Abstract
Two expression vectors utilizing the inducible taurine promoter (tauAp) were developed. Plasmid pLMB51 is a stable low-copy vector enabling expression in the environment and in planta. The higher copy number pLMB509 enables BD restriction-independent cloning, expression, and purification of polyhistidine-tagged proteins.
TEXT
The Alphaproteobacteria are an ecologically diverse class of bacteria that are a major constituent of both aquatic and terrestrial environments. Examples of this group include the extensively studied and agronomically important nitrogen-fixing rhizobia and the metabolically diverse Rhodobacter. For such an important group of bacteria, there is a need to develop tools to facilitate genetic analysis in the laboratory and in the environment.
The segregational stability of inducible vectors is particularly important when considering them for use in bacteria growing in the environment, where continuous selection is difficult to maintain. The copy number of the vector should also be low in order to prevent overexpression. Conversely, a higher copy number vector is more suitable for gene expression in the laboratory. In both instances, an important consideration when developing vectors is the selection of a highly inducible promoter to drive gene expression that has a low-uninduced background signal. The lacZ promoter from Escherichia coli is widely used to regulate gene expression and, along with its repressor lacI, has been used successfully in Alphaproteobacteria (6, 8). However, in a previous study in which the solute binding protein (SBP)-dependent transporters in the alphaproteobacterium Sinorhizobium meliloti strain 1021 (10) were characterized, we identified a tightly regulated and highly inducible promoter tauAp (SMb21526). This promoter is from an ATP binding cassette (ABC) transporter that is induced by the amino acid derivative taurine.
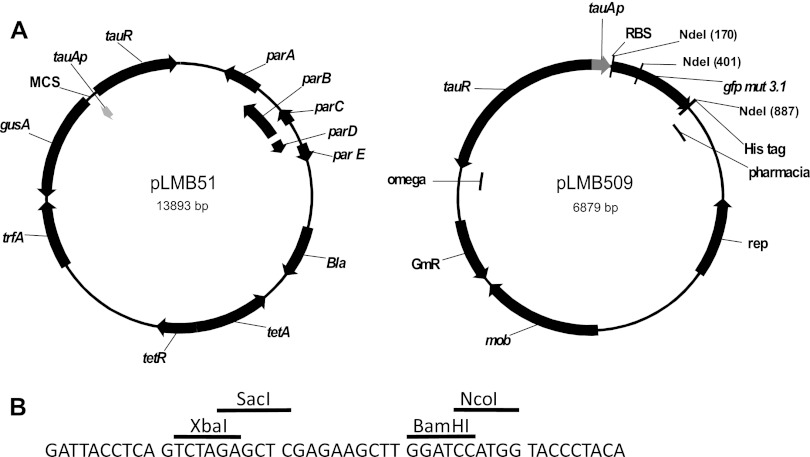
We developed two regulatable vectors for use in Alphaproteobacteria utilizing tauAp (Fig. 1A). The first vector (pLMB51) is based on the highly stable vector pJP2 (incP incompatibility group) (13) and has a copy number of 5 to 7 (4). The second vector (pLMB509) is based on pRU1097 (pBBR incompatibility group) (7) and has a copy number of around 10 (3). These vectors are available through Addgene (http://www.addgene.org/). The construction of the vectors is described in the supplemental material.
Fig 1.
Taurine inducible vectors. (A) Maps of pLMB51 and pLMB509. (B) Multiple-cloning site (MCS) of pLMB51 showing unique restriction sites.
Regulation of the tauA promoter.
Two plasmids, one with tauAp cloned upstream of a gfpmut3.1 reporter gene (pRU1357) (10) and the promoterless parent vector (pRU1097) (7), were conjugated (12) into Rhizobium leguminosarum strain Rlv3841 and S. meliloti 1021. When induced with 1 mM or 10 mM taurine added to tryptone-yeast extract (TY) agar (2) containing 20 μg ml−1 gentamicin (Fig. 2), expression was only observed in the S. meliloti background. This suggests positive regulation of tauAp by the endogenous TauR regulator, which is present in S. meliloti 1021 but not R. leguminosarum Rlv3841. Therefore, in the subsequent construction of vectors, tauR was cloned along with tauAp to enable expression in a wider range of nonnative alphaproteobacterial hosts.
Fig 2.
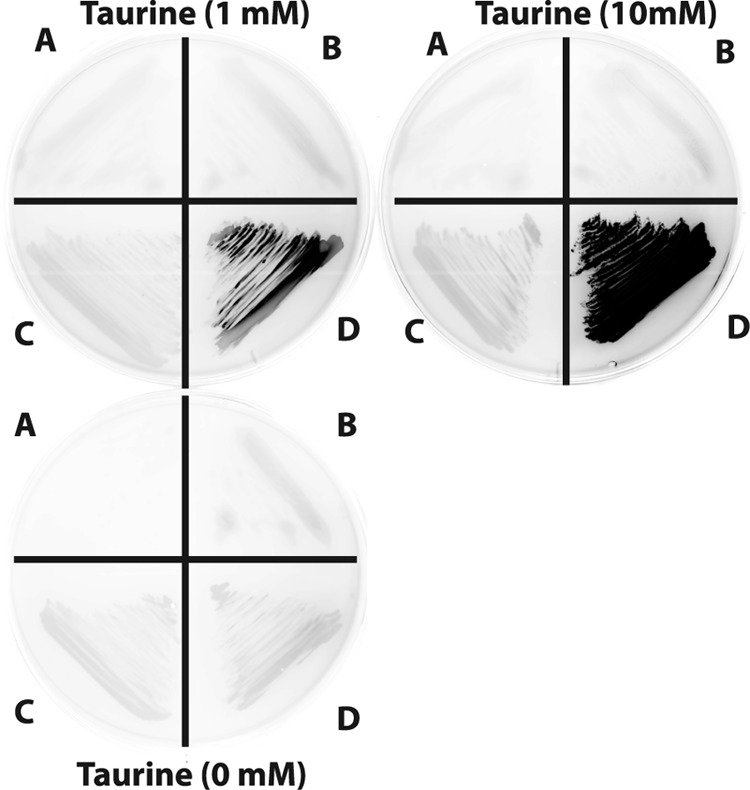
Green fluorescent protein (GFP) fluorescence of various fusions detected on a Fujifilm FLA-7000 image reader; darker shade indicates higher fluorescence. R. leguminosarum Rlv3841 and S. meliloti 1021 strains containing promoterless gfpmut3.1 parent plasmid (pRU1097) or its derivative with tauAp upstream of gfpmut3.1 (pRU1357), grown without (0 mM) or with (1 mM or 10 mM) taurine. Quadrant A, R. leguminosarum Rlv3841 pRU1097; quadrant B, R. leguminosarum Rlv3841 pRU1357; quadrant C, S. meliloti 1021 pRU1097; quadrant D, S. meliloti 1021 pRU1357.
Highly inducible and stable expression vector pLMB51.
Plasmid pJP2 is a stable low-copy gusA reporter probe vector that contains an active partitioning system (parABCDE genes), preventing segregational instability. The vector has been shown by histological staining to be environmentally stable, being retained in bacteroids in legume nodules over several weeks (13). To construct pLMB51, tauAp and tauR were cloned upstream of the gusA reporter in pJP2.
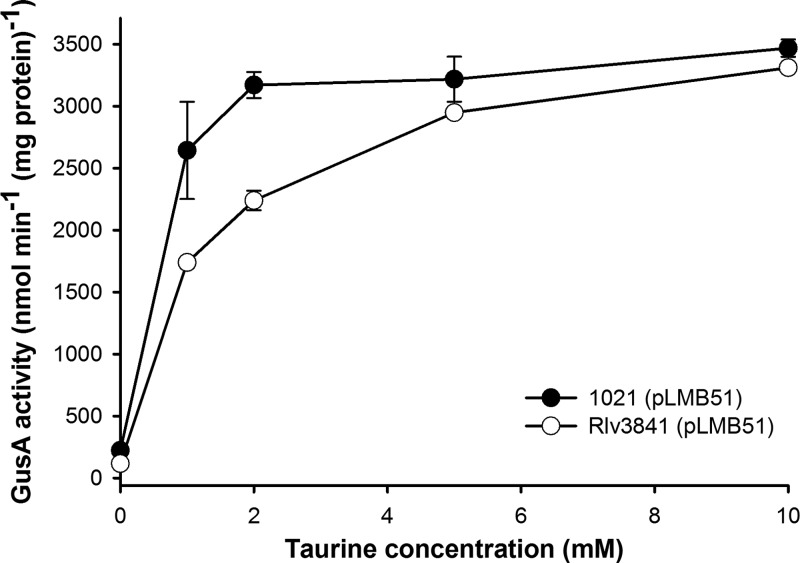
To test the inducibility of the expression vector, pLMB51 was conjugated into S. meliloti 1021 and R. leguminosarum Rlv3841 and grown with differing taurine concentrations in either general minimal salts (GMS) (10) or acid minimal salts (AMS) (11), respectively, supplemented with 2 μg ml−1 tetracycline, 10 mM d-glucose, and 10 mM ammonium chloride. The resulting GusA activity was measured (Fig. 3) as described previously (7). As expected, with a low concentration of taurine (1 mM), expression was greater in S. meliloti 1021 than in R. leguminosarum Rlv3841 [2,643 versus 1,739 nmol min−1 (mg protein)−1, respectively]. However, with a high concentration of taurine (10 mM), there was little difference in activity between S. meliloti 1021 and R. leguminosarum Rlv3841. This is likely to be due to the presence of a dedicated taurine uptake system in S. meliloti 1021, resulting in a higher intracellular concentration of taurine. In the absence of taurine, activity was indistinguishable from promoterless (pJP2) controls, demonstrating the high degree of promoter regulation. The available unique restriction sites for cloning downstream of tauAp are given in Fig. 1B; alternatively, additional sites within the gusA reporter gene can be utilized.
Fig 3.
GusA activity of R. leguminosarum Rlv3841 and S. meliloti 1021 carrying pLMB51 when induced with different concentrations of taurine.
Host range of tauR and tauAp.
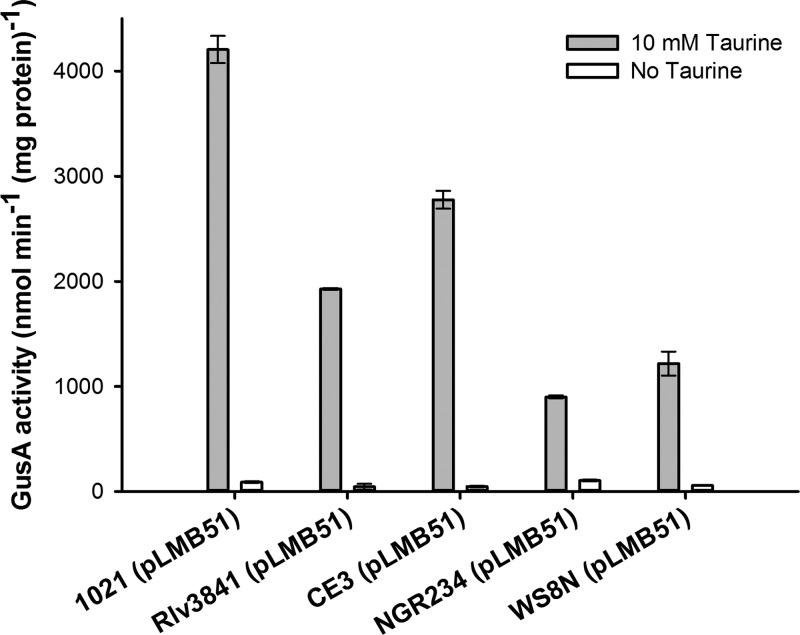
To further investigate the suitability of pLMB51 as an expression vector for use in other bacteria, it was introduced into a number of Proteobacteria. Cells were grown in the presence of 10 mM taurine, and GusA activity was measured. No activity was detected in either E. coli or Pseudomonas fluorescens strain SBW25 (data not shown). However, activity was detected in a range of Alphaproteobacteria (Fig. 4), including Rhizobium etli strain CE3, Rhizobium species strain NGR234 grown in AMS supplemented with 2 μg ml−1 tetracycline, 10 mM d-glucose, and 10 mM ammonium chloride, and Rhodobacter sphaeroides strain WS8N grown in TY containing 2 μg ml−1 tetracycline. This demonstrates the usefulness of pLMB51 as a broad-range alphaproteobacterial expression vector.
Fig 4.
GusA activity of different Alphaproteobacteria carrying pLMB51 when induced with taurine.
In planta expression.
R. leguminosarum Rlv3841 is a nitrogen-fixing symbiont that nodulates pea plants. To investigate if tauAp could be induced in planta, R. leguminosarum Rlv3841 carrying pLMB51 was inoculated on pea seeds (Pisum sativum cv. Avola) in 1-liter pots as described previously (1). Taurine was added to the growth medium as the plants matured on days 14 and 19. After 21 days, the nodules were crushed and bacteroids isolated as described previously (9), and GusA activity was determined (Fig. 5). Although activity was much weaker than in free-living cultures, significant expression was detected, even after 2 days of induction with as little as 1 mM taurine. This demonstrates the utility of pLMB51 as a vector for inducible in planta gene expression.
Fig 5.
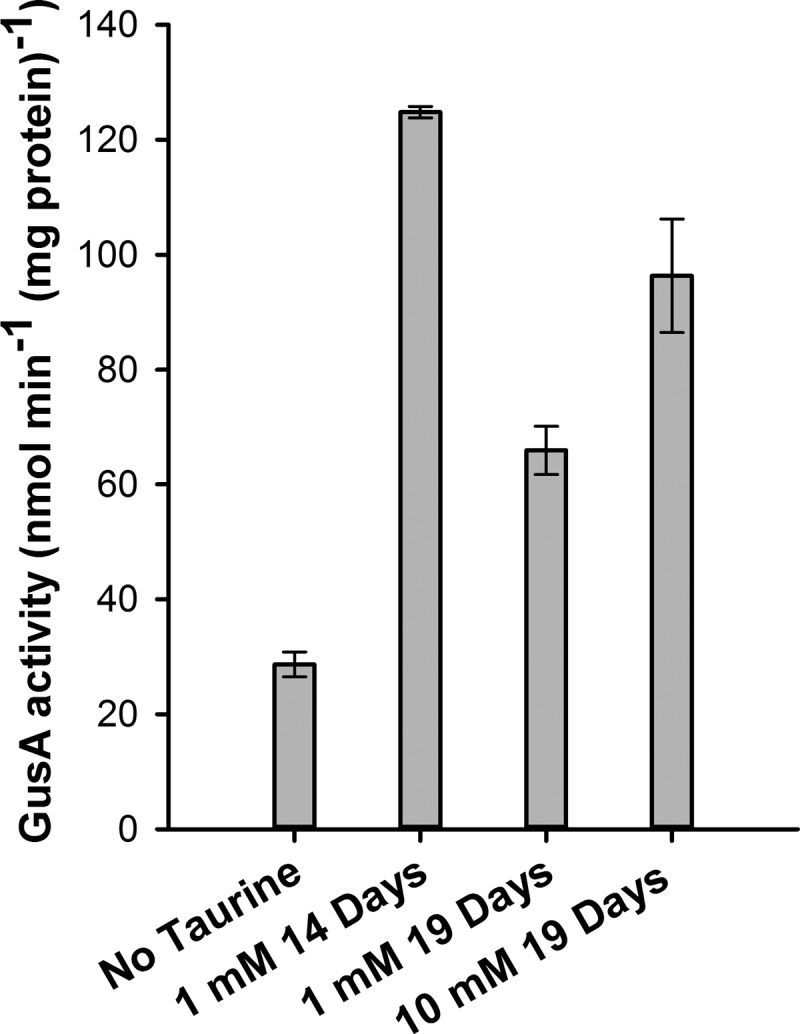
GusA activity of ground nodule extract after pea plants were induced with taurine after 14 and 19 days. Nodules were harvested after a total of 21 days of growth.
Highly inducible His tag expression vector pLMB509.
Polyhistidine tagging of proteins (to the C or N terminus) provides an easy and routine method of protein purification using nickel affinity chromatography. Vector pLMB509 was constructed to utilize tauAp for the controllable overexpression of His-tagged proteins in Alphaproteobacteria. The vector is a derivative of pRU1097 (pBBR incompatibility) and was chosen because it has a higher copy number than pJP2. The vector pLMB509 incorporates a gfpmut3.1 reporter gene downstream of tauAp and a ribosomal binding site (RBS), followed by a six-His tag and a transcriptional stop codon. NdeI restriction sites flank the reporter gene, enabling the easy removal of gfpmut3.1 and subsequent replacement with the desired gene in frame (Fig. 1). When pLMB509 was conjugated into R. leguminosarum Rlv3841 and grown on TY medium, fluorescence was only detected when taurine was added to the medium (data not shown).
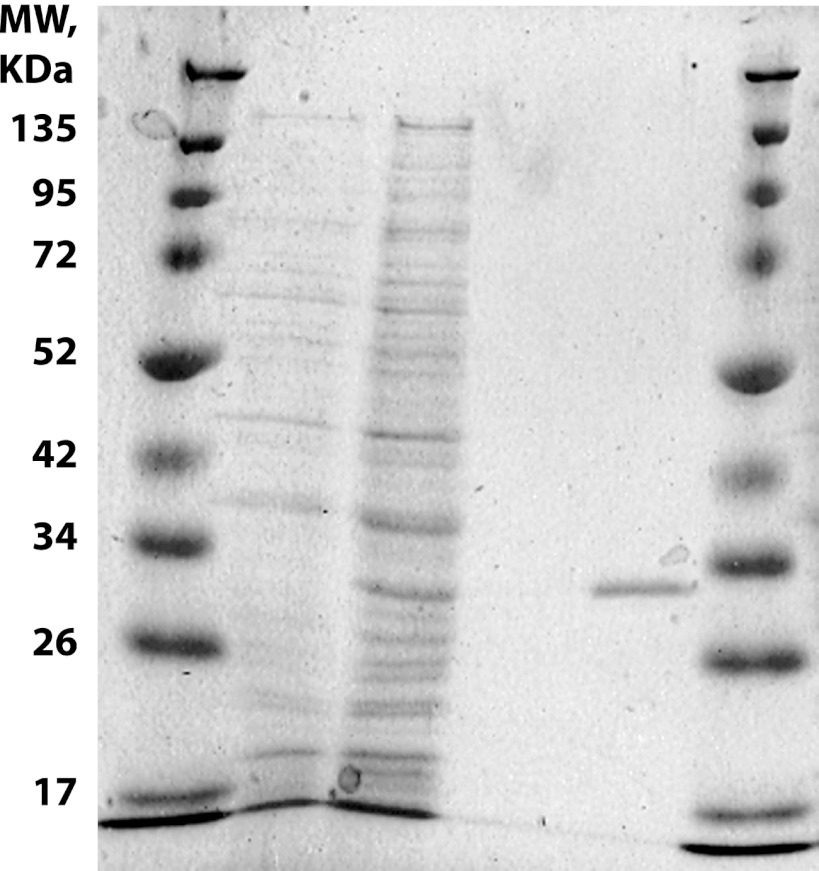
To further test the utility of the vector, gfpmut3.1 was removed by NdeI restriction digest and substituted with a characterized fucose periplasmic solute binding protein from S. meliloti 1021 (SMc02774), including the native leader sequence, to give pLMB548. Although it would be possible to NdeI digest a gene for cloning into pLMB509, we utilized BD In-Fusion cloning (Clontech). BD is a rapid method for cloning PCR amplified products without the need for restriction digest, ligation, or polishing of blunt ends. This facilitates high-throughput gene cloning, including the cloning of genes containing NdeI restriction sites. Therefore, the fucose solute binding protein was PCR amplified using BD primers pr1172 (AGGAGGAAGAACATATGATGAGCATAGCCAAGCGC) and pr1152 (TGGTGATGATGCATATGGTTGACCTTCGGATTCAGAAG). The sequences in bold are complementary to the vector digested with NdeI and should be added to any gene-specific sequence for BD cloning into pLMB548. To extract the fucose SBP, R. leguminosarum Rlv3841 cells containing pLMB548 were grown to mid-log phase in AMS containing 10 mM d-glucose and 10 mM ammonium chloride with or without the addition of 10 mM taurine. Periplasmic extraction was as previously described (5). Histidine-tagged protein (SMc02774) was purified from the periplasmic extraction using a 1-ml HisTrap FF column at 4°C on an AKTA basic 10 system (GE Life Sciences) (Fig. 6). R. leguminosarum was chosen as the host rather than S. meliloti 1021 (from which tauAp and the SBP gene derives) to illustrate the ease of overexpressing and purifying a heterologous protein.
Fig 6.
SDS-PAGE gel analysis of protein extractions. Lanes 1 and 6, protein ladder (Spectra; Fermentas). Lanes 2 and 4, total cell protein extraction and after nickel affinity chromatography purification are shown, respectively, when RLv3841 containing pLMB548 was uninduced and, in lanes 3 and 5, when induced with 10 mM taurine. The gel was stained with InstantBlue (Expedeon).
Vector uses.
Expression vectors pLMB51 and pLMB509 have a low background and are highly inducible in a broad range of Alphaproteobacteria. Plasmid pLMB51 is a low-copy vector that is stably maintained without the need for continuous selection and, as such, is a useful tool for gene expression in bacteria either cultured in the laboratory or introduced into the environment. Plasmid pLMB509 has a higher copy number than pLMB51 and permits high-throughput cloning and expression of His-tagged proteins for purification. This can be very valuable for proteins derived from Alphaproteobacteria whose expression proves problematic in hosts such as E. coli. Taurine is readily available and is less expensive than IPTG (isopropyl-β-d-thiogalactopyranoside), making it an excellent inducer.
Nucleotide sequence accession numbers.
The pLMB51 and pLMB509 sequences have been deposited in GenBank, National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/), under the accession numbers JQ895026 and JQ895027, respectively.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Biotechnology and Biological Sciences Research Council United Kingdom grant BB/F004753/1.
We thank Judith Armitage for providing R. sphaeroides WS8N.
Footnotes
Published ahead of print 20 July 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Allaway D, et al. 2000. Identification of alanine dehydrogenase and its role in mixed secretion of ammonium and alanine by pea bacteroids. Mol. Microbiol. 36: 508–515 [DOI] [PubMed] [Google Scholar]
- 2. Beringer JE. 1974. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84: 188–198 [DOI] [PubMed] [Google Scholar]
- 3. Elzer PH, Phillips RW, Kovach ME, Peterson KM, Roop RM. 1994. Characterization and genetic complementation of a Brucella abortus high-temperature-requirement A (htrA) deletion mutant. Infect. Immun. 62: 4135–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Figurski DH, Meyer RJ, Helinski DR. 1979. Suppression of ColE1 replication properties by the IncP-1 plasmid RK2 in hybrid plasmids constructed in vitro. J. Mol. Biol. 133: 295–318 [DOI] [PubMed] [Google Scholar]
- 5. Glenn AR, Dilworth MJ. 1979. Examination of Rhizobium leguminosarum for the production of extracellular and periplasmic proteins. J. Gen. Microbiol. 112: 405–409 [Google Scholar]
- 6. Ind AC, et al. 2009. Inducible-expression plasmid for Rhodobacter sphaeroides and Paracoccus denitrificans. Appl. Environ. Microbiol. 75: 6613–6615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Karunakaran R, Mauchline TH, Hosie AH, Poole PS. 2005. A family of promoter probe vectors incorporating autofluorescent and chromogenic reporter proteins for studying gene expression in Gram-negative bacteria. Microbiology 151: 3249–3256 [DOI] [PubMed] [Google Scholar]
- 8. Khan SR, Gaines J, Roop RM, Farrand SK. 2008. Broad-host-range expression vectors with tightly regulated promoters and their use to examine the influence of TraR and TraM expression on Ti plasmid quorum sensing. Appl. Environ. Microbiol. 74: 5053–5062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lodwig E, et al. 2004. Regulation of l-alanine dehydrogenase in Rhizobium leguminosarum bv. viciae and its role in pea nodules. J. Bacteriol. 186: 842–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mauchline TH, et al. 2006. Mapping the Sinorhizobium meliloti 1021 solute-binding protein-dependent transportome. Proc. Natl. Acad. Sci. U. S. A. 103: 17933–17938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Poole PS, Blyth A, Reid CJ, Walters K. 1994. myo-Inositol catabolism and catabolite regulation in Rhizobium leguminosarum bv. viciae. Microbiology 140: 2787–2795 [Google Scholar]
- 12. Poole PS, Schofield NA, Reid CJ, Drew EM, Walshaw DL. 1994. Identification of chromosomal genes located downstream of dctD that affect the requirement for calcium and the lipopolysaccharide layer of Rhizobium leguminosarum. Microbiology 140: 2797–2809 [DOI] [PubMed] [Google Scholar]
- 13. Prell J, Boesten B, Poole P, Priefer UB. 2002. The Rhizobium leguminosarum bv. viciae VF39 gamma-aminobutyrate (GABA) aminotransferase gene (gabT) is induced by GABA and highly expressed in bacteroids. Microbiology 148: 615–623 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.