Abstract
Alga-bacterium interactions are crucial for aggregate formation and carbon cycling in aquatic systems. To understand the initiation of these interactions, we investigated bacterial chemotaxis within a bilateral model system. Marinobacter adhaerens HP15 has been demonstrated to attach to the diatom Thalassiosira weissflogii and induce transparent exopolymeric particle and aggregate formation. M. adhaerens possesses one polar flagellum and is highly motile. Bacterial cells were attracted to diatom cells, as demonstrated by addition of diatom cell homogenate or diatom culture supernatant to soft agar, suggesting that chemotaxis might be important for the interaction of M. adhaerens with diatoms. Three distinct chemotaxis-associated gene clusters were identified in the genome sequence of M. adhaerens, with the clusters showing significant sequence similarities to those of Pseudomonas aeruginosa PAO1. Mutations in the genes cheA, cheB, chpA, and chpB, which encode histidine kinases and methylesterases and which are putatively involved in either flagellum-associated chemotaxis or pilus-mediated twitching motility, were generated and mutants with the mutations were phenotypically analyzed. ΔcheA and ΔcheB mutants were found to be swimming deficient, and all four mutants were impaired in biofilm formation on abiotic surfaces. Comparison of the HP15 wild type and its chemotaxis mutants in cocultures with the diatom revealed that the fraction of bacteria attaching to the diatom decreased significantly for mutants in comparison to that for the wild type. Our results highlight the importance of M. adhaerens chemotaxis in initiation of its interaction with the diatom. In-depth knowledge of these basic processes in interspecies interactions is pivotal to obtain a systematic understanding of organic matter flux and nutrient cycling in marine ecosystems.
INTRODUCTION
In the pelagic ocean, bacterial chemotaxis is considered an important selective advantage to compete for microscale nutrient patches, including marine snow and living organisms (11). Surfaces of phytoplankton cells represent microenvironments supplying bacteria with essential nutrients and sites for attachment, which greatly increase their capabilities to successfully compete with other microorganisms (7). Chemotaxis might initiate the interaction of bacterial cells with the phytoplankton organism, as it has been postulated for the paradigm of the so-called phycosphere (9). Indeed, attraction of marine bacteria to microalgae or their exudates and further bacterial utilization of attracting compounds have been predicted or demonstrated in previous studies (8, 11, 20, 44, 53, 54, 61). Besides the bacterial growth-promoting effect of phytoplankton exudates, bacteria in turn may support algal growth and reproduction by, e.g., remineralization of nutrients (15, 44).
Interactions of marine algae and bacteria may be envisioned to be by one of four major types: mutualism, parasitism, commensalism, or competition (25). These interactions are likely to be transient and highly dependent on micro- and macroecological environmental conditions. Different interaction types may exist simultaneously, and the balance between stimulatory and inhibitory effects ultimately determines the survival of particular organisms (15). The specific mechanisms by which those organisms interact and how factors such as motility or chemotaxis impact the fate of an interaction are largely unknown. Only a few previous studies have approached this topic by investigating specific interaction pathways. Two of those addressing bacterium-dinoflagellate interactions proposed that bacterial siderophore production may stimulate algal growth in a symbiotic manner (4, 5). Further, the algicidal activity of a Pseudoalteromonas sp. strain was suspected to be due to an extracellular serine protease produced by this bacterium (37). Additionally, bioactive polyunsaturated aldehydes produced by several marine phytoplankton organisms were shown to inhibit the growth of some bacterial strains but to stimulate that of others (48). A detailed biochemical mechanism for the interaction of Emiliania huxleyi with a Roseobacter clade organism, Phaeobacter gallaeciensis, was demonstrated by elicitation of roseobacticide production via algal aging-derived lignin breakdown products (51, 52). Those studies not only demonstrated the delicate balance between a symbiotic and parasitic interaction but also prompted the question of how interaction signals are perceived and transduced in either organism.
Despite the individual type of interaction, physical proximity and thus motility are thought to be prerequisites for cellular interactions in aquatic systems (26). Two independent types of bacterial motility were generally described for the model organism Pseudomonas aeruginosa, and these are also found in numerous other bacteria and have been shown to be regulated by distinct proteins encoded by different chemotaxis gene clusters (10, 16–18, 21, 32, 60). Bacterial flagellar movement is controlled via a methylation-dependent histidine-aspartate phosphorelay multienzyme che system (59). In contrast, twitching motility describes the bacterial movement on surfaces performed with the help of type IV pili (29) and mediated by the chp chemosensory system (10, 60), which controls the motor complex of the pili, causing extension and retraction of these cellular appendages. In P. aeruginosa, one of the five described chemotaxis gene clusters, namely, that comprised of the chp genes, is exclusively responsible for pilus chemotaxis (10, 16–18, 50, 60). In the present study, we investigated the chemotaxis gene clusters of Marinobacter adhaerens and the functions of certain individual chemotaxis genes by mutational analysis. This study describes the chemotaxis of a Marinobacter strain for the first time.
To understand the actual mechanisms by which diatom cells and bacteria interact and influence marine aggregate formation, a bilateral model system was established (22, 23, 31, 57). The bacterial strain M. adhaerens HP15 was demonstrated to attach to the diatom Thalassiosira weissflogii and to induce formation of transparent exopolymeric particles (TEPs) and aggregates in cocultures. Since the bacterium is genetically accessible (57), specific mechanisms of the interaction can now be studied on a molecular basis. Herein, we describe the chemotactic behavior of M. adhaerens toward the diatom T. weissflogii using functional knockouts of the genes coding for central histidine kinases and methyltransferases of two chemotaxis signaling cascades. Thereby, we aim to evaluate the importance of bacterial chemotaxis in alga-bacterium interactions and thus to extend our knowledge of its role in carbon flux and nutrient cycling in marine ecosystems.
MATERIALS AND METHODS
Microorganisms, plasmids, and growth conditions.
The bacterial strains, plasmids, and primers used are listed in Table 1. M. adhaerens was routinely grown in marine broth (MB) at 37°C and 250 rpm or on MB agar unless stated otherwise. Escherichia coli cells were grown in Luria-Bertani broth (LB) at 37°C and 250 rpm or on LB agar. The following antibiotics were added to the media when required: ampicillin (Amp), 50 μg ml−1; chloramphenicol (Cm), 25 μg ml−1; kanamycin (Km), 500 μg ml−1.
Table 1.
Bacterial strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Relevant characteristics or sequence (5′-3′)a | Source or reference |
|---|---|---|
| Strains | ||
| Marinobacter adhaerens | Wild-type DSM 23420 | 27 |
| Escherichia coli DH5α λ-pir | ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 hsdR17 deoR thi-1 supE44 gyrA96 relA1/λ-pir | 36 |
| chpA::Tn5 | chpA transposon insertion mutant; Kanr | 57 |
| ΔcheA | cheA deletion mutant; Cmr | This study |
| ΔcheB | cheB deletion mutant; Cmr | This study |
| ΔchpB | chpB deletion mutant; Cmr | This study |
| Plasmids | ||
| pGEM-T Easy | Linearized cloning vector; Ampr | Promega, Mannheim, Germany |
| pFCM1 | Carrier of Cmr with FRT sides | 14 |
| Primers | ||
| cheA_upstream_forward | AAGCTTGGTACCTTCAGAAACCGAGACTGG | This study |
| cheA_upstream_reverse | GTGATCTTGGCTTCACCA | This study |
| cheA_downstream_forward | ATCCGTCAGCCCTGCCTT | This study |
| cheA_downstream_reverse | AAGCTTGCCAGAGAAGGCCGAAG | This study |
| cheB_upstream_forward | TACGTTCTCGGCAGGCGC | This study |
| cheB_upstream_reverse | AAGCTTTCCTGAGCCTGCTTGGGG | This study |
| cheB_downstream_forward | AAGCTTGGTACCTGATCTGGCCGGAGCTGG | This study |
| cheB_downstream_reverse | CCACGCCAGGGAACGGTT | This study |
| chpB_upstream_forward | AAGCTTGGTACCTTCCCATGAGCGGACGGC | This study |
| chpB_upstream_reverse | GGAAGCCTTCGCCGGATG | This study |
| chpB_downstream_forward | GCATGGACGTGGTTGCCA | This study |
| chpB_downstream_reverse | AAGCTTTCCGGTCAGCCGCTCAAT | This study |
| ΔcheA_check_forward | AAGCTTCTTCGGCCTTCTCTGGCA | This study |
| ΔcheA_check_reverse | GAGCTCCCAGTCTCGGTTTCTGAA | This study |
| ΔcheB_check_forward | TTGACAGCGTTGTCCCGG | This study |
| ΔcheB_check_reverse | CACCAAGCGGCTCGATG | This study |
| ΔchpB_check_forward | GATCAGCGGCGAACTGGG | This study |
| ΔchpB_check_reverse | GGTGCGGGTAAGGGCAGA | This study |
Underlined bases indicate the positions of endonuclease recognition sites. Kanr, kanamycin resistant; Cmr, chloramphenicol resistant; Ampr, ampicillin resistant; FRT, FLP recombination target.
Cultures of T. weissflogii (CCMP 1336) were obtained from the Provasoli-Guillard National Center for Culture of Marine Phytoplankton (CCMP; ME). Diatom cultures were grown at 16°C in f/2 medium (28) using a 12-h light period at 115 μmol photons m−2 s−1. Diatom cell numbers were determined by cell counts in a Sedgewick Rafter Counting Chamber S50 (SPI Supplies, West Chester, PA).
DNA procedures.
Electrophoresis, electroporation, and PCR were performed by standard techniques (49). Plasmids were isolated using a NucleoSpin plasmid kit (Macherey-Nagel, Düren, Germany). Restriction enzymes and DNA-modifying enzymes were used as recommended by the manufacturer (Fermentas, St. Leon-Rot, Germany). DNA fragments were resolved in 1% agarose gels and extracted with a NucleoSpin extract kit (Macherey-Nagel). Electroporation of M. adhaerens was conducted as described previously (57).
Chemotactic behavior of M. adhaerens toward the diatom T. weissflogii.
For the chemotaxis plate assay, M. adhaerens was grown on an MB agar plate overnight at 37°C. A culture of T. weissflogii was grown to midexponential phase (∼50,000 cells ml−1). For preparation of diatom cell homogenates and supernatants, 150 ml of culture was harvested by centrifugation of 50 ml 3 times at 4°C and 4,000 rpm for 20 min. The supernatants were decanted and filter sterilized (pore size, 0.2 μm) before use. The cell pellets were resuspended in 1 ml of sterile ice-cold deionized water. Cells were homogenized by sonication, and successful cell disruption was assessed by microscopy.
Soft agar plates were prepared with agar (0.3%, wt/vol) and peptone (0.5 mg ml−1) in seawater. The suspension was autoclaved and cooled down to 60°C. Diatom homogenate, supernatant, or culture was added to reach three different concentrations corresponding to ∼10,000, ∼20,000, and ∼40,000 diatom cells ml−1. As a control, plates containing only agar and peptone in seawater without any version of diatom supplement were prepared. Twenty-five milliliters of suspension was poured per petri dish, the lid was closed, and the agar was solidified for 15 min. M. adhaerens cells were inoculated with sterile toothpicks in triplicate per setup. Plates were incubated at room temperature. The diameter of swimming (in mm) was measured daily for 7 days. One-tailed Student's t test was applied to access the significance of differences between the setups as well as the measurements on day 7.
Mutagenesis of M. adhaerens chemotaxis genes.
Identification of chemotaxis gene clusters in the M. adhaerens HP15 genome (GenBank accession no. CP001978) (23) was conducted by comparison to those of Pseudomonas aeruginosa PAO1 using BLASTP (2). PAO1 is a model organism for chemotaxis (33) and shares 90% 16S rRNA similarity to the rRNA sequence of M. adhaerens (62). Conserved domains of CheA, CheB, ChpA, and ChpB were analyzed using the Conserved Domain Database (CDD) (38). Transposon mutants of M. adhaerens were generated as described previously (38), and mutant chpA::Tn5 was identified from a pool of randomly generated transposon mutants by the soft agar swimming assay. Site-directed mutations in the genes cheA, cheB, and chpB were generated by homologous recombination. For this, up- and downstream gene regions of the respective genes were PCR amplified using the following primer pairs: cheA_upstream_forward/reverse, cheA_downstream_forward/reverse, cheB_upstream_forward/reverse, cheB_downstream_forward/reverse, chpB_upstream_forward/reverse, and chpB_downstream_forward/reverse (Table 1). Amplified DNA fragments were separately ligated into the pGEM-T Easy vector and transformed into Escherichia coli DH5α λ-pir. The chloramphenicol resistance cassette (Cmr) was excised from pFCM1 with KpnI and ligated into the upstream gene region vectors. The upstream gene region plus Cmr was brought into the pGEM-T Easy vector containing the downstream gene regions by HindIII/SpeI. The generated knockout constructs were introduced into M. adhaerens by electroporation, and colonies were screened on Cm-containing MB agar plates. Double crossover events were confirmed by PCR using the following primer pairs: ΔcheA_check_forward/reverse, ΔcheB_check_forward/reverse, and ΔchpB_check_forward/reverse (Table 1). Since the corresponding PCR fragments for the wild type and ΔcheB and for the wild type and ΔchpB were of similar size, restriction digests with KpnI were used to confirm successful mutagenesis.
Phenotypic characterization of M. adhaerens chemotaxis mutants.
The chemotactic mutant phenotypes were confirmed by light and transmission electron microscopy, soft agar swimming assay (57), and biofilm formation assay (43). For soft agar swimming, an overnight plate culture of M. adhaerens was inoculated with a toothpick in freshly prepared plates containing 10-fold-diluted MB and 0.3% agar. For biofilm formation, cells were precultured overnight in 5 ml of MB under standard conditions and inoculated at a final optical density (OD) of 0.1 in 150 μl MB in microtiter plates in five replicates per strain. After overnight incubation at 28°C without shaking, the optical densities of the cultures were determined and attached cells were visualized by crystal violet staining, as described previously (43), with elution in 150 μl of 96% ethanol. The absorbance was measured at 600 nm. One-tailed Student's t test was applied to assess the significance of differences between strains.
Attachment assay with the diatom T. weissflogii.
M. adhaerens wild type and its four chemotaxis mutants were grown in MB at 18°C to an OD of 0.6 (exponential growth phase). T. weissflogii cells were grown for 6 days to a cell concentration of ∼50,000 cells ml−1 (stationary growth phase). Aliquots of 10 ml f/2 medium were inoculated with bacterial and diatom cells at a final extrapolated OD of approximately 0.0001, corresponding to ∼1 × 105 bacterial cells ml−1 and a final diatom concentration of ∼5,000 diatom cells ml−1. Total bacterial cell numbers were determined by dilution plating on MB agar plates at the beginning of the experiments, and the suspensions were incubated at room temperature for 24 h without shaking. Attaching and nonattaching bacterial cells were separated by sieving through 10-μm-opening-size gauze (22). Both fractions were collected and thoroughly vortexed before the numbers of bacterial CFU ml−1 were determined by dilution plating on MB agar plates. One-tailed Student′s t test was applied to access the significance of differences.
Cell staining and microscopy.
Following the attachment assay, 20-μl aliquots of the attached fractions were fixed on a microscopy glass slide by gentle heating. Samples were stained with 20 μl of alcian blue in 3% acetic acid solution (Carl Roth, Karlsruhe, Germany) for 30 min. After rinsing with water and drying, 20 μl of carbol fuchsin (ProLab Diagnostics, Merseyside, United Kingdom) solution was added as the second stain for 1 min. After washing with water and drying, microscopic pictures were taken using a light microscope with ×1,000 magnification and oil immersion. All samples were prepared and analyzed with 6 to 10 replicates. Each replicate was derived from a different attachment assay.
RESULTS
Migration behavior of M. adhaerens toward the diatom T. weissflogii.
When bacterial cells are inoculated into the center of a soft agar plate, they generate a concentration gradient by consumption of nutrients. By sensing the generated gradient, they swim outwards following the increase in nutrient concentration. In seawater soft agar with any version of diatom supplement, M. adhaerens showed a swimming behavior which was significantly stronger than that in the respective control soft agar (n = 3, P < 0.05) (Fig. 1), indicating that the cells were attracted to diatom exudates as well as cellular components. Attraction was highest to secreted products, as the diameter of migration was larger for the diatom culture and supernatant than for the diatom homogenate. The migration behavior toward diatom culture and the supernatant did not differ significantly on day 7 (n = 3, P > 0.05). Comparing three different concentrations of cell-free diatom culture supernatant, all concentration values yielded significantly different results (n = 3, P < 0.05) (Fig. 2). Hence, a significant trend between diameter of migration and supernatant content can be observed.
Fig 1.
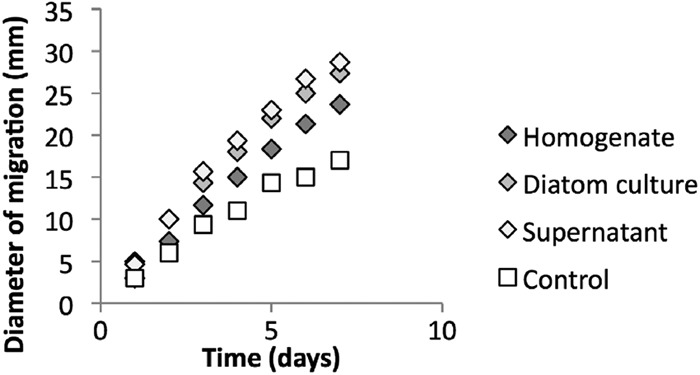
Migration behavior of M. adhaerens wild type in seawater soft agar (0.3%) supplemented with peptone (0.5 g ml−1) and the highest concentration of diatom cell homogenate, undiluted diatom culture, or cell-free diatom culture supernatant, shown in mm of diameter of migration. Seawater soft agar plates with peptone served only as controls. Three replicates were conducted per setup. Error bars show the standard deviation (n = 3, P < 0.05 for all comparisons on day 7, but P = 0.11 for diatom culture versus supernatant).
Fig 2.
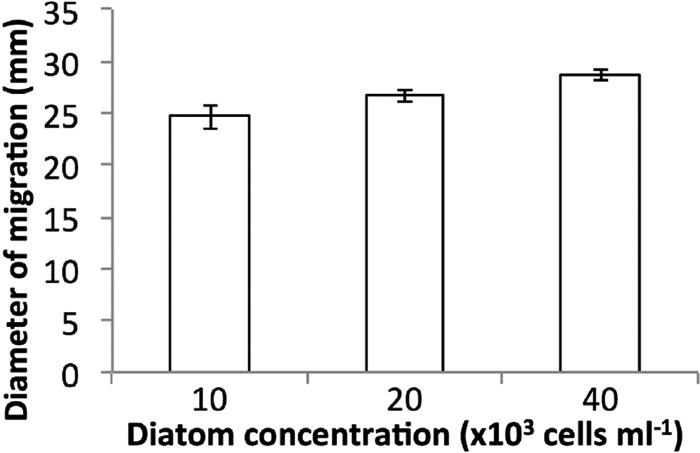
Migration behavior of M. adhaerens in seawater soft agar (0.3%) supplemented with peptone (0.5 mg ml−1) and cell-free supernatants derived from diatom cultures containing different cell concentrations at day 7, demonstrated in mm of diameter of migration. Three replicates were conducted per setup. Error bars show the standard deviation (n = 3, P < 0.05 for all comparisons, but P > 0.05 for a high versus a low concentration of diatom supernatant).
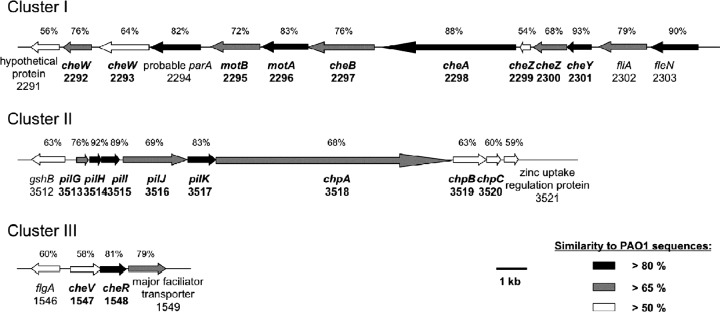
Chemotaxis gene clusters in M. adhaerens.
Three distinct chemotaxis gene clusters were identified in the genome sequence of M. adhaerens compared with the genome sequence information of P. aeruginosa PAO1 (Fig. 3). The respective protein sequences of both strains were closely related to each other, with the general similarity being higher than 65% and reaching up to 93% in particular proteins. Chemotaxis gene clusters I and III comprise the chemotaxis genes putatively responsible for the regulation of flagellar movement. They were found to be homologous to clusters I and V of P. aeruginosa PAO1 (21, 58), respectively. The M. adhaerens chemotaxis gene cluster II corresponded to cluster IV of strain PAO1, which controls twitching motility via pili (10, 32, 39). No significant homologs of chemotaxis gene clusters II and III of PAO1 were identified in M. adhaerens. All three chemotaxis clusters of the M. adhaerens strain were found to be located in close vicinity to genes encoding the respective motility appendages.
Fig 3.
Schematic presentation of the three chemotaxis gene clusters of M. adhaerens HP15 based on protein similarities to the corresponding gene products of P. aeruginosa PAO1. The PAO1 equivalents to M. adhaerens gene clusters I and III represent genes involved in flagellar chemotaxis. The PAO1 counterpart of gene cluster II is regulating twitching motility. All three gene clusters are in close proximity to the associated motility appendage genes. Mutations in the genes cheA, cheB (cluster I), chpA, and chpB (cluster II) were constructed in this study. Similarities to PAO1 of >80% (black), >65% (gray), and >50% (white) are shown. Size marker, 1 kb.
By comparison to the CDD (38, 39), CheA contained a regulatory domain acting as a histidine kinase, which has been proposed to interact with the regulator CheW; a histidine kinase-like ATPase; a signal-transducing histidine kinase; and a histidine phosphotransfer domain. The same domains were found in ChpA. However, an additional signal receiver domain and in total six histidine phosphotransfer domains were detected. A methylesterase domain was present in CheB as well as in ChpB. However, CheB contained a CheY-like receiver domain, which was absent in ChpB.
Migration behavior and surface attachment of M. adhaerens mutants.
Mutants were generated for the genes cheA, cheB, chpA, and chpB, which encode the initiating enzymes of the chemotaxis signaling cascade. The M. adhaerens mutants ΔcheA and ΔcheB carry mutations in chemotaxis gene cluster I and were generally nonchemotactic in 10-fold-diluted MB soft agar (Fig. 4). Transmission electron microscopy pictures showed that the mutant cells formed proper flagella identical to those of the wild type of M. adhaerens (data not shown), as previously demonstrated for P. aeruginosa PAO1 (39). In contrast, the M. adhaerens mutants chpA::Tn5 and ΔchpB, carrying mutations in chemotaxis gene cluster II, exhibited a wild-type swimming behavior. This demonstrated that these genes were not essential for flagellum-mediated chemotaxis.
Fig 4.
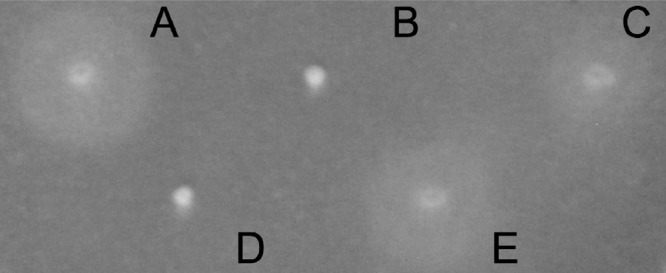
Phenotypes of M. adhaerens wild type and its chemotaxis mutants in 0.3% soft agar swimming assay. (A) Wild type; (B) ΔcheA; (C) chpA::Tn5; (D) ΔcheB; (E) ΔchpB.
The ability of all four M. adhaerens mutants to attach to an abiotic surface and form biofilms was assayed in microtiter plates. The wild type and all four mutants reached exponential growth overnight. Staining with crystal violet revealed that the four chemotaxis mutants showed a significantly decreased ability to attach to the wells of the microtiter plate, in contrast to the wild type of M. adhaerens (Fig. 5) (n = 5, P < 0.001 for wild type versus mutant). These results indicated that chemotaxis mediated by both the polar flagellum and putative pili is required for surface attachment and biofilm formation of M. adhaerens.
Fig 5.
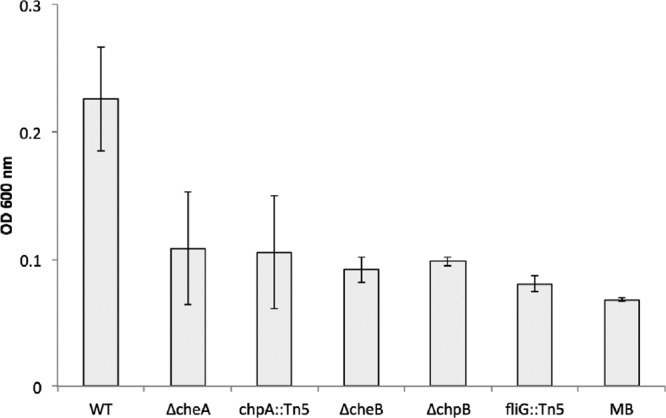
Bacterial surface attachment of M. adhaerens wild type (WT) and its chemotaxis mutants to an abiotic surface, visualized with crystal violet staining after washing off of planktonic cells and determined by absorbance measurement at 600 nm (n = 5, P < 0.001 for wild type versus mutant). Wells filled with MB served as controls.
Attachment of M. adhaerens chemotaxis mutants to diatom cells.
The role of chemotaxis in the interaction of M. adhaerens with diatom cells was assessed by comparing the wild type and chemotaxis mutants in regard to their ability to attach to cells of T. weissflogii (Table 2). The average total cell numbers, determined as numbers of CFU ml−1 of the M. adhaerens wild type and its mutants, were statistically similar at the start of the cocultures (time zero). However, after 24 h of incubation with diatoms, the calculated total cell numbers as well as the experimentally determined numbers of nonattached cells of the strain HP15 wild type were significantly lower than those of the mutants (Table 2). Comparative determination of the numbers of CFU ml−1 of the attached cells revealed that the wild type had slightly higher numbers attached to diatom cells than any of the mutants, but this was not statistically significant.
Table 2.
Cell dynamics of M. adhaerens HP15 and its chemotaxis mutants in the T. weissflogii attachment assay observed after dilution platinga
| Group | Avg no. of bacterial CFU ml−1 (105) ± SD |
||||
|---|---|---|---|---|---|
| Wild type | ΔcheA mutant | ΔcheB mutant | chpA::Tn5 mutant | ΔchpB mutant | |
| Total at time zero | 2.41 ± 0.49 | 1.92 ± 0.82 | 2.83 ± 0.46 | 2.33 ± 0.17 | 2.03 ± 0.36 |
| Total at 24 h | 1.44 ± 0.86 | 4.31 ± 0.37b | 4.74 ± 0.76b | 2.69 ± 0.66b | 2.51 ± 0.39b |
| Attached cells | 0.34 ± 0.13 | 0.25 ± 0.17 | 0.27 ± 0.03 | 0.29 ± 0.13 | 0.28 ± 0.04 |
| Free-living cells | 1.10 ± 0.78 | 4.06 ± 0.48b | 4.47 ± 0.76b | 2.40 ± 0.54b | 2.23 ± 0.38b |
Total cell numbers at the start of the experiment (time zero) and after 24 h of incubation as well as numbers of free-living bacterial cells and those attached to diatoms were quantified.
n = 6, P < 0.05 for wild type versus mutants.
In order to address the apparent paradox that decreased numbers of nonattached wild-type cells were not reflected in significantly increased numbers of attached cells (Table 2), microscopic analyses after sequential staining of the attached fractions with carbol fuchsin (for bacterial and diatom cells) and alcian blue (for TEPs) were conducted (Fig. 6). Interestingly, and in contrast to the results for samples of the ΔcheA and ΔchpB mutants, considerably more wild-type cells were found to be attached to diatom cells or alcian blue-stainable TEP aggregates. These results suggest that determination of the numbers of CFU ml−1 for the attached fractions did not reveal the true numbers of attached cells but that any observed CFU might have been the result of single-colony growth derived from multiple proximate diatom-attached wild-type cells. Interestingly and as reproducibly found (data not shown), diatom cells coincubated with the M. adhaerens HP15 wild type were stained with alcian blue, while those incubated with the ΔcheA and ΔchpB mutants were preferentially stained with carbol fuchsin only (Fig. 6A). This notion suggests a differential physiological state or exopolymer secretion of the diatom in association with the coincubated bacteria. Alcian blue-visualized TEPs were more populated by wild-type cells than samples from treatments with the ΔcheA or ΔchpB mutant (Fig. 6B). Microscopic analysis and staining of the fractions of free-living, nonattached bacterial cells revealed that these cells did not form aggregates (data not shown), suggesting that their numbers of CFU ml−1 most probably reflected true cell numbers.
Fig 6.
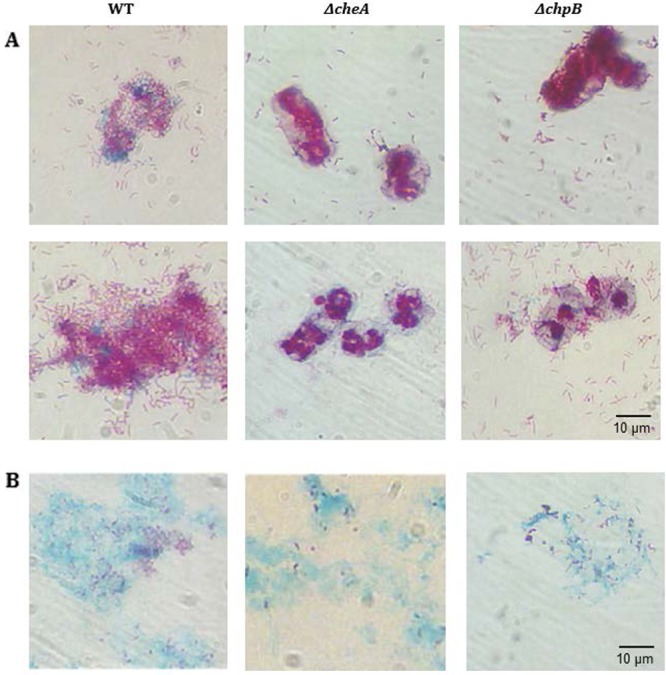
Microscopic analysis of carbol fuchsin- and alcian blue-stained samples of the diatom-attached fractions following a 24-h coincubation of T. weissflogii and M. adhaerens wild type or its ΔcheA and ΔchpB mutants. Carbol fuchsin stains bacteria and diatom cells, whereas alcian blue detects TEPs. Diatom cells may be covered by bacteria in some images. (A) Typical images of bacterial cells attached to diatom cells; (B) representative images of alcian blue-stained aggregates populated by carbol fuchsin-stained bacterial cells.
Altogether, our results indicate that chemotaxis-deficient mutants of M. adhaerens HP15 exhibited a decreased diatom attachment, while the HP15 wild type possibly impacted the physiology of the diatom cells, hinting at an important role of chemotaxis in bacterial attachment to the surface of cells or exudates of T. weissflogii.
DISCUSSION
The phycosphere of a phytoplankton cell is an important area of nutrient uptake for bacteria in the oceans (24). Various types of interactions between those two groups of organisms might be beneficial or detrimental for one or both partners. To identify genes and gene products which are essential for the interaction, a bilateral model system consisting of the diatom T. weissflogii and the bacterial strain M. adhaerens was previously established (22, 23, 31, 57). This interaction promotes the growth of both the bacterium and the diatom. Moreover, M. adhaerens attached to the diatom and the coculture resulted in increased TEP synthesis and aggregation of the diatom cells (22). Processes required for initiation of this interaction might be bacterial motility, chemotaxis, and surface attachment (15). It was hypothesized that bacterial chemotaxis may be required for diatom-bacterium interaction since M. adhaerens is highly motile and possesses distinct chemotaxis gene clusters. Importantly, M. adhaerens was found to be attracted to diatom-derived supplements in soft agar. Migration diameter formation solely due to growth was excluded by a direct comparison of the wild type and chemotaxis-deficient mutants in the same soft agar plate (data not shown). Furthermore, these mutants were not impaired in growth, as validated by liquid culture (data not shown), but nevertheless, they did not form the circle generated by outward-migrating cells. A significant difference between the amount of added cell-free spent diatom supernatant and the diameter of migration of the wild type was observed. Since bacterial attraction to diatom homogenate was less than that to diatom culture or culture supernatant, it is intriguing to speculate that this might be due to yet unknown attracting substances secreted by T. weissflogii. Alternatively, the less pronounced effects of diatom homogenates might be due to the simultaneous presence of various potentially toxic and thus repelling substances formed during the breakage of diatom cells, as described previously (48). Testing fractionated cell-derived or supernatant-based attractant-containing solutions might elucidate this type of interaction. In particular, an intense biochemical analysis of the metabolites secreted by the diatom might reveal certain beneficial or detrimental allelochemicals acting toward M. adhaerens. Since interactions are generally not stable in a changing environment like aquatic systems, T. weissflogii might produce molecules to attract the bacterium to initiate a symbiosis but might switch to production of deterring substances under specific conditions.
Interestingly, M. adhaerens does not utilize as the sole carbon source any sugars that had previously been shown to be produced by T. weissflogii (3, 31). However, as shown in the same previous study (31), M. adhaerens is able to grow on specific amino acids as the sole carbon source. By consuming specific protein or unknown carbohydrate components of diatom exudates, M. adhaerens might likely modify the chemical structure of the algal exudates, thereby potentially impacting aggregation of diatom cells, e.g., by enzymatic hydrolysis, as proposed previously for phytoplankton-bacterium interaction (45, 46, 56). To evaluate this hypothesis, time-dependent testing of aggregate formation and TEP synthesis during cocultures in so-called rolling tank experiments (22) comparing the wild type and the chemotaxis mutants will potentially reveal the actual mechanism of metabolite exchange and/or modification during the diatom-bacterium interaction.
Following the nutrient gradient established during bacterial growth (41), chpA and chpB mutants of M. adhaerens showed a swimming phenotype indistinguishable from that of the wild type, whereas mutants with defects in cheA and cheB showed a clearly decreased migration behavior. These observations indicate that the Chp chemosensory pathway might not be involved in the swimming-mediated chemotaxis of M. adhaerens. These findings are supported by the proposed twitching motility function of the corresponding genes in P. aeruginosa (21, 32, 39, 60). The conditioned agar matrix might require utilization of the flagellum for motility but not that of putative pili by M. adhaerens. As shown for PAO1, twitching motility mutants can be observed only on very thin agar medium (19). However, application of this method to demonstrate twitching motility of M. adhaerens in the current study was not successful (data not shown). Additional analysis tools, such as time-lapse microscopy of flow chambers, are required to fully understand the phenotype of chp mutants (35, 40, 43). However, all four chemotaxis-deficient mutants of HP15 showed a decreased ability to attach to an abiotic surface and to diatom cells. The latter finding raises the interesting question of how flagella and putative pili contribute individually to the attachment process, which needs to be addressed in future studies.
Previously, the impact of bacterial chemotaxis on attachment to abiotic surfaces has been described for the environmental organism Aeromonas spp., the plant pathogen Agrobacterium tumefaciens, and the soilborne diazotroph Azospirillum brasilense (34, 40, 55). For all three organisms, the attachment of chemotaxis-deficient mutants to abiotic surfaces was notably compromised. In Vibrio cholerae, chemotaxis was proposed to play a stage-specific role in attachment to a polystyrene microtiter dish, since a cheY3 mutant was deficient in generating a monolayer (42). In contrast, chemotaxis-deficient mutants of E. coli appear to form biofilms indistinguishable from the biofilm formed by the wild type (47). The opportunistic human pathogen P. aeruginosa has a rather complex chemotaxis system, in which one of the chemotaxis gene clusters exclusively regulates twitching motility (10, 33). Even though sequence analysis revealed homologies of the twitching motility gene cluster of P. aeruginosa in the genome of M. adhaerens, the two bacteria seem to greatly differ in their ability to attach to surfaces and to form biofilms. Although the respective P. aeruginosa mutants form a more robust biofilm than the wild type (13), chp mutants of M. adhaerens have a significantly decreased ability to attach to an abiotic surface and, hence, to form biofilms. Notably, most of the previously reported studies were based on tests with abiotic surfaces. In contrast, the current study demonstrated a chemotaxis-mediated attachment of a bacterial species to the environmentally relevant biotic surface of marine diatoms.
A somewhat puzzling outcome of the current study was the apparent discrepancy in the total numbers of CFU ml−1 of HP15 wild-type cells determined after the 24-h coincubation with T. weissflogii cells compared to the corresponding total cell numbers at the start of the experiments. Subsequent microscopic analyses, however, revealed that despite thorough vortexing of samples, a significant portion of wild-type cells stayed attached to either diatom cells or alcian blue-stainable aggregates. This suggests a potential limitation of the CFU method used herein and in many other studies. Following sample treatment and dilution plating, any single bacterial colony might thus have originated from multiple diatom-aggregated bacterial cells, thereby falsifying the actual cell numbers and causing a decrease in total cell numbers for the HP15 wild type. This assumption is supported by the converse results for the four tested mutants, for which total cell numbers predominantly derived from free-living, nonattached cells increased over the course of the experiments.
On the one hand, these findings indicate a relatively strong binding of HP15 wild-type cells to biotic surfaces, and on the other hand, these results demonstrate that determinations of the numbers of CFU alone are not sufficient to investigate the interaction of M. adhaerens and T. weissflogii. However, concluding from the attachment assay as well as microscopic observations, an interaction pathway between bacteria and diatoms involving bacterial migration toward and attachment to the diatom, followed by a yet unknown sequence of events leading to TEP formation, might be cautiously proposed. TEP formation can in turn be associated with aggregation (22). A continuation of this study aims to understand which partner in this specific interaction actually secretes and/or modifies the TEP leading to marine snow formation.
In summary, although a wealth of knowledge on mechanisms of chemotaxis in various bacterial species has been acquired in vitro (1, 6, 12, 33, 59), the individual effects of bacterial chemotaxis in environmentally relevant habitats are still poorly understood. For the first time and using genetic tools, this study demonstrated a potentially important role of chemotaxis for initiation of phytoplankton-bacterium interactions.
Our future research will focus on the identification of the chemotactic signal(s) of this interaction; the nutrient exchange between M. adhaerens and T. weissflogii, including TEP and aggregate formation; and the effects of bacterial chemotaxis on TEP formation by the diatom under various environmental conditions. Consequently, future experiments will elucidate the importance of molecular interaction mechanisms on global processes (30), particularly on the efficiency of the oceanic carbon pump.
ACKNOWLEDGMENTS
This work was financially supported by the International Max Planck Research School for Marine Microbiology and Jacobs University Bremen. The study was supported by grants of the Deutsche Forschungsgemeinschaft (GR1540/15-1 and UL169/6-1).
Footnotes
Published ahead of print 20 July 2012
REFERENCES
- 1. Adler J. 1975. Chemotaxis in bacteria. Annu. Rev. Biochem. 44:341–356 [DOI] [PubMed] [Google Scholar]
- 2. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 3. Aluwihare L, Repeta D. 1999. A comparison of the chemical characteristics of oceanic DOM and extracellular DOM produced by marine algae. Mar. Ecol. Prog. Ser. 186:105–117 [Google Scholar]
- 4. Amin S, Green D, Küpper F, Carrano C. 2009. Vibrioferrin, an unusual marine siderophore: iron binding, photochemistry, and biological implications. Inorg. Chem. 48:11451–11458 [DOI] [PubMed] [Google Scholar]
- 5. Amin SA, et al. 2012. Siderophore-mediated iron uptake in two clades of Marinobacter spp. associated with phytoplankton: the role of light. Biometals 25:181–192 [DOI] [PubMed] [Google Scholar]
- 6. Armitage J, Schmitt R. 1997. Bacterial chemotaxis: Rhodobacter sphaeroides and Sinorhizobium meliloti—variations on a theme. Microbiology 143:3671–3682 [DOI] [PubMed] [Google Scholar]
- 7. Azam F. 1998. Microbial control of oceanic carbon flux: the plot thickens. Science 280:694–695 [Google Scholar]
- 8. Bell W, Mitchell R. 1972. Chemotactic and growth responses of marine bacteria to algal extracellular products. Biol. Bull. 143:265–277 [Google Scholar]
- 9. Bell W, Lang J, Mitchell R. 1974. Selective stimulation of marine bacteria by algal extracellular products. Limnol. Oceanogr. 19:833–839 [Google Scholar]
- 10. Bertrand JJ, West JT, Engel JN. 2010. Genetic analysis of the regulation of type IV pilus function by the Chp chemosensory system of Pseudomonas aeruginosa. J. Bacteriol. 192:994–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Blackburn N, Fenchel T, Mitchell J. 1998. Microscale nutrient patches in planktonic habitats shown by chemotactic bacteria. Science 282:2254–2256 [DOI] [PubMed] [Google Scholar]
- 12. Boin MA, Austin MJ, Häse CC. 2004. Chemotaxis in Vibrio cholerae. FEMS Microbiol. Lett. 239:1–8 [DOI] [PubMed] [Google Scholar]
- 13. Caiazza NC, Merritt JH, Brothers KM, O'Toole GA. 2007. Inverse regulation of biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 189:3603–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Choi K-H, Schweizer HP. 2005. An improved method for rapid generation of unmarked Pseudomonas aeruginosa deletion mutants. BMC Microbiol. 5:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cole J. 1982. Interactions between bacteria and algae in aquatic ecosystems. Annu. Rev. Ecol. Syst. 13:291–314 [Google Scholar]
- 16. Darzins A. 1994. Characterization of a Pseudomonas aeruginosa gene cluster involved in pilus biosynthesis and twitching motility: sequence similarity to the chemotaxis proteins of enterics and the gliding bacterium Myxococcus xanthus. Mol. Microbiol. 11:137–153 [DOI] [PubMed] [Google Scholar]
- 17. Darzins A. 1993. The pilG gene product, required for Pseudomonas aeruginosa pilus production and twitching motility, is homologous to the enteric, single-domain response regulator CheY. J. Bacteriol. 175:5934–5944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Darzins A. 1995. The Pseudomonas aeruginosa pilK gene encodes a chemotactic methyltransferase (CheR) homologue that is translationally regulated. Mol. Microbiol. 15:703–717 [DOI] [PubMed] [Google Scholar]
- 19. Deziel E, Comeau Y, Villemur R. 2001. Initiation of biofilm formation by Pseudomonas aeruginosa 57RP correlates with emergence of hyperpiliated and highly adherent phenotypic variants deficient in swimming, swarming, and twitching motilities. J. Bacteriol. 183:1195–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fenchel T. 2002. Microbial behavior in a heterogeneous world. Science 296:1068–1071 [DOI] [PubMed] [Google Scholar]
- 21. Ferrandez A, Hawkins A, Summerfield D, Harwood C. 2002. Cluster II che genes from Pseudomonas aeruginosa are required for an optimal chemotactic response. J. Bacteriol. 184:4374–4383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gärdes A, Iversen MH, Grossart H-P, Passow U, Ullrich MS. 2011. Diatom-associated bacteria are required for aggregation of Thalassiosira weissflogii. ISME J. 5:436–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gärdes A, et al. 2010. Complete genome sequence of Marinobacter adhaerens type strain (HP15), a diatom-interacting marine microorganism. Stand. Genomic Sci. 3:97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goldman J. 1984. Conceptual role for microaggregates in pelagic waters. Bull. Mar. Sci. 35:462–476 [Google Scholar]
- 25. Grossart H. 1999. Interactions between marine bacteria and axenic diatoms (Cylindrotheca fusiformis, Nitzschia laevis, and Thalassiosira weissflogii) incubated under various conditions in the lab. Aquat. Microb. Ecol. 19:1–11 [Google Scholar]
- 26. Grossart H, Riemann L, Azam F. 2001. Bacterial motility in the sea and its ecological implications. Aquat. Microb. Ecol. 25:247–258 [Google Scholar]
- 27. Grossart H, Schlingloff A, Bernhard M, Simon M, Brinkhoff T. 2004. Antagonistic activity of bacteria isolated from organic aggregates of the German Wadden Sea. FEMS Microbiol. Ecol. 47:387–396 [DOI] [PubMed] [Google Scholar]
- 28. Guillard R, Ryther J. 1962. Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt and Detonula confervacea Cleve. Can. J. Microbiol. 8:229–239 [DOI] [PubMed] [Google Scholar]
- 29. Henrichsen J. 1983. Twitching motility. Annu. Rev. Microbiol. 37:81–93 [DOI] [PubMed] [Google Scholar]
- 30. Ianora A, et al. 2011. The relevance of marine chemical ecology to plankton and ecosystem function: an emerging field. Marine Drugs 9:1625–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kaeppel EC, Gärdes A, Seebah S, Grossart H-P, Ullrich MS. 2012. Marinobacter adhaerens sp. nov., prominent in aggregate formation with the diatom Thalassiosira weissflogii. Int. J. Syst. Evol. Microbiol. 62:124–128 [DOI] [PubMed] [Google Scholar]
- 32. Kato J, Nakamura T, Kuroda A, Ohtake H. 1999. Cloning and characterization of chemotaxis genes in Pseudomonas aeruginosa. Biosci. Biotechnol. Biochem. 63:155–161 [DOI] [PubMed] [Google Scholar]
- 33. Kato J, Kim H-E, Takiguchi N, Kuroda A, Ohtake H. 2008. Pseudomonas aeruginosa as a model microorganism for investigation of chemotactic behaviors in ecosystem. J. Biosci. Bioeng. 106:1–7 [DOI] [PubMed] [Google Scholar]
- 34. Kirov S, Castrisios M, Shaw J. 2004. Aeromonas flagella (polar and lateral) are enterocyte adhesins that contribute to biofilm formation on surfaces. Infect. Immun. 72:1939–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Klausen M, et al. 2003. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 48:1511–1524 [DOI] [PubMed] [Google Scholar]
- 36. Larsen RA, Wilson MM, Guss AM, Metcalf WW. 2002. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch. Microbiol. 178:193–201 [DOI] [PubMed] [Google Scholar]
- 37. Lee SO, et al. 2000. Involvement of an extracellular protease in algicidal activity of the marine bacterium Pseudoalteromonas sp. strain A28. Appl. Environ. Microbiol. 66:4334–4339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Marchler-Bauer A, et al. 2011. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 39:D225–D229 doi:10.1093/nar/gkq1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Masduki A, Nakamura J, Ohga T, Umezaki R. 1995. Isolation and characterization of chemotaxis mutants and genes of Pseudomonas aeruginosa. J. Bacteriol. 177:948–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Merritt P, Danhorn T, Fuqua C. 2007. Motility and chemotaxis in Agrobacterium tumefaciens surface attachment and biofilm formation. J. Bacteriol. 189:8005–8014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Miller T, Hnilicka K, Dziedzic A, Desplats P, Belas R. 2004. Chemotaxis of Silicibacter sp. strain TM1040 toward dinoflagellate products. Appl. Environ. Microbiol. 70:4692–4701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Moorthy S, Watnick PI. 2005. Identification of novel stage-specific genetic requirements through whole genome transcription profiling of Vibrio cholerae biofilm development. Mol. Microbiol. 57:1623–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. O'Toole G, Kolter R. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295–304 [DOI] [PubMed] [Google Scholar]
- 44. Paerl H, Pinckney J. 1996. A mini-review of microbial consortia: their roles in aquatic production and biogeochemical cycling. Microb. Ecol. 31:225–247 [DOI] [PubMed] [Google Scholar]
- 45. Passow U. 2002. Transparent exopolymer particles (TEP) in aquatic environments. Prog. Oceanogr. 55:287–333 [Google Scholar]
- 46. Passow U. 2002. Production of transparent exopolymer particles (TEP) by phyto- and bacterioplankton. Mar. Ecol. Prog. Ser. 236:1–12 [Google Scholar]
- 47. Pratt L, Kolter R. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285–293 [DOI] [PubMed] [Google Scholar]
- 48. Ribalet F, Intertaglia L, Lebaron P, Casotti R. 2008. Differential effect of three polyunsaturated aldehydes on marine bacterial isolates. Aquat. Toxicol. 86:249–255 [DOI] [PubMed] [Google Scholar]
- 49. Sambrook J, Fritsch E, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 50. Schmidt J, et al. 2011. The Pseudomonas aeruginosa chemotaxis methyltransferase CheR1 impacts on bacterial surface sampling. PLoS One 6:e18184 doi:10.1371/journal.pone.0018184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Seyedsayamdost MR, Carr G, Kolter R, Clardy J. 2011. Roseobacticides: small molecule modulators of an algal-bacterial symbiosis. J. Am. Chem. Soc. 133:18343–18349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Seyedsayamdost MR, Case RJ, Kolter R, Clardy J. 2011. The Jekyll-and-Hyde chemistry of Phaeobacter gallaeciensis. Nat. Chem. 3:331–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Seymour J, Ahmed T, Durham W, Stocker R. 2010. Chemotactic response of marine bacteria to the extracellular products of Synechococcus and Prochlorococcus. Aquat. Microb. Ecol. 59:161–168 [Google Scholar]
- 54. Seymour J, Ahmed T, Stocker R. 2009. Bacterial chemotaxis towards the extracellular products of the toxic phytoplankton Heterosigma akashiwo. J. Plankton Res. 31:1557–1561 [Google Scholar]
- 55. Siuti P, Green C, Edwards AN, Doktycz MJ, Alexandre G. 2011. The chemotaxis-like Che1 pathway has an indirect role in adhesive cell properties of Azospirillum brasilense. FEMS Microbiol. Lett. 323:105–112 [DOI] [PubMed] [Google Scholar]
- 56. Smith DC, Steward GF, Long RA, Azam F. 1995. Bacterial mediation of carbon fluxes during a diatom bloom in a mesocosm. Deep Sea Res. Part II Top. Stud. Oceanogr. 42:75–97 [Google Scholar]
- 57. Sonnenschein EC, et al. 2011. Development of a genetic system for Marinobacter adhaerens HP15 involved in marine aggregate formation by interacting with diatom cells. J. Microbiol. Methods 87:176–183 [DOI] [PubMed] [Google Scholar]
- 58. Stover CK, et al. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964 [DOI] [PubMed] [Google Scholar]
- 59. Wadhams GH, Armitage JP. 2004. Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell Biol. 5:1024–1037 [DOI] [PubMed] [Google Scholar]
- 60. Whitchurch C, et al. 2004. Characterization of a complex chemosensory signal transduction system which controls twitching motility in Pseudomonas aeruginosa. Mol. Microbiol. 52:873–893 [DOI] [PubMed] [Google Scholar]
- 61. Willey J, Waterbury J. 1989. Chemotaxis toward nitrogenous compounds by swimming strains of marine Synechococcus spp. Appl. Environ. Microbiol. 55:1888–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhang Z, Schwartz S, Wagner L, Miller W. 2000. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 7:203–214 [DOI] [PubMed] [Google Scholar]