown to an OD600 of ≈1.0 in 2-(N-morpholino)ethanesulfonic acid (MES)-buffered magnesium minimal medium (MgM), were used to extract RNA and for subsequent gene-specific amplification. To determine the levels of specific genes inside Caco-2 cells, the Caco-2 cells (1 × 106) were infected with S. Typhimurium LT2 at a multiplicity of infection (MOI) of 100 and incubated at 37°C and 5% CO2 in a humidified chamber. Cells were washed with Dulbecco's phosphate-buffered saline (DPBS) after 2 h, and total RNA was extracted using TRIzol (Invitrogen Life Technologies, Carlsbad, CA), for adhesion-mimicking conditions. For invasion-mimicking conditions, initial medium was replaced with gentamicin-containing medium after 1 h and plates were further incubated for 90 min. For all experiments, first-strand synthesis was carried out using murine leukemia virus (MuLV) reverse transcriptase enzyme and random hexamer (Applied Biosystems, Foster City, CA) for 60 min at 42°C followed by 5 min at 99°C. The expression levels of specific genes were quantified by amplifying 25 ng cDNA with 10 pmol primers using SYBR Green PCR mix (Applied Biosystems, Foster City, CA) on an ABI-Prism 7000 HT (Applied Biosystems, Foster City, CA). The amplifications were carried out with the following cycle parameters: one cycle of 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. After completion of amplification cycles, melt curve data were generated to determine the amplification quality. The relative expression levels for obacunone/DMSO were calculated by the threshold cycle (2−ΔΔCT) method (24). The means ± standard deviations (SDs) of three biological replicates are presented.
Adhesion and gentamicin protection (invasion) assays.
The adhesion assay was performed as described previously (50). Colon adenocarcinoma epithelial Caco-2 cells (ATCC CCL-228) were purchased from the ATCC (Manassas, VA) and routinely maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS). For adhesion assay, Caco-2 cells were plated in 6-well plates at a density of 1 × 105 cells/well. S. Typhimurium LT2 was grown overnight in a 25-ml culture. The Salmonella cells were centrifuged at 5,000 × g for 10 min and resuspended in 0.9% saline solution. The cells were counted by dilution in 0.9% saline solution and plating on tryptic soy agar (TSA) plates. The Caco-2 cells were infected with S. Typhimurium LT2 cells at a multiplicity of infection (MOI) of 10, in the presence of 100 μg/ml obacunone or an equivalent amount of DMSO. The plates were incubated for 3 h at 37°C and 5% CO2. The excess S. Typhimurium LT2 cells were washed three times with PBS. The Caco-2 cells were lysed with 0.1% Triton X-100 solution. The resulting lysate was then diluted using 0.9% saline solution in 6 serial dilutions and plated on the tryptic soy agar plates. Colonies were counted after 24 h and recorded as CFU.
The invasion assay was performed as described before (45). For the gentamicin protection assay, 1 × 105 Caco-2 cells were infected with S. Typhimurium LT2 at an MOI of 10 in the presence of 100 μg/ml obacunone or DMSO. One hour after infection at 37°C and 5% CO2, the medium was replaced with DMEM plus 10% FBS and 100 μg/ml gentamicin to kill noninvasive cells, and incubation was continued for an additional 90 min. After incubation, the Caco-2 cells were washed three times with PBS and lysed with 0.1% Triton X-100 solution. The CFU were enumerated by plating the appropriate dilutions in 0.9% saline solution on TSA.
β-Galactosidase assays.
Overnight cultures of S. Typhimurium strains EE658 and RL829 were diluted (1:100) in LB medium and grown to an OD600 of ≈1.0 in the presence of 100 μg/ml obacunone or DMSO. The β-galactosidase assay was performed as described previously (26, 35). Briefly, 1 ml cells was pelleted and resuspended in chilled Z-buffer. A600 for resuspended cells was recorded, and 100 μl of cells was further diluted with Z-buffer and permeabilized with 0.1% SDS and chloroform. O-Nitrophenyl-β-galactoside (4 mg/ml) was added to diluted culture and incubated at room temperature. Sodium carbonate (1 M) was added to stop the reaction. A420 and A550 were measured spectrophotometrically. The Miller units were calculated using the following equation: Miller units = 1,000 × [(OD420 − 1.75 × OD550)/(T × V × OD600)]. The data were analyzed with an analysis of variance (ANOVA) followed by Tukey's pairwise multiple comparison test on SPSS 16.0 (SPSS Inc., Chicago, IL). The effect was considered significant at P < 0.01.
Maltose utilization assay.
To test for the ability to use maltose as a carbon source, S. Typhimurium was grown on morpholinepropanesulfonic acid (MOPS) minimal medium (36). The overnight culture was diluted 100-fold in MOPS minimal medium supplemented with 0.2% maltose or glucose as the sole source of carbon, and S. Typhimurium growth was measured in the presence of 100 μg/ml obacunone as described under “Growth and metabolic activity.” In addition, M63 medium was prepared containing 2 g (NH4)2SO4, 13.6 g KH2PO4, 0.5 mg FeSO4 · 7H2O, and 1 mM MgSO4 · 7H2O but without Casamino Acids and vitamins. The S. Typhimurium growth was measured in M63 medium supplemented with 0.2% maltose or glucose in the presence of 100 μg/ml obacunone as described above.
Microarray data accession number.
The microarray data set is available at the NCBI Gene Expression Omnibus database (accession no. GSE37234).
RESULTS
Effect of obacunone on Salmonella growth.
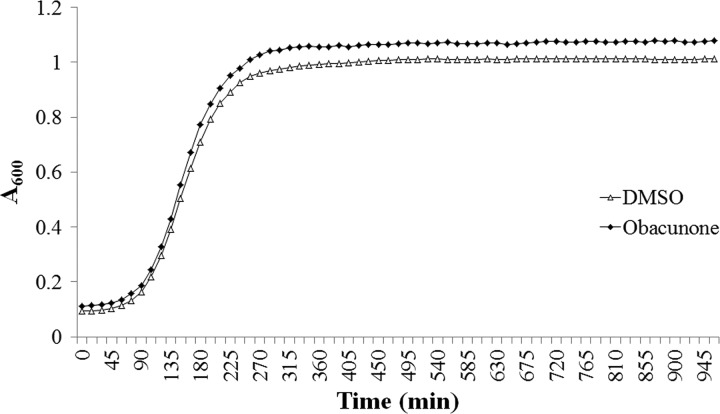
Growth of S. Typhimurium LT2 in the presence of 100 μg/ml obacunone or an equivalent amount of DMSO was measured up to 16 h. Obacunone did not seem to affect S. Typhimurium growth at the tested concentrations (Fig. 2). The mean generation times for DMSO and obacunone treatments were 22.07 and 22.03 min, respectively.
Fig 2.
Growth curve of S. Typhimurium LT2 in the presence of 100 μg/ml obacunone and DMSO.
Microarray expression profile.
To understand the response of S. Typhimurium LT2 to obacunone, cDNA microarrays from PFGRC were used. Total RNA samples prepared from three different obacunone- and DMSO-treated cultures of S. Typhimurium were converted to cDNA and labeled with Cy3/Cy5 monoreactive dyes. Equal quantities of Cy3- and Cy5-labeled cDNAs from DMSO and obacunone treatments were mixed and hybridized to 6 Salmonella Typhimurium LT2 version 4 microarrays. The arrays contain open reading frames (ORFs) from strain LT2 SGSC1412 including plasmid, Salmonella enterica serovar Typhi CT18, and Salmonella enterica serovar Ty2. The data were lowess normalized and evaluated for quality. Student's t test with the Benjamini-Hochberg correction was employed to determine the ORFs differentially expressed between control and obacunone treatments. Upon exposure to 100 μg/ml obacunone, a total of 175 genes (3.2%) were differentially expressed (P < 0.01) out of 5,462 ORFs present. However, only 95 genes demonstrated a >2-fold change in the expression in either direction. Several regulons were downregulated by obacunone exposure, including Salmonella pathogenicity island 1 (SPI1), the maltose regulon, and the hydrogenase 1 operon (Table 2).
Table 2.
Differentially regulated gene/ORF of S. Typhimurium LT2 upon exposure to 100 μg/ml obacunone
| Gene or ORF | Putative identification | Fold change (±SD) |
|---|---|---|
| Virulence | ||
| prgK | Needle complex inner membrane lipoprotein | −9.9 (0.5) |
| prgJ | Needle complex minor subunit | −11.3 (0.8) |
| hilA | Invasion protein regulator | −8.2 (0.7) |
| iagB | Invasion protein precursor | −7.3 (1.0) |
| sicP | Secretion chaperone | −9.0 (0.7) |
| sopE2 | Invasion-associated secreted protein | −10.2 (0.5) |
| sipD | Translocation machinery component | −3.5 (0.7) |
| sipC | Translocation machinery component | −3.9 (0.8) |
| sicA | Surface presentation of antigen secretory proteins | −2.4 (0.6) |
| spaR | Needle complex export protein | −7.3 (0.5) |
| spaP | Surface presentation of antigen protein SpaP | −7.3 (0.8) |
| invJ | Needle length control protein | −2.7 (0.4) |
| invI | Needle complex assembly protein | −4.2 (0.8) |
| invB | Secretion chaperone | −8.7 (0.6) |
| invA | Needle complex export protein | −5.1 (0.6) |
| invE | Invasion protein | −3.2 (0.6) |
| invG | Outer membrane secretin precursor | −7.5 (0.4) |
| invF | Invasion regulatory protein | −5.6 (0.7) |
| invH | Needle complex outer membrane lipoprotein precursor | −2.6 (0.9) |
| Hydrogenase | ||
| hyaA | Hydrogenase 1 small subunit | −9.4 (0.6) |
| hyaB | Hydrogenase 1 large subunit | −8.7 (0.9) |
| hyaC | Hydrogenase 1 b-type cytochrome subunit | −9.1 (0.8) |
| hyaD | Hydrogenase 1 maturation protease | −8.9 (1.2) |
| hyaE | Hydrogenase 1 operon protein HyaE | −7.9 (0.8) |
| hyaF | Putative hydrogenase 1 protein | −8.9 (0.7) |
| hycH | Hydrogenase 3 large subunit processing protein | −9.7 (0.6) |
| Maltose | ||
| malF | Maltose transporter membrane protein | −5.4 (0.6) |
| malK | Maltose/maltodextrin transporter ATP-binding protein | −6.1 (0.4) |
| lamB | Maltoporin | −6.2 (0.6) |
| malM | Maltose regulon periplasmic protein | −5.3 (0.6) |
| malE | Maltose ABC transporter periplasmic protein | −8.8 (0.8) |
Validation of microarray data by qRT-PCR.
In order to verify the results obtained from the microarray experiment, qRT-PCR was performed on the selected genes. Based on the interest, one gene from each affected operon was selected (Table 1). The housekeeping gene rpoA was used as a control. The threshold cycle (CT) values were normalized to levels of rrsH, and fold change was calculated by the 2(−ΔΔCT) method (24). A comparison of microarray data and qRT-PCR data is presented in Table 3. In general, the results of qRT-PCR analysis were consistent with the microarray results. However, the numerical values obtained from qRT-PCR differed from the values in microarray analysis. This difference was possibly a result of the different sensitivities of the two assays and because of the fact that different biological samples were used for qRT-PCR analysis.
Table 3.
Validation of microarray-based expression profiles of selected genes by qRT-PCR
| Gene | Relative expression level |
|
|---|---|---|
| Microarray | qRT-PCR | |
| hilA | −8.2 | −8.02 (±0.4) |
| prgK | −9.9 | −8.91 (±0.3) |
| invB | −8.7 | −6.83 (±0.1) |
| sopE2 | −10.2 | −5.90 (±0.5) |
| hyaA | −9.4 | −6.09 (±0.3) |
| malF | −5.4 | −2.21 (±0.1) |
| malK | −6.1 | −2.78 (±0.2) |
| rpoA | 1.10 (±0.1) | |
Regulation of SPI1 by obacunone.
A downregulation of about 19 genes of the SPI1-carried genes by 2.4- to 11.3-fold (Table 2) was detected using microarray analysis. Among these was the central transcriptional regulator of SPI1, hilA (downregulated by 8.2-fold), in response to obacunone. Needle complex structural protein genes prgK and prgJ (carried in the prg/org operon) and invG were repressed by 9.9-, 11.3-, and 7.9-fold, respectively. Furthermore, export apparatus protein genes spaR and spaP were downregulated. The iagB gene, encoding a muramidase, speculated to help in the passage of needle through the peptidoglycan layer, was also repressed (7.3-fold). The chaperones sicP and invB were downregulated by 9.0- and 8.7-fold, respectively. In addition to the repression of SPI1-carried genes, obacunone also downregulated the effector sopE2, carried outside SPI1. It is pertinent to note that InvF/SicA, encoded in SPI1, regulates the expression of sopE2 (10).
The repression of SPI1 would be predicted to show defects in adhesion and invasion of epithelial cells. To confirm the findings of the microarray experiment, the effect of obacunone on the adhesion to and invasion of epithelial cell line Caco-2 was examined. Caco-2 cells were grown in DMEM and infected with S. Typhimurium LT2 in the presence of 100 μg/ml obacunone. The obacunone exposure resulted in a significantly reduced number of adhesive Salmonella cells (P < 0.01). In the presence of obacunone, 4.85 log CFU Salmonella cells were recovered compared to 7.34 log CFU in DMSO treatment (Fig. 3). Furthermore, obacunone reduced the number of Salmonella cells by ≈1.84 log units (P < 0.01) in a gentamicin protection assay (Fig. 3). These results were consistent with the downregulation of SPI1.
Fig 3.
Effect of 100 μg/ml obacunone on adhesion and invasion by Salmonella Typhimurium LT2 on Caco-2 cells. The Caco-2 cells (1 × 105) were seeded in six-well plates and infected with S. Typhimurium LT2 at a multiplicity of infection of 10. The number of S. Typhimurium cells attached to Caco-2 cells was counted by lysing Caco-2 cells 3 h postinfection, plating them on LB agar plates, and counting the colonies after 24 h. The gentamicin protection assay as described in the text was used to count the number of invasive S. Typhimurium cells. The asterisk denotes a significant difference (P < 0.01) from DMSO treatment.
Regulation of maltose operon.
MalEFG (49) and MalK-LamB-MalM (44) constitute the ABC transport complex, which facilitates the uptake of maltose and maltodextrins (4). Obacunone exposure resulted in downregulation of malEFG and malK-lamB operons. Specifically, malE and malF were downregulated by 8.8- and 5.4-fold, respectively. The malK-lamB-malM genes, which are transcribed in the opposite direction from the malEFG operon, were downregulated by 6.1-, 6.2-, and 5.3-fold, respectively.
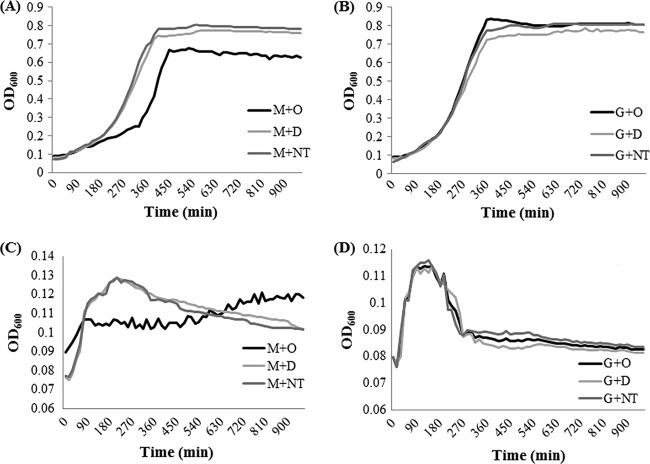
Repression of the mal operon would then suggest that obacunone might impair the uptake of maltose and consequently reduce the growth rate of S. Typhimurium on maltose. To investigate if obacunone impairs maltose transport, growth of S. Typhimurium in the MOPS minimal medium and M63 medium with maltose (0.2%) as sole carbon source was measured. Glucose (0.2%) was used as a positive control. In MOPS medium, the growth rates of S. Typhimurium were similar for obacunone and DMSO treatments when glucose was used as a carbon source (Fig. 4B). However, suppressed growth of S. Typhimurium was observed in the presence of obacunone when maltose was used as a carbon source (Fig. 4A). These results indicate a maltose uptake impaired by obacunone, resulting in a lower growth rate. To more clearly discern the effect of obacunone on maltose uptake, M63 medium deprived of Casamino Acids and vitamins was prepared. The medium was expected to be stringently limited in its resources and ability to sustain growth. The prepared medium was supplemented with maltose or glucose (0.2% in each case) and used to study the growth pattern of S. Typhimurium. The S. Typhimurium growth pattern showed a typical response in M63 minimal medium with glucose as a sole carbon source, where the OD600 reached its maximum at 210 min and decreased thereafter, giving rise to a bell-shaped curve (Fig. 4D, G+NT). Additionally, the growth curves were similar for obacunone, DMSO, and medium without any treatment, when glucose was used as a carbon source. For maltose-supplemented cultures, the growth curves for medium without treatment and DMSO treatment simulated the patterns of glucose-treated cultures (Fig. 4C), suggesting that in DMSO-treated cultures the bacteria exhausted the available carbon source in the first 210 min and the population faced a nutrient limitation afterwards. In contrast, obacunone-treated cultures demonstrated a lower but steady growth rate, indicating a reduced rate of maltose uptake, which might have prevented the early exhaustion of carbon source and maintained a steady state. Taken together, the data indicate that obacunone downregulates the mal operon and impairs the maltose uptake.
Fig 4.
Growth of Salmonella Typhimurium LT2 in the presence of 100 μg/ml obacunone as determined on MOPS minimal medium using maltose (A) and glucose (B) as the sole carbon source. M63 minimal medium was prepared without Casamino Acids and vitamins and used to elucidate S. Typhimurium growth in the presence of 100 μg/ml obacunone using maltose (C) and glucose (D) as the sole carbon source. The data represent the means of three biological replicates. M, maltose; G, glucose; O, obacunone; D, DMSO; NT, no treatment.
Regulation of hydrogenase operon.
The nomenclature of S. Typhimurium hydrogenases is not well established. Therefore, the annotation assignment presented by Zbell et al. (59) was followed in the current study. Specifically, symbols hyaABCDEF were used for the genes with locus tags STM1786 to STM1791. Obacunone seems to repress the entire operon hyaABCDEF by 7.9- to 9.4-fold (Table 2). In addition, downregulation of hycH (9.7-fold), which encodes the large subunit of hydrogenase 3, was observed.
Obacunone influences hilA expression in a dose-dependent fashion.
HilA is a central regulator for genes carried in SPI1 (1, 2). To determine if obacunone exerts a dose-dependent effect on hilA expression, S. Typhimurium was grown in the presence of 6.25, 12.5, 25, 50, and 100 μg/ml obacunone, and hilA expression was measured using qRT-PCR. The results of the study indicate that obacunone influences the expression of hilA in a concentration-dependent fashion (Fig. 5). Exposure of 12.5 μg/ml obacunone suppressed hilA expression by 2.34-fold. The lowest tested concentration, 6.25 μg/ml, produced a change of −1.99-fold. These data indicate that obacunone at a low dose of 12.5 μg/ml may be able to alter the gene expression pattern of hilA significantly and possibly also that of hilA-dependent genes.
Fig 5.
Expression of hilA in the presence of different concentrations of obacunone. Salmonella Typhimurium LT2 was grown in the presence of different concentrations of obacunone, and qRT-PCR was conducted as described in Materials and Methods. The data are presented as means of three biological replicates ± SDs. The change in gene expression was considered significant if the fold change was >2-fold.
Effect of obacunone on genes carried in SPI2.
Expression of SPI2 is induced inside macrophages and epithelial cells, primarily when Salmonella is present in specialized phagosomes, Salmonella-containing vacuoles (40, 53). SsrAB is a two-component system encoded within SPI2 which controls the expression of SPI2 genes encoding TTSS and effectors, while SpiC is an effector encoded within SPI2 (13, 33, 38). Additionally, ssrAB is transcribed in the opposite direction from spiC. In order to understand the effect of obacunone on SPI2, relative expression levels of ssrA, ssrB, and spiC in MES-buffered magnesium minimal medium (MgM), which was reported to stimulate SPI2 TTSS gene expression (11, 13), were measured. We first measured the induction of ssrAB and spiC in MES-buffered MgM over LB medium in S. Typhimurium grown to an OD600 of ∼1.0. ssrA, ssrB, and spiC were induced by 7.7-, 6.0-, and 8.6-fold, respectively, in MES-buffered MgM compared to LB medium (Fig. 6A). To determine the effect of obacunone, S. Typhimurium was grown to an OD600 of ∼1.0 in MES-buffered MgM in the presence of 100 μg/ml obacunone or DMSO. The pH of the cultures was maintained at 7.5 and did not change during the course of investigation. Obacunone seems to repress the expression of ssrA, ssrB, and spiC by 9.7-, 11.8-, and 6.8-fold, respectively, in MES-buffered MgM (Fig. 6B), while a +1.1-fold change was recorded for rpoA.
Fig 6.
(A) Expression of ssrA, ssrB, and spiC was measured in MES-buffered magnesium minimal medium. The fold change was calculated over LB medium-grown cultures. The cultures were grown to an OD600 of ≈1.0 before RNA was extracted and qRT-PCR was performed. (B) Expression of ssrA, ssrB, and spiC upon exposure to 100 μg/ml obacunone in MES-buffered magnesium minimal medium. The cultures were grown to an OD600 of ≈1.0 in the presence of obacunone or DMSO before RNA was extracted and qRT-PCR was performed. The fold change was calculated as obacunone/DMSO.
Effect of obacunone on expression of SPI1- and SPI2-carried genes in Caco-2 cells.
SPI1 and SPI2 are implicated in initial attachment, subsequent internalization, and systemic infection of S. Typhimurium in host cells. Moreover, both SPI1 and SPI2 are reportedly induced inside the mammalian epithelial cells 2 to 4 h postinfection (16). We were interested in understanding if obacunone has an effect on expression of SPI1 and SPI2 under conditions mimicking the infection process. Expression of hilA and ssrA was measured as a surrogate to understand the expression pattern of SPI1 and SPI2. We first determined the expression of hilA and ssrA at 2 and 2.5 h postinfection under conditions mimicking adhesion and invasion assays, respectively. The expression levels were calculated over S. Typhimurium grown in DMEM plus 10% FBS under the same conditions as those for S. Typhimurium-infected Caco-2 cells. An induction of ≈3.4- and 2.3-fold under conditions mimicking adhesion at 2 h was observed in ssrA and hilA expression, respectively (Fig. 7A), whereas, under conditions mimicking invasion at 2.5 h, ssrA and hilA were upregulated by ≈7.4- and 3.3-fold, respectively (Fig. 7B). The effect of obacunone on SPI1 and SPI2 was then determined under adhesion- and invasion-mimicking conditions at 2 and 2.5 h. Exposure of obacunone resulted in repression of hilA and ssrA by 2.55- and 3.16-fold at 2 h (Fig. 7C) and by 3.9- and 4.4-fold at 2.5 h (Fig. 7D), respectively.
Fig 7.

(A) Expression of SPI1- and SPI2-carried hilA and ssrA under adhesion-mimicking conditions. To determine the expression, Caco-2 cells were infected with S. Typhimurium at a multiplicity of infection of 100. Total RNA was isolated 2 h postinfection using TRIzol, and qRT-PCR was performed to determine the relative expression of hilA and ssrA compared to that in S. Typhimurium grown in DMEM plus 10% FBS used to culture Caco-2 cells. (B) Expression of hilA and ssrA under invasion-mimicking conditions (2.5 h after infection). The infected Caco-2 cells were treated with 100 μg/ml gentamicin by replacing the medium after 1 h. RNA was extracted from cells after 2.5 h from the beginning of the experiment. The fold change was calculated over S. Typhimurium culture grown in DMEM plus 10% FBS. (C and D) Expression of hilA and ssrA under adhesion-mimicking conditions similar to those in panel A (C) and invasion-mimicking conditions similar to those in panel B (D). Infection was carried out in the presence of 100 μg/ml obacunone or an equivalent amount of DMSO. The fold change was calculated as obacunone/DMSO.
Obacunone represses hilA expression in an envZ-dependent manner.
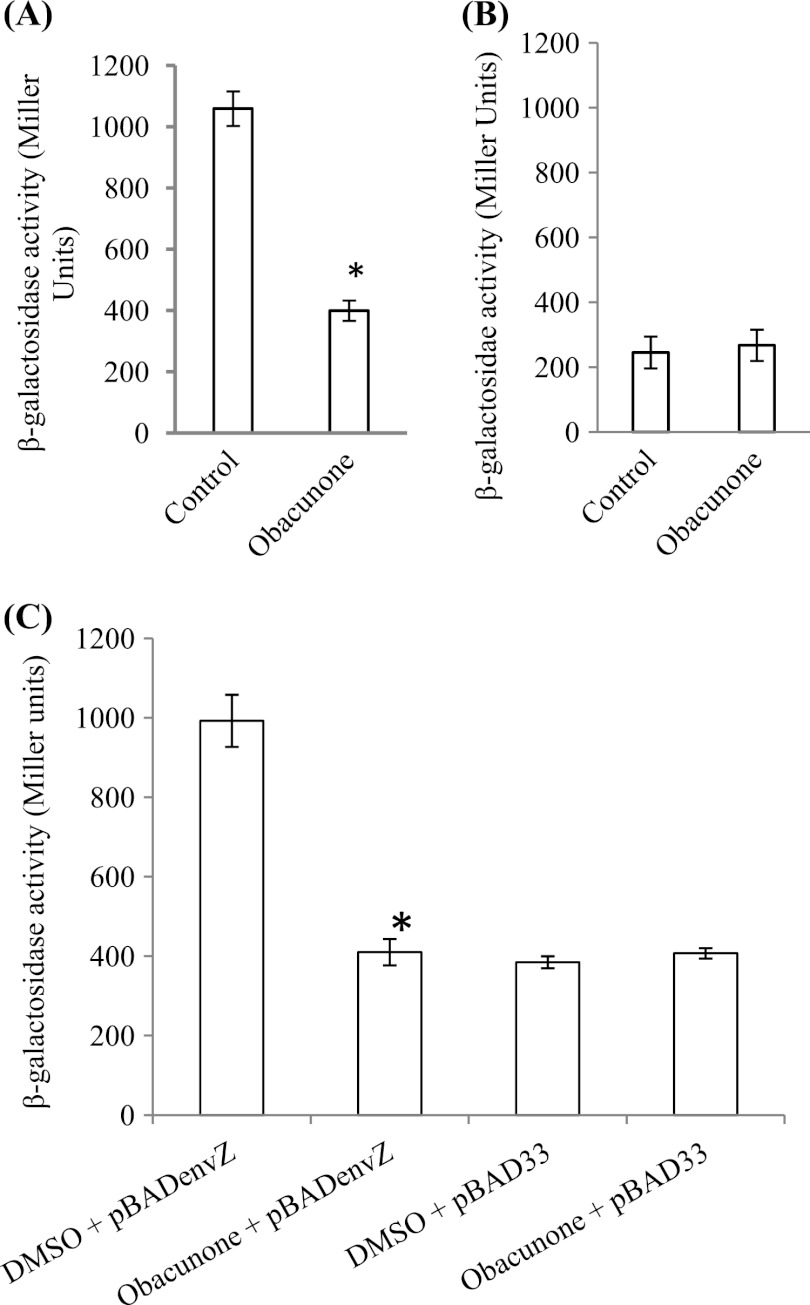
Several environmental and genetic factors regulate expression of SPI1, SPI2, and mal operons; however, EnvZ-OmpR is reported to directly regulate the three operons (7, 22, 27). To determine the role of EnvZ-OmpR, hilA chromosomal fusion reporter strains EE658 and RL829 (2, 26) (kindly provided by C. A. Lee, Department of Biology, Massachusetts Institute of Technology, Cambridge, MA) were employed. First, the expression of hilA::Tn5lacZ in a chromosomal fusion reporter in S. Typhimurium SL1344 strain EE658 in response to obacunone and DMSO was measured. Obacunone repressed the expression of hilA080::Tn5lacZ by ≈3-fold (P < 0.001) (Fig. 8A). To determine the role of EnvZ-OmpR in obacunone-mediated repression of hilA, S. Typhimurium SL1344 strain RL829 (26, 27) was used. Strain RL829 has a disruption in envZ and is defective in envZ-mediated induction of hilA. Obacunone did not affect (fold change, 0.92) the expression of hilA (P > 0.05) in an envZ mutant background (Fig. 8B). To further verify, envZ was cloned in plasmid pBAD33 (pBADenvZ) and electroporated into RL829. The expression of pBADenvZ was induced with 0.02% arabinose, and β-galactosidase activity was measured as readout for hilA expression. Obacunone repressed the expression of hilA080::Tn5lacZ by 2.42-fold (P < 0.01) under experimental conditions (Fig. 8C), whereas no difference was observed in obacunone- and DMSO-treated RL829 containing only vector. Additionally, envZ expression in the presence of 100 μg/ml obacunone calculated over DMSO was 1.5 (±0.4)-fold (data not shown).
Fig 8.
Expression of hilA080::Tn5lacZ upon exposure of 100 μg/ml obacunone or an equal amount of DMSO in EE658 (A), S. Typhimurium strain RL829 (B), and S. Typhimurium strain RL829 supplemented with envZ in pBAD33 and induced with 0.02% arabinose (C). The data were analyzed using one-way ANOVA followed by Tukey's test. Asterisks denote significance at P < 0.01.
DISCUSSION
Citrus fruits are consumed very widely in the United States and worldwide. According to the Economic Research Service, USDA (12), the per capita consumption of total citrus fruits and juices has varied between 84 and 114 lb (fresh weight equivalent) since the year 2000. Furthermore, citrus is a significant contributor and a rich source of dietary phytochemicals such as flavonoids and limonoids (29). Obacunone is an important constituent of citrus fruits and juices and likely to be consumed in significant quantities. Since our previous studies demonstrated that obacunone potentially inhibits enterohemorrhagic E. coli (EHEC) virulence (56), we were further interested in its effect on S. Typhimurium, another important intestinal pathogen. As with EHEC, obacunone did not affect the growth of S. Typhimurium.
To elucidate the effect of obacunone, the gene expression profile of S. Typhimurium in the presence of 100 μg/ml obacunone was studied using cDNA microarrays. A concentration of 100 μg/ml obacunone was used based on our previous studies, to obtain a measurable response in S. Typhimurium (55, 56). Obacunone exposure resulted in repression of three distinct functional systems, SPI1, the hydrogenase operon, and the maltose transport system. HilA is the transcriptional regulator of SPI1 and is regulated by a variety of genetic and environmental factors (1, 2, 27). In particular, HilA regulates the expression of inv, prg/org, and spa operons in SPI1 (25). Obacunone repressed hilA as well as hilA-dependent genes carried in prg/org, inv, and spa operons. In addition, the effector sopE2, which is carried outside SPI1 but regulated by SPI1-encoded InvF/SicA (10), was repressed. Furthermore, independent measurement of selected genes confirmed the general results obtained from microarray studies (Table 3). Previously, we have demonstrated that a citrus flavonoid, naringenin, also has an antivirulence effect on S. Typhimurium LT2 by repressing SPI1 (54). However, the modes of action of naringenin and obacunone seem to be different. For instance, naringenin seems to repress SPI1 in a HilD-dependent manner, represses the flagellar operon, and induces the RND efflux pump, whereas obacunone seems to repress SPI1 in an envZ-dependent manner (discussed below) and does not seem to affect flagella or the RND efflux system. The different modes of action may likely arise from the fact that obacunone and naringenin belong to two different chemical classes and differ very widely in terms of structure.
The SPI1-encoded needle complex is essential for initial attachment and entry of the pathogen into the host cells (46), whereas SPI2-encoded effectors are required for intracellular survival (5). Repression of various operons inside SPI1 indicated two possibilities: (i) impaired assembly of needle complex and export apparatus will result in reduced adhesion and invasion of animal cells and (ii) repression of SPI1 by obacunone is mediated through hilA. The hilA repression by obacunone was dose dependent (Fig. 6), suggesting that the repression of SPI1 by obacunone is mediated through hilA. To determine that obacunone treatment may reduce the attachment and subsequent internalization of S. Typhimurium, Caco-2 cell models of adhesion and invasion were employed. Consistent with the observed hilA and SPI1 repression, obacunone significantly reduced the number of S. Typhimurium cells adhering to and internalized in Caco-2 cells (Fig. 3). Furthermore, obacunone did not affect the viability of Caco-2 cells during the time period used for the assay (41; also unpublished data).
The second system affected by obacunone was the maltose transporter. The maltose transporter facilitates the uptake and efficient catabolism of maltose and maltodextrins (4). In E. coli O157:H7 (EHEC), maltose metabolism was demonstrated to be necessary to initiate infection in a mixed culture model (19). To demonstrate the effect on the maltose transporter, maltose utilization assays were conducted under two different medium conditions. The MOPS medium is well defined in composition, and growth is dependent upon the supply of carbon source. Additionally, MOPS medium provides buffering action, thereby eliminating differences due to pH. The M63 minimal medium that was prepared contained only the salts but no energy source except the added sugar (glucose or maltose at 0.2%); therefore, growth was solely dependent upon carbon source and was very restricted for a short time. Obacunone repressed the maltose transporter, suggesting a reduced uptake of maltose and maltodextrin. Furthermore, a suppressed growth rate of S. Typhimurium indicated impaired maltose utilization, plausibly due to repressed transporter. Altogether, the data suggest that obacunone exposure resulted in a reduced rate of maltose uptake, which may be attributed to downregulation of the maltose transporter.
The third system repressed by obacunone was hydrogenase. Salmonella contains three hydrogenase enzymes (28), which are important for cellular metabolism. Although the E. coli hya operon, a homologue of S. Typhimurium LT2 hyaABCDEF, was reported to be transcribed as a polycistronic message (32), the polycistronic nature of the S. Typhimurium hya operon is not established. Repression of the entire hya operon by obacunone indicates a possibility that S. Typhimurium hya is also coded as a polycistronic message. However, this hypothesis needs further experimental work to be established.
The enzyme Hyc together with formate dehydrogenase oxidizes formate to produce CO2 and H2 (47) during fermentative growth of bacteria. The majority of H2 produced by Hyc is recycled by the enzyme Hya (60). Obacunone exposure affected the genes carried in the hydrogenase 1 (hya) operon and resulted in repression of the entire hya operon and hycH of the hyc operon. Although the overall hydrogen uptake activity was reported to be higher under rich medium and aerobic conditions such as LB medium (28), regulation of hydrogenases appears to be complex and not well understood. Additionally, we did not observe any differences in pH due to obacunone during the experiments, ruling out any regulation due to pH. Therefore, at this point the significance of hydrogenase regulation by obacunone is unclear. Furthermore, hydrogenases seem to play a role in S. Typhimurium virulence (28), which might have an additive effect with reduced virulence due to repression of SPI1 and SPI2 and requires further study.
The reduced number of S. Typhimurium cells recovered from epithelial cells indicated a possibility that obacunone might affect the expression or function of the SPI2-encoded TTSS in addition to observed repression of SPI1. SPI2 was reported to be induced in MES-buffered MgM. Therefore, S. Typhimurium was grown in MES-buffered MgM in the presence of 100 μg/ml obacunone and relative changes in expression of ssrA, ssrB, and spiC were measured using qRT-PCR. The results suggested that obacunone repressed the expression of ssrA, ssrB, and spiC, suggesting regulation of SPI2. We were further interested in determination of the effect of obacunone on the expression of SPI1 and SPI2 under adhesion and invasive model conditions. Both SPI1 and SPI2 are essential for S. Typhimurium pathogenicity. SPI1 is induced during early invasion stages, whereas SPI2 gets induced during late stages of S. Typhimurium infection (40, 53). Obacunone exposure resulted in repression of hilA and ssrA measured under adhesion and invasive conditions, suggesting that obacunone may affect S. Typhimurium virulence under these model conditions. Furthermore, it is possible that obacunone may demonstrate antivirulence potential under in vivo conditions.
Global regulators EnvZ/OmpR, BarA/SirA, and CsrA in S. Typhimurium regulate SPI1 and the mal regulon (7, 22, 27). In addition, ompR was reported to regulate the SsrA-SsrB two-component system encoded in SPI2 (23). CsrA is a posttranscriptional regulator and therefore was ruled out of the current study (22). SirA is reported to activate SPI1 and SPI2 in a growth-phase-dependent fashion, i.e., at late stationary phase (10 h) in LB medium (6, 31), by inducing CsrB and CsrC, which counteract the repressive effect of CsrA. Theoretically, repression of SirA is not likely to enhance the repressive effect of CsrA. Moreover, the relevance of SirA for SPI2 regulation under in vivo conditions is currently unknown. Therefore, SirA was not further studied. Since EnvZ/OmpR is the two-component system which regulates SPI1, SPI2, and the maltose regulon at early to late stationary phase and in epithelial cell culture models, similarly to conditions used in our studies, EnvZ/OmpR was a possible candidate under tested experimental conditions.
If obacunone alters hilA expression by modulating EnvZ/OmpR, the differential expression of hilA would be abrogated in the absence of EnvZ/OmpR. The dependence of hilA regulation on EnvZ was tested by measuring the expression of hilA in an EnvZ mutant background. S. Typhimurium strain RL829 is arabinose insensitive, contains chromosomal fusion hilA080::Tn5lacZY, and carries a mutation in envZ (26). The levels of β-galactosidase activity were similar in DMSO- and obacunone-treated samples. Additionally, restoration of the repressive effect of obacunone upon expression of envZ from the plasmid in RL829 suggested that the repression of hilA by obacunone is mediated via EnvZ. Altogether, the data seem to suggest that obacunone represses at least SPI1 and possibly SPI2 and the mal regulon in an EnvZ-dependent fashion (Fig. 9). In the present study, involvement of BarA/SirA was not investigated; therefore, a possible role of this two-component system in obacunone activity could not be ruled out at present and needs further investigation. S. Typhimurium strain LT2 carries a mutation in the rpoS allele and is less virulent than strains carrying the wild-type allele. Furthermore, rpoS is shown to regulate spvABCD and spvR genes, which are essential for systemic infection but do not affect the ability of S. Typhimurium to infect epithelial cells in tissue culture (58). Additionally, rpoS does not seem to activate hilA (21). Our results indicate that obacunone did not affect the spv gene cluster but repressed hilA; therefore, the antivirulence activity of obacunone may not depend on ropS.
Fig 9.

Speculative model of action of obacunone on S. Typhimurium LT2. Obacunone seems to repress hilA and possibly SPI1 in an EnvZ-dependent fashion (solid line). Additionally, obacunone is speculated to repress ssrAB (SPI2) and the mal operon in an EnvZ-dependent manner (dashed line). The mode of action of obacunone on EnvZ/OmpR is unknown.
In a nutshell, obacunone exposure repressed SPI1 and SPI2, which resulted in reduced numbers of S. Typhimurium cells attached to and internalized in Caco-2 cells. Furthermore, the presented data seem to suggest that obacunone exerts its action in an EnvZ-dependent fashion. However, EnvZ also regulates the expression of porins encoded by ompC and ompF. One possible scenario is that obacunone is transported inside the cell through porins OmpC and OmpF and that the actual target is different than EnvZ/OmpR. Another possibility is that obacunone regulates SPI1 and SPI2 in a SirA/BarA-dependent fashion. These possibilities require further investigation and will be helpful in deducing the mode of action of obacunone. Additionally, structure-function studies are needed to determine the structural features responsible for antivirulence activity. Furthermore, it is also possible that obacunone may affect expression of additional Salmonella pathogenicity islands. Such a possibility requires further investigation. We previously reported that obacunone represses TTSS in E. coli O157:H7 (56); it is possible that obacunone represses E. coli O157:H7 TTSS in a fashion similar to that for S. Typhimurium. In conclusion, the data suggest that obacunone exerts an antivirulence effect on S. Typhimurium and may serve as a lead compound in development of antivirulence strategies to counter S. Typhimurium infection.
ACKNOWLEDGMENTS
This project is based upon the work supported by the USDA CSREES IFAFS 2009-34402-19831 “Designing Foods for Health” through the Vegetable and Fruit Improvement Center.
We also extend our heartfelt thanks to C. A. Lee (Department of Biology, Massachusetts Institute of Technology, Cambridge, MA) for providing the Salmonella Typhimurium mutant strains.
Footnotes
Published ahead of print 20 July 2012
REFERENCES
- 1. Bajaj V, Hwang C, Lee CA. 1995. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol. Microbiol. 18:715–727 [DOI] [PubMed] [Google Scholar]
- 2. Bajaj V, Lucas RL, Hwang C, Lee CA. 1996. Co-ordinate regulation of Salmonella Typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol. Microbiol. 22:703–714 [DOI] [PubMed] [Google Scholar]
- 3. Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B Methodol. 57:289–300 [Google Scholar]
- 4. Boos W, Shuman H. 1998. Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol. Mol. Biol. Rev. 62:204–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brawn LC, Hayward RD, Koronakis V. 2007. Salmonella SPI1 effector SipA persists after entry and cooperates with a SPI2 effector to regulate phagosome maturation and intracellular replication. Cell Host Microbe 1:63–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bustamante VH, et al. 2008. HilD-mediated transcriptional cross-talk between SPI-1 and SPI-2. Proc. Natl. Acad. Sci. U. S. A. 105:14591–14596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Case CC, Bukau B, Granett S, Villarejo MR, Boos W. 1986. Contrasting mechanisms of envZ control of mal and pho regulon genes in Escherichia coli. J. Bacteriol. 166:706–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention 2010. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food—10 states, 2009. MMWR Morb. Mortal. Wkly. Rep. 59:418–430 [PubMed] [Google Scholar]
- 9. Clatworthy AE, Pierson E, Hung DT. 2007. Targeting virulence: a new paradigm for antimicrobial therapy. Nat. Chem. Biol. 3:541–548 [DOI] [PubMed] [Google Scholar]
- 10. Darwin KH, Miller VL. 2001. Type III secretion chaperone-dependent regulation: activation of virulence genes by SicA and InvF in Salmonella Typhimurium. EMBO J. 20:1850–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Deiwick J, Nikolaus T, Erdogan S, Hensel M. 1999. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol. Microbiol. 31:1759–1773 [DOI] [PubMed] [Google Scholar]
- 12. Economic Research Service 2012. Food availability (per capita) data system. Economic Research Service, US Department of Agriculture, Washington, DC. http://www.ers.usda.gov/Data/FoodConsumption/ [Google Scholar]
- 13. Garmendia J, Beuzon CR, Ruiz-Albert J, Holden DW. 2003. The roles of SsrA-SsrB and OmpR-EnvZ in the regulation of genes encoding the Salmonella Typhimurium SPI-2 type III secretion system. Microbiology 149:2385–2396 [DOI] [PubMed] [Google Scholar]
- 14. Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hasegawa S, Bennett RD, Verdon CP. 1980. Limonoids in citrus seeds: origin and relative concentration. J. Agric. Food Chem. 28:922–925 [Google Scholar]
- 16. Hautefort I, et al. 2008. During infection of epithelial cells Salmonella enterica serovar Typhimurium undergoes a time-dependent transcriptional adaptation that results in simultaneous expression of three type 3 secretion systems. Cell. Microbiol. 10:958–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Herman Z, Fong CH, Ou P, Hasegawa S. 1990. Limonoid glucosides in orange juices by HPLC. J. Agric. Food Chem. 38:1860–1861 [Google Scholar]
- 18. Hsu W-J, Berhow M, Robertson GH, Hasegawa S. 1998. Limonoids and flavonoids in juices of Oroblanco and Melogold grapefruit hybrids. J. Food Sci. 63:57–60 [Google Scholar]
- 19. Jones SA, et al. 2008. Glycogen and maltose utilization by Escherichia coli O157:H7 in the mouse intestine. Infect. Immun. 76:2531–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Keyser P, Elofsson M, Rosell S, Wolf-Watz H. 2008. Virulence blockers as alternatives to antibiotics: type III secretion inhibitors against Gram-negative bacteria. J. Intern. Med. 264:17–29 [DOI] [PubMed] [Google Scholar]
- 21. Kosarewicz A, Königsmaier L, Marlovits TC. 2012. The blueprint of the type-3 injectisome. Philos. Trans. R. Soc. B Biol. Sci. 367:1140–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lawhon SD, et al. 2003. Global regulation by CsrA in Salmonella typhimurium. Mol. Microbiol. 48:1633–1645 [DOI] [PubMed] [Google Scholar]
- 23. Lee AK, Detweiler CS, Falkow S. 2000. OmpR regulates the two-component system SsrA-SsrB in Salmonella pathogenicity island 2. J. Bacteriol. 182:771–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 25. Lostroh CP, Lee CA. 2001. The HilA box and sequences outside it determine the magnitude of HilA-dependent activation of PprgH from Salmonella pathogenicity island 1 (SPI1). J. Bacteriol. 183:4876–4885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lucas RL, Lee CA. 2001. Roles of hilC and hilD in regulation of hilA expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 183:2733–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lucas RL, et al. 2000. Multiple factors independently regulate hilA and invasion gene expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:1872–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maier RJ, Olczak A, Maier S, Soni S, Gunn J. 2004. Respiratory hydrogen use by Salmonella enterica serovar Typhimurium is essential for virulence. Infect. Immun. 72:6294–6299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Manach C, Scalbert A, Morand C, Rémésy C, Jiménez L. 2004. Polyphenols: food sources and bioavailability. Am. J. Clin. Nutr. 79:727–747 [DOI] [PubMed] [Google Scholar]
- 30. Manners GD, Jacob RA, Breksa AP, III, Schoch TK, Hasegawa S. 2003. Bioavailability of citrus limonoids in humans. J. Agric. Food Chem. 51:4156–4161 [DOI] [PubMed] [Google Scholar]
- 31. Martínez LC, et al. 2011. Integration of a complex regulatory cascade involving the SirA/BarA and Csr global regulatory systems that controls expression of the Salmonella SPI-1 and SPI-2 virulence regulons through HilD. Mol. Microbiol. 80:1637–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Menon NK, et al. 1990. Cloning and sequencing of a putative Escherichia coli [NiFe] hydrogenase-1 operon containing six open reading frames. J. Bacteriol. 172:1969–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Miao EA, Miller SI. 2000. A conserved amino acid sequence directing intracellular type III secretion by Salmonella Typhimurium. Proc. Natl. Acad. Sci. U. S. A. 97:7539–7544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Miller EG, Porter JL, Binnie WH, Guo IY, Hasegawa S. 2004. Further studies on the anticancer activity of citrus limonoids. J. Agric. Food Chem. 52:4908–4912 [DOI] [PubMed] [Google Scholar]
- 35. Miller J. 1972. Assay of β-galactosidase, p 352–355 In Miller JH. (ed), Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 36. Neidhardt FC, Bloch PL, Smith DF. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. O'Callaghan D, Charbit A. 1990. High efficiency transformation of Salmonella typhimurium and Salmonella typhi by electroporation. Mol. Gen. Genet. 223:156–158 [DOI] [PubMed] [Google Scholar]
- 38. Ochman H, Soncini FC, Solomon F, Groisman EA. 1996. Identification of a pathogenicity island required for Salmonella survival in host cells. Proc. Natl. Acad. Sci. U. S. A. 93:7800–7804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ozaki Y, et al. 1995. Limonoid glucosides in fruit, juice and processing by-products of Satsuma mandarin (Citrus unshiu Marcov.). J. Food Sci. 60:186–189 [Google Scholar]
- 40. Pfeifer CG, Marcus SL, Steele-Mortimer O, Knodler LA, Finlay BB. 1999. Salmonella typhimurium virulence genes are induced upon bacterial invasion into phagocytic and nonphagocytic cells. Infect. Immun. 67:5690–5698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Poulose SM, Harris ED, Patil BS. 2006. Antiproliferative effects of citrus limonoids against human neuroblastoma and colonic adenocarcinoma cells. Nutr. Cancer 56:103–112 [DOI] [PubMed] [Google Scholar]
- 42. Poulose SM, Harris ED, Patil BS. 2005. Citrus limonoids induce apoptosis in human neuroblastoma cells and have radical scavenging activity. J. Nutr. 135:870–877 [DOI] [PubMed] [Google Scholar]
- 43. Quackenbush J. 2002. Microarray data normalization and transformation. Nat. Genet. 32:496–501 [DOI] [PubMed] [Google Scholar]
- 44. Raibaud O, Roa M, Braun-Breton C, Schwartz M. 1979. Structure of the malB region in Escherichia coli K12. I. Genetic map of malK-lambdaB operon. Mol. Gen. Genet. 174:241–248 [DOI] [PubMed] [Google Scholar]
- 45. Rakeman JL, Bonifield HR, Miller SI. 1999. A HilA-independent pathway to Salmonella Typhimurium invasion gene transcription. J. Bacteriol. 181:3096–3104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rhen M, Maskell D, Mastroeni P, Threlfall J. (ed). 2007. Salmonella: molecular biology and pathogenesis. Horizon Press, Norwich, Norfolk, United Kingdom [Google Scholar]
- 47. Rossmann R, Sawers G, Böck A. 1991. Mechanism of regulation of the formate-hydrogenlyase pathway by oxygen, nitrate, and pH: definition of the formate regulon. Mol. Microbiol. 5:2807–2814 [DOI] [PubMed] [Google Scholar]
- 48. Rozen S, Skaletsky HJ. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365–386 [DOI] [PubMed] [Google Scholar]
- 49. Silhavy TJ, et al. 1979. Structure of the malB region in Escherichia coli K12. II. Genetic map of malE, F, G operon. Mol. Gen. Genet. 174:249–259 [DOI] [PubMed] [Google Scholar]
- 50. Sittka A, Pfeiffer V, Tedin K, Vogel J. 2007. The RNA chaperone Hfq is essential for the virulence of Salmonella Typhimurium. Mol. Microbiol. 63:193–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sperandio V. 2007. Novel approaches to bacterial infection therapy by interfering with bacteria-to-bacteria signaling. Expert Rev. Anti Infect. Ther. 5:271–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tanaka T, et al. 2001. Inhibition of azoxymethane-induced colon carcinogenesis in male F344 rats by the citrus limonoids obacunone and limonin. Carcinogenesis 22:193–198 [DOI] [PubMed] [Google Scholar]
- 53. Valdivia RH, Falkow S. 1997. Fluorescence-based isolation of bacterial genes expressed within host cells. Science 277:2007–2011 [DOI] [PubMed] [Google Scholar]
- 54. Vikram A, et al. 2011. Citrus flavonoid represses Salmonella pathogenicity island 1 and motility in S. Typhimurium LT2. Int. J. Food Microbiol. 145:28–36 [DOI] [PubMed] [Google Scholar]
- 55. Vikram A, Jesudhasan PR, Jayaprakasha GK, Pillai SD, Patil BS. 2011. Citrus limonoids interfere with Vibrio harveyi cell-cell signaling and biofilm formation by modulating response regulator luxO. Microbiology 157:99–110 [DOI] [PubMed] [Google Scholar]
- 56. Vikram A, Jesudhasan PR, Jayaprakasha GK, Pillai SD, Patil BS. 2010. Grapefruit bioactive limonoids modulate E. coli O157:H7 TTSS and biofilm. Int. J. Food Microbiol. 140:109–116 [DOI] [PubMed] [Google Scholar]
- 57. Voetsch AC, et al. 2004. FoodNet estimate of the burden of illness caused by nontyphoidal Salmonella infections in the United States. Clin. Infect. Dis. 38:S127–S134 [DOI] [PubMed] [Google Scholar]
- 58. Wilmes-Riesenberg MR, Foster JW, Curtiss R. 1997. An altered rpoS allele contributes to the avirulence of Salmonella typhimurium LT2. Infect. Immun. 65:203–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zbell AL, Benoit SL, Maier RJ. 2007. Differential expression of NiFe uptake-type hydrogenase genes in Salmonella enterica serovar Typhimurium. Microbiology 153:3508–3516 [DOI] [PubMed] [Google Scholar]
- 60. Zbell AL, Maier RJ. 2009. Role of the Hya hydrogenase in recycling of anaerobically produced H2 in Salmonella enterica serovar Typhimurium. Appl. Environ. Microbiol. 75:1456–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhao S, et al. 2009. β-Lactam resistance in Salmonella strains isolated from retail meats in the United States by the National Antimicrobial Resistance Monitoring System between 2002 and 2006. Appl. Environ. Microbiol. 75:7624–7630 [DOI] [PMC free article] [PubMed] [Google Scholar]