Abstract
Three vip3 genes were identified in two Bacillus thuringiensis Spanish collections. Sequence analysis revealed a novel Vip3 protein class (Vip3C). Preliminary bioassays of larvae from 10 different lepidopteran species indicated that Vip3Ca3 caused more than 70% mortality in four species after 10 days at 4 μg/cm2.
TEXT
Vegetative insecticidal proteins (Vip) are secretable proteins from Bacillus thuringiensis (4, 19) which do not share sequence homology with known Cry proteins and display insecticidal activity against a wide variety of lepidopterans (5, 21) and coleopterans (19) and some sap-sucking insect pests (17). The members of the Vip3 family characterized to date exhibit activity against lepidopterans, and several of them do not compete with Cry proteins for binding sites (12, 18). They are classified into two subfamilies (Vip3A and Vip3B), and some are especially toxic for species with little susceptibility to several Cry proteins (4, 14). All of these features have made Vips a research target for broadening the host-range of B. thuringiensis-based biopesticides and for the management of insect resistance to B. thuringiensis proteins (2).
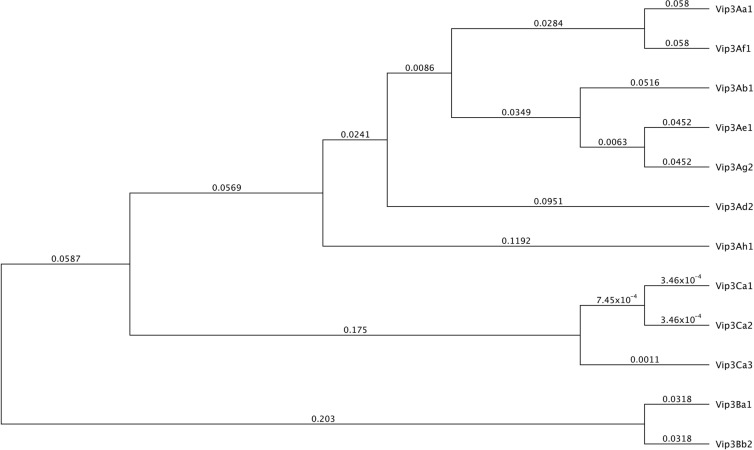
This work aimed to identify novel vip3-like genes from two Spanish B. thuringiensis collections. Around 800 strains were previously screened by PCR with primers from conserved vip3 gene sequences (vip3-sc.fw and vip3-sc.rev) (7). Most of the genes identified showed very high similarity to those previously described (7). Strains NaB8.1 and NaB8.3 were isolated from barley husk in Miranda de Arga (Navarra, Spain) and maize dust in Allo (Navarra, Spain), respectively, two localities located 30 km apart (11). Strain V-MU2.21 was isolated from a storehouse dust sample collected 810 km away from the other two, in Murcia (Spain) (7). Each of the three strains yielded a single amplicon using the vip3-specific primers and showed Sau3AI and AluI restriction endonuclease profiles distinct from those of other known vip3 genes (7). Amplicons were cloned into pGEM-T Easy (Promega) and sequenced. Full-length open reading frames (ORFs) were obtained by a genome walking strategy, completely amplified from the original strains, and cloned into expression vectors pET-28b(+) (for strains NaB8.1 and NaB8.3) and pMaab10 (for strain V-MU2.21). Each full-length ORF was 2,412 bp long and encoded 803-residue proteins with 90.8-kDa predicted molecular masses and 99.8% identity among them. Sequence alignment with known Vip3 proteins revealed identities within the 45 to 78% range. They therefore constituted a new Vip3 subfamily designated Vip3C by the B. thuringiensis Nomenclature Committee (http://www.lifesci.sussex.ac.uk/Home/Neil_Crickmore/Bt/), and the proteins (from strains NaB8.1, V-MU2.21, and NaB8.3, respectively) were named Vip3Ca1 (ADZ46178), Vip3Ca2 (AEE98106), and Vip3Ca3 (HQ876489). In the new Vip3 protein dendrogram, the novel Vip3Ca proteins were grouped in a separate branch (Fig. 1).
Fig 1.
Dendrogram of Vip3 proteins from different subfamilies, including novel Vip3Ca1, Vip3Ca2, and Vip3Ca3 proteins. Branch lengths represent the number of substitutions per site of the multiple-sequence alignment as a measure of divergence.
Differences between the Vip3Ca and Vip3Aa1 amino acid sequences were found over the entire length but were more frequent toward the C terminus, as was previously reported for differences between Vip3Ba1 and Vip3Aa1 (15). The signal peptide (SP), potentially involved in Vip3 secretion (4, 15), remained highly conserved and differed in only two amino acids. It is not clear whether these few changes modify the insecticidal properties of Vip3, but the N terminus seems to be required for the correct formation of the trypsin-resistant core fragment in some Vip3 proteins (13). The two proteolytic processing sites (PPSs), PPS1 and PPS2, were less conserved. Protease treatment of Vip3Aa1 has been reported to give rise to four major protein products with molecular masses of approximately 22, 33, 45, and 66 kDa (5). The 22-kDa fragment constitutes the N-terminal portion of Vip3Aa1, whereas the 66-kDa fragment comprises the rest of the protein. This fragment may vary in size from 62 to 66 kDa among different Vip3 proteins and is occasionally known as the “trypsin-resistant core” (13). Proteolytic processing of the 66-kDa fragment renders both the 33- and 45-kDa fragments (4, 5, 15). How the few changes in the Vip3Ca PPSs may modify either the protein activity or the host range is uncertain, but different Vip3 proteins have shown very distinct insecticidal properties (4, 15). Other remarkable differences were the presence of two insertions immediately downstream of PPS1 and PPS2 that, together with another one at the C terminus, may also increase the size of the trypsin-resistant core fragment described for Vip3Aa1 (4, 5, 15) from 66 kDa to approximately 69 kDa. Finally, Vip3C proteins maintained only one of the three residues of the C terminus stabilizing domain described for Vip3Aa1 (5). This higher divergence toward the C terminus might indicate lower functional constraints and, consequently, more permissibility to nonsynonymous substitutions (20).
The only differences among the three novel Vip3Ca proteins consisted of two point mutations rendering nonsynonymous substitutions. The first one, at position 3, modified the predicted secondary structure of the putative SP from a turn (Vip3Ca1 and Vip3Ca3) to an alpha-helix (Vip3Ca2). The second one, at position 215, caused a slight enlargement of the predicted secondary structure (alpha-helix) of the trypsin-resistant core in Vip3Ca3. The three Vip3Ca proteins had identical PPSs. These few differences are not attributable to laboratory contamination or PCR failure since they were cloned in two different laboratories and obtained in independent replications.
For a preliminary screening of the insecticidal properties of Vip3Ca (host range and toxicity), Vip3Ca3 was picked because of its high yields, expressed in Escherichia coli, column purified, quantified by densitometry, and used to challenge larvae of 10 different lepidopteran species (Table 1). Bioassays were conducted with neonate larvae that were placed over a surface-contaminated artificial diet. Two protein concentrations (0.4 and 4 μg/cm2) were tested for all species except Lobesia botrana, which was challenged with 10 and 100 μg/ml of diet (16). Water was used as a negative control. Sixteen larvae were used for each protein concentration, and each bioassay was repeated twice. Bioassays were conducted at 25°C, 60% ± 5% relative humidity, and a 16:8-h (light/dark) photoperiod. Absolute and functional mortality rates (dead larvae plus larvae remaining in the first instar) were scored after 10 days. Chrysodeixis chalcites, Helicoverpa armigera, Mamestra brassicae, Trichoplusia ni, and Spodoptera littoralis seemed to be moderately to highly susceptible to the toxin, particularly C. chalcites. Compared with previously characterized Vip3 or Cry proteins, several differences in their insecticidal activity could be detected (4, 15; http://www.glfc.forestry.ca/bacillus/BtSearch.cfm). For example, in H. armigera, higher mortality rates have been obtained with other Vip3 or Cry toxins (3, 8), whereas M. brassicae mortality rates obtained with Vip3Ca3 were in a range similar to that of the mean lethal concentration (LC50) of several Cry1 proteins (9). Nevertheless, determination of the LC50 of the three Vip3Ca proteins will allow more reliable comparisons among the Vip3C proteins and between Vip3C proteins and other B. thuringiensis toxins.
Table 1.
Effect of toxin Vip3Ca3 on larvae of 10 lepidopteran species
| Insect species | Mean mortality rateb (%) ± SD |
|||
|---|---|---|---|---|
| Absolute |
Functional |
|||
| 0.4 μg/cm2 | 4 μg/cm2 | 0.4 μg/cm2 | 4 μg/cm2 | |
| A. ipsilon | 16 ± 1.5 | 22 ± 3.5 | 16 ± 1.5 | 22 ± 3.5 |
| C. chalcites | 73 ± 2.9 | 79 ± 5.9 | 73 ± 2.9 | 79 ± 5.9 |
| H. armigera | 15 ± 2.9 | 65 ± 2.9 | 38 ± 5.9 | 83 ± 5.9 |
| L. botranaa | 5 ± 0.00 | 12 ± 4.5 | 5 ± 0.00 | 16 ± 4.9 |
| M. brassicae | 42 ± 2.9 | 80 ± 2.9 | 44 ± 5.9 | 85 ± 5.9 |
| O. nubilalis | 3 ± 0.0 | 6 ± 0.0 | 3 ± 0.0 | 6 ± 0.0 |
| S. exigua | 8 ± 2.5 | 19 ± 11.2 | 21 ± 0.7 | 86 ± 13.4 |
| S. frugiperda | 10 ± 9.6 | 27 ± 7.1 | 20 ± 9.5 | 54 ± 2.7 |
| S. littoralis | 19 ± 2.9 | 73 ± 2.9 | 21 ± 5.9 | 88 ± 5.9 |
| T. ni | 35 ± 3.4 | 94 ± 1.0 | 35 ± 3.4 | 94 ± 1.0 |
For L. botrana bioassays, the Vip3Ca toxin was incorporated into the diet at 10 and 100 μg protein/ml of diet.
Absolute mortality rates were corrected for the background mortality rate (control).
Strong growth inhibition was observed in S. exigua and S. frugiperda, as reflected by the effective mortality rates. This outcome has already been reported for other B. thuringiensis toxins (1, 6, 10) but seems to be a particular response in certain host-toxin combinations and does not always occur. Actually, Vip3Ca3 had little or no effect on the growth (of surviving larvae) of 7 of the 10 species analyzed here.
Finally, two of the species tested showed very low susceptibility to Vip3Ca3. One of them was Ostrinia nubilalis, for which no toxic effects of any of the Vip3Aa proteins assayed so far have been reported (4, 12, 21). The other one was L. botrana, which has been challenged with Vip3 for the first time.
ACKNOWLEDGMENTS
We acknowledge the technical support of N. Gorría and R. González-Martínez for their help in insect rearing.
This research was supported by the Spanish Ministry of Science and Innovation (grant AGL2009-13340-C02), by grant ACOMP/2009/313 from the Generalitat Valenciana, by European FEDER funds, and by Bayer BioScience N.V.
Footnotes
Published ahead of print 3 August 2012
REFERENCES
- 1. Avilla C, Vargas-Osuna E, González-Cabrera J, Ferré J, González-Zamora JE. 2005. Toxicity of several delta-endotoxins of Bacillus thuringiensis against Helicoverpa armigera (Lepidoptera: Noctuidae) from Spain. J. Invertebr. Pathol. 90:51–54 [DOI] [PubMed] [Google Scholar]
- 2. Bravo A, Likitvivatanavong S, Gill SS, Soberón M. 2011. Bacillus thuringiensis: a story of a successful bioinsecticide. Insect Biochem. Mol. Biol. 41:423–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Doss VA, Kumar KA, Jayakumar R, Sekar V. 2002. Cloning and expression of the vegetative insecticidal protein (vip3V) gene of Bacillus thuringiensis in Escherichia coli. Protein Expr. Purif. 26:82–88 [DOI] [PubMed] [Google Scholar]
- 4. Estruch JJ, et al. 1996. Vip3A, a novel Bacillus thuringiensis vegetative insecticidal protein with a wide spectrum of activities against lepidopteran insects. Proc. Natl. Acad. Sci. U. S. A. 93:5389–5394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Estruch JJ, et al. December 1998. Plant pest control. Patent WO 9844137 [Google Scholar]
- 6. Hernández-Martínez P, Ferré J, Escriche B. 2008. Susceptibility of Spodoptera exigua to 9 toxins from Bacillus thuringiensis. J. Invertebr. Pathol. 97:245–250 [DOI] [PubMed] [Google Scholar]
- 7. Hernández-Rodríguez CS, Boets A, Van Rie J, Ferré J. 2009. Screening and identification of vip genes in Bacillus thuringiensis strains. J. Appl. Microbiol. 107:219–225 [DOI] [PubMed] [Google Scholar]
- 8. Hernández-Rodríguez CS, Van Vliet A, Bautsoens N, Van Rie J, Ferré J. 2008. Specific binding of Bacillus thuringiensis Cry2A insecticidal proteins to a common site in the midgut of Helicoverpa species. Appl. Environ. Microbiol. 74:7654–7659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Höfte H, Whiteley HR. 1989. Insecticidal crystal proteins of Bacillus thuringiensis. Microbiol. Rev. 53:242–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ibargutxi MA, Muñoz D, Ruiz de Escudero I, Caballero P. 2008. Interactions between Cry1Ac, Cry2Ab, and Cry1Fa Bacillus thuringiensis toxins in the cotton pests Helicoverpa armigera (Hübner) and Earias insulana (Boisduval). Biol. Control 47:89–96 [Google Scholar]
- 11. Iriarte J, et al. 1998. Environmental distribution and diversity of Bacillus thuringiensis in Spain. Syst. Appl. Microbiol. 21:97–106 [DOI] [PubMed] [Google Scholar]
- 12. Lee MK, Walters FS, Hart H, Palekar N, Chen JS. 2003. The mode of action of the Bacillus thuringiensis vegetative insecticidal protein Vip3A differs from that of Cry1Ab delta-endotoxin. Appl. Environ. Microbiol. 69:4648–4657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li C, et al. 2007. Bacillus thuringiensis Vip3 mutant proteins: insecticidal activity and trypsin sensitivity. Biocontrol Sci. Technol. 17:699–708 [Google Scholar]
- 14. MacIntosh SC, et al. 1990. Specificity and efficacy of purified Bacillus thuringiensis proteins against agronomically important insects. J. Invertebr. Pathol. 56:258–266 [DOI] [PubMed] [Google Scholar]
- 15. Rang C, Gil P, Neisner N, Van Rie J, Frutos R. 2005. Novel Vip3-related protein from Bacillus thuringiensis. Appl. Environ. Microbiol. 71:6276–6281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ruiz de Escudero I, et al. 2006. Molecular and insecticidal characterization of a Cry1I protein toxic to insects of the families Noctuidae, Tortricidae, Plutellidae, and Chrysomelidae. Appl. Environ. Microbiol. 72:4796–4804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sattar S, Maiti MK. 2011. Molecular characterization of a novel vegetative insecticidal protein from Bacillus thuringiensis effective against sap-sucking insect pest. J. Microbiol. Biotechnol. 21:937–946 [DOI] [PubMed] [Google Scholar]
- 18. Sena JA, Hernández-Rodríguez CS, Ferré J. 2009. Interaction of Bacillus thuringiensis Cry1 and Vip3A proteins with Spodoptera frugiperda midgut binding sites. Appl. Environ. Microbiol. 75:2236–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Warren GW. 1997. Vegetative insecticidal proteins: novel proteins for control of corn pests, p 109–121. In Carozzi N, Koziel M. (ed), Advances in insect control: role of transgenic plants. Taylor and Francis, London, United Kingdom [Google Scholar]
- 20. Wu J, et al. 2007. Evidence for positive Darwinian selection of Vip gene in Bacillus thuringiensis. J. Genet. Genomics 34:649–660 [DOI] [PubMed] [Google Scholar]
- 21. Yu CG, Mullins MA, Warren GW, Koziel MG, Estruch JJ. 1997. The Bacillus thuringiensis vegetative insecticidal protein Vip3A lyses midgut epithelium cells of susceptible insects. Appl. Environ. Microbiol. 63:532–536 [DOI] [PMC free article] [PubMed] [Google Scholar]