Abstract
The twin-arginine transport (Tat) system is essential for cell viability in Sinorhizobium meliloti and may play a role during the development of root nodules. Utilizing an in vivo recombination strategy, we have constructed 28 strains that contain deletions in predicted Tat substrates. Testing of these mutations for symbiotic proficiency on the plant hosts alfalfa and sweet clover shows that some of these mutations affect associations with these hosts differentially.
TEXT
Sinorhizobium meliloti is an alphaproteobacterium capable of forming nitrogen-fixing nodules on the roots of leguminous plants, including alfalfa, sweet clover, and fenugreek. The interaction of S. meliloti with its host has been used as a model system to study plant-microbe interactions. Completion of the S. meliloti genome sequence has provided an invaluable tool to help study plant-microbe interactions and to gain insight into how it functions as an organism (2, 6, 15, 16). As a result, a large number of previously uncharacterized genes have been examined using genome-based analysis (3, 10, 23–25, 27, 44).
The ability to transport and target proteins to the inner membrane is an important process involved in many cellular events. The twin-arginine transport (Tat) system is a highly conserved translocation pathway that transports proteins across the cytoplasmic membrane. These protein substrates are typically inserted into the inner membrane or shuttled into the periplasmic space; many of these substrates are redox proteins containing Fe-S or molybdopterin cofactors. Unlike the general secretory (Sec) system, Tat substrates are prefolded and often require cofactor insertion for proper enzymatic function. An important characteristic of Tat substrates is a leader signal motif at the N terminus (S/TRRxFLK), which directs transport through the Tat pathway. The number of substrates transported through the Tat pathway varies, depending upon the organism. This can range from very few proteins to the majority of proteins, such as in the haloarchaeal bacteria (38).
The Tat system has been implicated in pathogenesis of bacteria, including Agrobacterium tumefaciens, Pseudomonas aeruginosa, and many others (9, 13, 31). It has been previously reported that in Rhizobium leguminosarum, the twin-arginine transport pathway was required for correct association with its host plant (29). A mutation in the tatC operon resulted in Fix− plants producing empty nodules. The authors proposed that a developmental breakdown occurred during the infection process due to the loss of the twin-arginine transport system and, therefore, protein substrates exported via the twin-arginine transport mechanism may be required for development.
Bioinformatic data predict that the S. meliloti genome contains at least 100 putative Tat substrates (1, 29). Since the Tat transport system is necessary for cell viability in S. meliloti (35), little is known about the precise role some of these proteins may play during the symbiotic process. To address this, we initiated a systematic approach to delete putative genes which are predicted to encode Tat substrates, utilizing the ORFeome platform that was developed for the S. meliloti genome (22, 40).
Targeting of putative Tat substrates in S. meliloti.
When this work was initiated, TATFIND analyses (38) had been carried out on the S. meliloti genome, identifying 94 putative proteins with Tat-like motifs (29). With the release of TatP (4), the number of predicted proteins encoded in the genome of S. meliloti with Tat-like motifs was 128. Fifty-four of these proteins scored above the suggested trusted cutoff. A comparison of the outputs showed that in addition to proteins that were common, each program had identified proteins that were not recognized by the other program; 26 of the proteins identified by TATFIND were not found by TatP. In addition, by manual scanning of the annotated genome, other candidates that were not identified by either program were also identified. Taken altogether, a list of 154 proteins with Tat-like motifs was generated. Other than fbcF (SMc00187), which encodes Rieske protein involved in electron transport and known to affect nitrogen fixation (43, 45), no other obvious candidates that might affect symbiotic development were found.
To generate a list of genes for deletion, the following criteria were used. First, genes encoding proteins that were identified by TatP and TATFIND below the trusted cutoff value were eliminated. Second, genes at the beginning or at the end of operons were also eliminated because the in vivo deletion strategy employs the recombination of frt sequences into sites adjacent to genes flanking the target; final products of these could potentially delete intergenic promoter regions, yielding pleiotropic mutations. The remaining genes, listed in Table S1 in the supplemental material, were targeted for deletion.
More recently a new program, PRED-TAT (1), predicted 142 putative Tat substrates with cleavage sites. Using the list of GI numbers, the predicted proteins were retrieved from GenBank, and each sequence was reanalyzed. It was found that 13 of the 142 proteins did not contain Tat-like leaders (see Table S1 in the supplemental material). As well, of the 10 proteins manually predicted to be Tat substrates, 5 were corroborated by PRED-TAT, whereas 4 were predicted to have transmembrane helices in the N-terminal region, and the remaining protein contained a leader that was below the trusted cutoff (see Table S1).
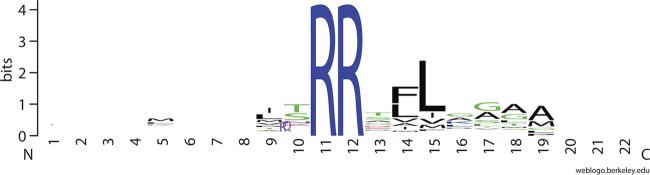
To determine if S. meliloti leaders carried any characteristic signatures, the remaining 129 leaders were aligned using ClustalX (42). The final alignments were manually inspected and optimized by removing 8 sequences. The remaining 121 were used to generate an S. meliloti Tat leader consensus using WebLogo (11) (Fig. 1). The logo is similar to the previously published logos; however, it may be of note that our analysis shows that a number of the putative leaders seem to contain a methionine 6 amino acids prior to the canonical twin-arginine motif. The significance of this observation at this time is not clear.
Fig 1.
Consensus sequence of twin-arginine transport motif. The N-terminal amino acids of predicted S. meliloti Tat substrates were aligned around the twin arginines using ClustalX. Conservation of sequence is shown in bits as previously described (39).
Construction of in vivo deletions using the S. meliloti ORFeome.
To generate targeted in vivo deletions, we employed the ORFeome system with some modifications as previously described (17, 22, 40). Following the final recombination between the frt sites that flanked the targeted gene, each junction was PCR amplified and sequenced to ensure that the proper mutation was constructed. The strains, plasmids, and primers that were used or generated are listed in Tables S2 and S3 in the supplemental material.
Evidence for host-dependent phenotypes.
To address the hypothesis that Tat-dependent substrates may play a role in symbiosis, mutants were screened for symbiotic capabilities by inoculation onto two host plants—alfafa and sweet clover. The inoculated plants were assessed qualitatively by their appearance, as well as the number and positioning of nodules. Plant dry matter accumulation was used to assay symbiotic efficiency, as previously described, and independent trials were normalized on the basis of plant dry matter accumulation from plants inoculated with the wild type (32, 36, 37).
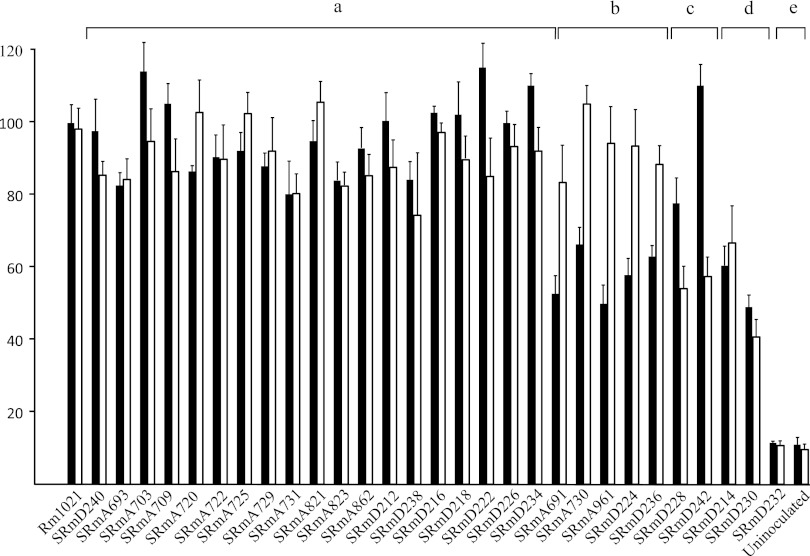
Except for SRmD232 (fbcF), which was obviously not providing fixed nitrogen, most plants did not appear to be drastically different with respect to the appearance or the number, shape, or color of nodules on each root system compared with the wild type. However, based on dry matter accumulation, the mutants fell into 4 distinct classes (Fig. 2). The majority of the mutants did not have different dry matter accumulation on either host (Fig. 2). SRmD232 (fbcF), did not show any dry matter accumulation greater than that of the uninoculated plant controls. Two strains—SRmD230 (SMb21380) and SRmD214 (SMb21067)—showed reduced dry matter accumulation on both hosts (Fig. 2). Five strains—SRmA691, SRmD224, SRmD236, SRmD228, and SRmD242—showed significantly reduced dry matter accumulation on alfalfa but not on sweet clover.
Fig 2.
Plant phenotypes exhibited by putative Tat substrate deletions. Shown are plant dry weights of alfalfa (black bars) or sweet clover (white bars) inoculated with the strain indicated. The data represent between 4 and 6 independent experiments, each consisting of 30 plants, and are presented as percentages of the wild type. Wild-type plant weights (100%) were 57 ± 6 mg/plant (alfalfa, n = 6) and 85 ± 11 mg/plant (sweet clover, n = 6). Differences between the plants inoculated with mutant and the wild type are based on Student's t test at the 95% confidence level (P < 0.0001).
The mutants that displayed phenotypes were generally those whose genes encode proteins that putatively affect either metabolism or metabolite transport. Two of these mutant genes, SMa02021 and SMb20671, encode sugar-binding proteins that are associated with ABC-type transporters. Whereas a deletion of SMb20671 displays an effect on both alfalfa and sweet clover, a deletion of SMa02021 appears to only show an affect on alfalfa (Fig. 2). Both of these genes have been previously screened for expression against a bank of 117 carbon substrates (27). Neither of these was strongly induced by any of the conditions assayed; however, it may be of note that each did show a modest 3-fold induction with lyxose. The significance of this observation is unclear at this time.
Based on the annotation, as well as rudimentary sequence analysis, three of the remaining mutants that displayed a plant-dependent phenotype appear to affect carbon catabolism. Both SRmD224, carrying a deletion in SMc00817, and SRmD214, carrying a deletion of SMb21380, putatively affect the degradation of aromatic compounds. Whereas some plant-derived aromatic compounds have been identified as being important to the physiology of S. meliloti, our knowledge is limited by the characterization of relatively few compounds (19, 26, 30, 34, 41). In addition, the mutation in SRmD242 (SMb20342) also falls into the category of an enzyme that is likely a redox enzyme that can potentially oxidize compounds within the periplasmic space and contribute to the overall energetics of the organism.
Two of the mutants tested, SRmA691, carrying a deletion of napA, and SRmD236, carrying a deletion of nosZ, affect denitrification (Fig. 2). The existence of enzymes involved in denitrification has long been known in S. meliloti (7, 8, 20). The existence of a complete denitrification pathway became clear following the complete sequencing of the genome (16).
The involvement of nitric oxide (NO) in symbiotic and pathogenic associations is well known (5, 14). The production of NO has clearly been shown in nodule tissue and infection threads (12). Microarray analyses have shown that both nos and nap are upregulated in response to nitric oxide in a FixL-independent manner and that NO within nodules can be synthesized by both the bacteria and the plant and used as a source of electrons (and thus an energy source) in Medicago truncatula nodules (21, 28). More recently it has been shown that an hmp mutant in S. meliloti both is affected in competition for nodule occupancy (12) and has lower levels of nitrogenase activity (28). The finding that nos and nap mutations have differential symbiotic phenotypes adds to this body of evidence.
This work was initiated to identify putative Tat-dependent substrates that have a role in symbiosis. Based on the previous findings in R. leguminosarum (29), we had anticipated finding mutants with severe developmental phenotypes. Based on simple BLAST searches, only the genes associated with denitrification had E values of zero in R. leguminosarum. As the work proceeded, it became clear that the strategy we had employed was not yielding the expected results; however, additional screening on a second host, sweet clover, revealed a number of host-dependent effects (Fig. 2). Taken together, the results presented here identify a number of genes that have not been previously implicated in having a role in symbiosis, as well as genes that appear to have roles that are host dependent.
At present, our data do not allow us to distinguish whether we have caused a subtle developmental delay or affected nitrogen fixation or whether the phenotypes are due to a combination of events. It is important to remember that although our results are reproducible, these mutants have not been complemented. Many of these mutations are in complex operons, and their phenotypes do not lend themselves to simple isolation of complementing cosmids, as has been previously described (18, 32, 33, 36). In this aspect, it is noteworthy that both mutations affecting denitrification yielded the same plant phenotype (Fig. 2).
To our knowledge, the systematic deletion of genes encoding putative twin-arginine transport substrates to find physiological function is unique. The data show that Tat substrates may play a more complex role in symbiosis than previously envisaged. To properly define the role that these proteins play within the plant-microbe interaction is going to be dependent on careful physiological, biochemical, and microscopic approaches.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by an NSERC Discovery grant to I.J.O. B.S.P. was partially supported by a University of Manitoba Faculty of Science postgraduate award. H.Y. was partially funded by an undergraduate University of Manitoba Faculty of Science summer award.
Footnotes
Published ahead of print 27 July 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Bagos PG, Nikolaou EP, Liakopoulos TD, Konstantionos TD. 2010. Combined prediction of Tat and Sec signal peptides with hidden Markov models. Bioinformatics 26: 2811–2817 [DOI] [PubMed] [Google Scholar]
- 2. Barnett MJ, et al. 2001. Nucleotide sequence and predicted functions of the entire Sinorhizobium meliloti pSymA megaplasmid. Proc. Natl. Acad. Sci. U. S. A. 98: 9883–9888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Becker A, et al. 2009. A portal for rhizobial genomes: RhizoGATE integrates a Sinorhizobium meliloti genome annotation update with postgenome data. J. Biotechnol. 140: 45–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bendtsen JD, Nielsen H, Widdick D, Palmer T, Brunak S. 2005. Prediction of twin-arginine signal peptides. BMC Bioinformatics 6: 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cabrera JJ, et al. 2011. The nitric oxide response in plant-associated endosymbiotic bacteria. Biochem. Soc. Trans. 39: 1880–1885 [DOI] [PubMed] [Google Scholar]
- 6. Capela D, et al. 2001. Analysis of the chromosome sequence of the legume symbiont Sinorhizobium meliloti strain 1021. Proc. Natl. Acad. Sci. U. S. A. 98: 9877–9882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chan Y-K, Barran L, Bromfield ESP. 1989. Denitrification activity of phage types of representative of two populations of indigenous Rhizobium meliloti. Can. J. Microbiol. 35: 737–740 [Google Scholar]
- 8. Chan YK, Wheatcroft R. 1993. Detection of nitrous oxide structural gene in Rhizobium meliloti strains and its location on the nod megaplasmid of JJc10 and SU47. J. Bacteriol. 175: 19–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ciapina LP, Picchi SC, Lacroix J, de Macedo Lemos EG, Odberg-Ferragut C. 2011. A putative twin-arginine translocation system in the phytopathogenic bacterium Xylella fastidosa. Can. J. Microbiol. 57: 149–154 [DOI] [PubMed] [Google Scholar]
- 10. Cowie A, et al. 2006. An integrated approach to functional genomics: construction of a novel reporter gene fusion library for Sinorhizobium meliloti. Appl. Environ. Microbiol. 72: 7156–7167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Crooks GE, Hon G, Chandonia J, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res. 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. del Giudice J, et al. 2011. Nitric oxide is required for an optimal establishment of the Medicago truncatula-Sinorhizobium meliloti symbiosis. New Phytologist 191: 405–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ding A, Christie PJ. 2003. Agrobacterium tumefaciens twin-arginine-dependent translocation is important for virulence, flagellation, and chemotaxis but not type IV secretion. J. Bacteriol. 185: 760–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fang FC. 2004. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat. Rev. Microbiol. 2: 820–832 [DOI] [PubMed] [Google Scholar]
- 15. Finan TM, et al. 2001. The complete sequence of the 1,683-kb pSymB megaplasmid from the N2-fixing endosymbiont S. meliloti. Proc. Natl. Acad. Sci. U. S. A. 98: 9889–9894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Galibert F, et al. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293: 668–672 [DOI] [PubMed] [Google Scholar]
- 17. Geddes BA, Oresnik IJ. 18 May 2012. Genetic characterization of a complex locus necessary for the transport and catabolism of erythritol, adonitol, and l-arabitol in Sinorhizobium meliloti. Microbiology [Epub ahead of print.] doi:10.1099/mic.0.057877-0 [DOI] [PubMed] [Google Scholar]
- 18. Geddes BA, et al. 2010. A locus necessary for the transport and catabolism of erythritol in Sinorhizobium meliloti. Microbiology 156: 2970–2981 [DOI] [PubMed] [Google Scholar]
- 19. Gordon DM, Ryder MH, Heinrich K, Murphy PJ. 1996. An experimental test of the rhizopine concept in Rhizobium meliloti. Appl. Environ. Microbiol. 62: 3991–3996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Holloway P, McCormick W, Watson RJ, Chan Y-K. 1996. Identification and analysis of the dissimilatory nitrous oxide reduction genes, nosRZDFY of Rhizobium meliloti. J. Bacteriol. 178: 1505–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Horchani F, et al. 2011. Both plant and bacterial nitrate reductases contribute to nitric oxide production in Medicago truncatula nitrogen-fixing nodules. Plant Physiol. 155: 1023–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. House BL, Mortimer MW, Kahn ML. 2004. New recombination methods for Sinorhizobium meliloti genetics. Appl. Environ. Microbiol. 70: 2806–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Humann JL, et al. 2008. Construction and expression of sugar kinase transciptional gene fusions by using the Sinorhizobium meliloti ORFeome. Appl. Environ. Microbiol. 74: 6756–6765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jacob AI, et al. 2008. Mutational analysis of the Sinorhizobium meliloti short-chain dehydrogenase/reductase family reveals substantial contribution to symbiosis and catabolic diversity. Mol. Plant Microbe Interact. 21: 979–989 [DOI] [PubMed] [Google Scholar]
- 25. Luo L, et al. 2005. Two new Sinorhizobium meliloti LysR-type transcriptional regulators required for nodulation. J. Bacteriol. 187: 4562–4572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. MacLean AM, MacPherson G, Aneja P, Finan TM. 2006. Characterization of the β-ketoadipate pathway in Sinorhizobium meliloti. Appl. Environ. Microbiol. 72: 5403–5413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mauchline TH, et al. 2006. Mapping the Sinorhizobium meliloti 1021 solute-binding protein-dependent transportome. Proc. Natl. Acad. Sci. U. S. A. 103: 17933–17938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Meilhoc E, Skapski CCA, Bruand C. 2010. The response to nitric oxide of the nitrogen-fixing symbiont Sinorhizobium meliloti. Mol. Plant Microbe Interact. 23: 748–759 [DOI] [PubMed] [Google Scholar]
- 29. Meloni S, et al. 2003. The twin-arginine translocation (Tat) system is essential for Rhizobium-legume symbiosis. Mol. Microbiol. 48: 1195–1207 [DOI] [PubMed] [Google Scholar]
- 30. Murphy PJ, Wexler W, Grzemski W, Rao JP, Gordon D. 1995. Rhizopines—their role in symbiosis and competition. Soil Biol. Biochem. 27: 525–529 [Google Scholar]
- 31. Ochsner UA, Snyder A, Vasil AI, Vasil ML. 2002. Effects of the twin-arginine translocase on the secretion of virulence factors, stress response, and pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 99: 8312–8317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Oresnik IJ, Charles TC, Finan TM. 1994. Second site mutations specifically suppress the Fix− phenotype of Rhizobium meliloti ndvF mutations on alfalfa: identification of a conditional ndvF-dependent mucoid colony phenotype. Genetics 136: 1233–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Oresnik IJ, Pacarynuk LA, O'Brien SAP, Yost CK, Hynes MF. 1998. Plasmid encoded catabolic genes in Rhizobium leguminosarum bv. trifolii: evidence for a plant-inducible rhamnose locus involved in competition for nodulation. Mol. Plant Microbe Interact. 11: 1175–1185 [Google Scholar]
- 34. Parke D, Ornston LN. 1984. Nutritional diversity of Rhizobiaceae revealed by auxonography. J. Gen. Microbiol. 130: 1743–1750 [Google Scholar]
- 35. Pickering BS, Oresnik IJ. 2010. The twin arginine transport system appears to be essential for viability in Sinorhizobium meliloti. J. Bacteriol. 193: 6409–6418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Poysti NJ, Loewen ED, Wang Z, Oresnik IJ. 2007. Sinorhizobium meliloti pSymB carries genes necessary for arabinose transport and catabolism. Microbiology 153: 727–736 [DOI] [PubMed] [Google Scholar]
- 37. Poysti NJ, Oresnik IJ. 2007. Characterization of Sinorhizobium meliloti triose phosphate isomerase genes. J. Bacteriol. 189: 3445–3451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rose RW, Bruser T, Kissinger JC, Pohlschroder M. 2002. Adaptation of protein secretion to extremely high-salt conditions by extensive use of the twin-arginine translocation pathway. Mol. Microbiol. 45: 943–950 [DOI] [PubMed] [Google Scholar]
- 39. Schneider TD, Stephens RM. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18: 6097–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schroeder BK, et al. 2005. Development of a functional genomics platform for Sinorhizobium meliloti: construction of an ORFeome. Appl. Environ. Microbiol. 71: 5858–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tepfer D, et al. 1988. A plasmid of Rhizobium meliloti 41 encodes catabolism of two compounds from root exudate of Calystegium sepium. J. Bacteriol. 170: 1153–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL-X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Thony-Meyer L, Stax D, Hennecke H. 1989. An unusual gene cluster for the cytochrome bc1 complex in Bradyrhizobium japonicum and its requirement for effective root nodule symbiosis. Cell 57: 683–697 [DOI] [PubMed] [Google Scholar]
- 44. Valverde C, et al. 2008. Prediction of Sinorhizobium meliloti sRNA genes and experimental detection in strain 2011. BMC Genomics 9: 416 doi:10.1186/1471-2164-9-416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wu G, et al. 1996. The cytochrome bc1 complex but not CycM is necessary for symbiotic nitrogen fixation by Rhizobium leguminosarum. Microbiology 142: 3381–3388 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.