Abstract
The respiratory chain of Escherichia coli contains three different cytochrome oxidases. Whereas the cytochrome bo oxidase and the cytochrome bd-I oxidase are well characterized and have been shown to contribute to proton translocation, physiological data suggested a nonelectrogenic functioning of the cytochrome bd-II oxidase. Recently, however, this view was challenged by an in vitro biochemical analysis that showed that the activity of cytochrome bd-II oxidase does contribute to proton translocation with an H+/e− stoichiometry of 1. Here, we propose that this apparent discrepancy is due to the activities of two alternative catabolic pathways: the pyruvate oxidase pathway for acetate production and a pathway with methylglyoxal as an intermediate for the production of lactate. The ATP yields of these pathways are lower than those of the pathways that have so far always been assumed to catalyze the main catabolic flux under energy-limited growth conditions (i.e., pyruvate dehydrogenase and lactate dehydrogenase). Inclusion of these alternative pathways in the flux analysis of growing E. coli strains for the calculation of the catabolic ATP synthesis rate indicates an electrogenic function of the cytochrome bd-II oxidase, compatible with an H+/e− ratio of 1. This analysis shows for the first time the extent of bypassing of substrate-level phosphorylation in E. coli under energy-limited growth conditions.
INTRODUCTION
The branched nature of the respiratory chain of Escherichia coli and of many other microbial species has been the subject of many studies at both the physiological (3–5, 7, 35, 45) and the biochemical (9, 10, 36, 41, 43) levels. As the respiratory chain constitutes an essential part of the machinery of living cells for conserving energy under nonfermentative conditions, understanding the mechanisms that underlie proton pumping, as well as quantification of the efficiency of these processes, is paramount to grasp the essence of their bioenergetics. In addition, since bacteria must cope with ever-changing environments, it is not surprising that adaptive strategies evolved that are directly related to the processes involved in energy conservation (3, 5, 26). Under respiratory conditions, the catabolic machinery of E. coli is made up of two tightly linked biochemical networks: the cytoplasmic glycolytic pathway and the tricarboxylic acid (TCA) cycle on one hand and the membrane-embedded respiratory chain on the other. Glycolysis and the TCA cycle provide the respiratory chain with electrons (mostly as NADH and reduced flavin adenine dinucleotide [FADH2]) and, in addition, add directly to the overall ATP yield of catabolism via the phosphoglycerate kinase, the pyruvate kinase, and the succinyl-coenzyme A (CoA) synthetase reactions. The membrane-bound respiratory module allows the cell flexibility in the H+/e− ratio (the number of protons delivered to the periplasmic side of the membrane per electron) and rapid responsiveness in the rate of ATP synthesis (Fig. 1).
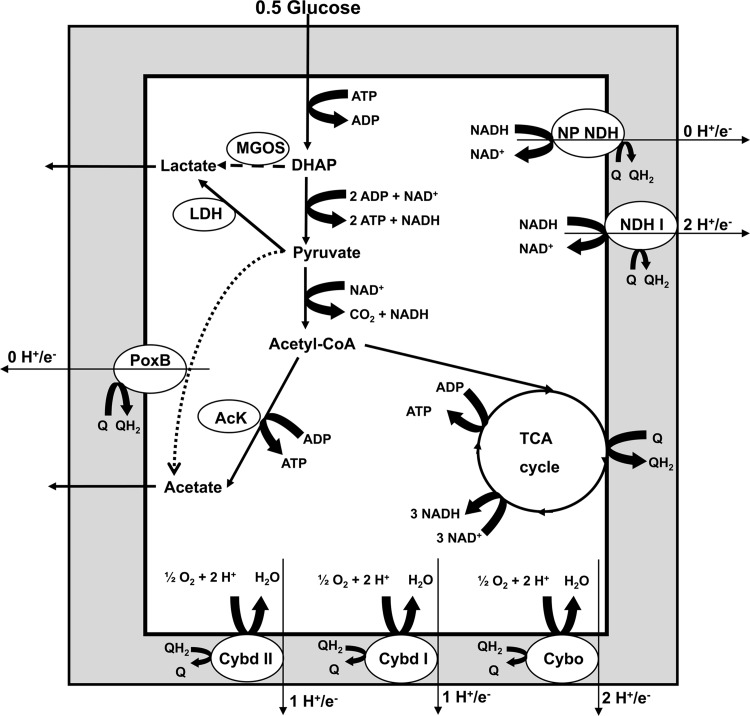
Fig 1.
Scheme of the E. coli aerobic respiratory chain (adapted from reference 39 with permission from the publisher). Enzyme bioenergetic efficiency is indicated as the number of protons delivered to the periplasmic side of the membrane per electron (the H+/e− ratio). NDH I is the proton-translocating NADH-quinone oxidoreductase, and NDH II, WrbA, QOR, QOR2, and YhDH are the non-proton-translocating NADH-quinone oxidoreductases (NP NDHs). NDH I and the NP NDHs transfer electrons to the quinones (Q) to yield reduced quinones (QH2). Three quinol-oxygen oxidoreductases, cytochromes bo3 (Cybo), bd-I (Cybd I), and bd-II (Cybd II), oxidize QH2 and reduce O2 to H2O. Acetate formation via acetate kinase (AcK) leads to ATP formation, in contrast to acetate synthesized by PoxB. Similarly, lactate formed by one of the three lactate dehydrogenases results in ATP formation (LDH here represents Dld, LdhA, and LldD), while lactate synthesized via methylglyoxal synthase (MGOS) from DHAP leads to ATP consumption via the activity of the phosphotransferase system (PTS) and 6-phosphofructokinase (PFK). The gray area indicates the cytoplasmic membrane. The dashed and dotted lines indicate uncoupling at the level of substrate phosphorylation catabolic pathways by MGOS and PoxB, respectively.
Thus, E. coli contains an NADH dehydrogenase (11, 32, 40), NDH I, that translocates protons with an H+/e− stoichiometry of 2 (8, 17, 18, 22, 49), although a recent study proposed an H+/e− stoichiometry of 1.5 (50). It also expresses functional, membrane-associated NADH dehydrogenases that do not show this activity (e.g., NDH II and WrbA [11, 33, 51]). Similarly, E. coli is known to have three different cytochrome oxidases (35, 36, 42), among which cytochrome bd-I oxidase has an H+/e− ratio of 1 (36) and cytochrome bo oxidase has an H+/e− ratio of 2 (36, 37).
Recently (7), we proposed that the H+/e− stoichiometry of the third oxidase, cytochrome bd-II (42), was 0. This was based on a quantitative physiological analysis of a series of strains with modified compositions of the respiratory chain. In the analysis, specific ATP synthesis rates were calculated on the assumption that the energy source, glucose (which was limiting growth), was dissimilated to acetyl-CoA using fully coupled pathways (i.e., the pathways that synthesize the most ATP from a single glucose molecule), and thus, product formation (i.e., lactate and acetate) was assumed to be catalyzed by lactate dehydrogenase (LDH) and acetate kinase, respectively. However, a recent biochemical characterization of the cytochrome bd-II oxidase showed this complex to be electrogenic, with an H+/e− stoichiometry of 1 (10).
Here, we present a physiological analysis that accounts for the discrepancy between our results (7) and those of Borisov et al. (10). We show that the presence and activity of two alternative dissimilatory pathways that had not been included in the quantitative analysis presented previously resulted in an overestimation of the ATP produced by substrate-level phosphorylation in our study (7). These pathways are the pyruvate oxidase system (PoxB) (2, 12, 28), which produces acetate but bypasses ATP synthesis (compared to acetate formation catalyzed through the activity of pyruvate dehydrogenase), and the methylglyoxal pathway (MGOP) to lactate, which in effect consumes ATP during lactate production rather than producing it (14, 25, 30, 43). Physiological analysis of a relevant set of mutant strains, enzyme assays, and quantitative assessment of flux distributions based on calculations of the ATP flux (qATP) provide arguments to reevaluate the proton-translocating capability of the cytochrome bd-II oxidase. Accordingly, a value for its H+/e− ratio of 1 is calculated in vivo, consistent with the in vitro biochemical observations obtained (10).
MATERIALS AND METHODS
Construction of deletion mutants.
E. coli single-deletion strains were ordered from the Keio collection (6). In order to construct strains with multiple deletions, the kanamycin marker was first removed, as described by Datsenko and Wanner (15). Mutants with double and triple deletions were constructed by P1 phage transduction of the desired mutations (Table 1). The mutants were checked by PCR.
Table 1.
E. coli strains used in this study
| Straina | Genotype | Cytochrome(s) remaining | Reference(s) |
|---|---|---|---|
| BW25113 | K-12 wild type | All cytochromes | 6 |
| MB20 | JW0421 (ΔcyoB), kanamycin marker removed | Cytochrome bd-I, bd-II | 6, 7 |
| MB21 | JW0961 (ΔappB), kanamycin marker removed | Cytochrome bd-I, bo | 6, 7 |
| MB28 | JW0723 (ΔcydB), kanamycin marker removed | Cytochrome bo, bd-II | 6, 7 |
| MB30 | BW25113, ΔcyoB ΔappB ΔnuoB, kanamycin marker removed | Cytochrome bd-I | 7 |
| MB34 | BW25113, ΔcydB ΔappB ΔnuoB, kanamycin marker removed | Cytochrome bo | 7 |
| MB37 | BW25113, ΔcyoB ΔcydB ΔnuoB, kanamycin marker removed | Cytochrome bd-II | 7 |
| PS27 | BW25113, ΔpoxB::kan ΔcydB ΔappB ΔnuoB | Cytochrome bo | This work |
| PS28 | BW25113, ΔpoxB::kan ΔcyoB ΔappB ΔnuoB | Cytochrome bd-I | This work |
| PS29 | BW25113, ΔpoxB::kan ΔcyoB ΔcydB ΔnuoB | Cytochrome bd-II | This work |
Strains MB30, MB34, MB37, PS27, PS28, and PS29 do not contain the proton translocating NADH dehydrogenase NDH-I.
Continuous cultures.
E. coli K-12 strain BW25113 and various deletion derivatives (Table 1) were grown in an Applikon 2-liter fermentor at a constant dilution rate of 0.15 ± 0.01 h−1. The steady state was verified by two consecutive organic acid measurements with at least a 2-h interval. All values represent experiments performed at least in triplicate. For MB37, an additional duplicate was performed in addition to the previously performed triplicate (7), and the averaged quintuplicate data set was used for this report. A defined simple salts medium, described previously (16), with nitrilotriacetic acid (2 mM) rather than citrate as a chelator was used. Selenite (30 μg/liter) and thiamine (15 mg/liter) were added to the medium. For strains PS27, PS28, and PS29, tryptone (100 mg/liter), yeast extract (50 mg/liter), and NaCl (50 mg/liter) were additionally supplied to support growth. Glucose was used as the main carbon and energy source (>98% of total carbon) at a final concentration of 50 mM. The dilution rate was set by adjusting the medium supply rate. The pH was maintained at 7.0 ± 0.1 by titration with sterile 4 M NaOH, and the temperature was controlled at 37°C with a stirring rate of 600 rpm. The air supply rate was set at 0.5 liter/min. In all cultures, the steady-state specific rates of fermentation product formation and of glucose, CO2, and O2 consumption were measured as described by Alexeeva et al. (3).
Analysis of carbon fluxes.
Steady-state bacterial dry weights were determined as described previously (4, 5). Glucose, pyruvate, lactate, formate, acetate, succinate, and ethanol contents were determined by high-performance liquid chromatography (HPLC) (LKB) with a Rezex organic acid analysis column (Phenomenex) at 45°C with 7.2 mM H2SO4 as the eluent, using an RI 1530 refractive index detector (Jasco) and AZUR chromatography software for data integration. All data collected showed a carbon balance of 94% ± 13% as calculated from the glucose consumption and CO2 and fermentation product formation rates. Undefined components (e.g., from tryptone and yeast extract) were added in such small amounts that they could be neglected.
Enzyme assays for d- and l-lactate and LDH synthase and methylglyoxal (MGO) synthase activities.
The lactate assay kit from Megazyme was used to measure the amounts of d-lactate and l-lactate produced in the batch and chemostat cultures. The assay for measuring LDH activity was adopted (and modified) from Garrigues et al. and Neves at al. (19, 31). The assay was performed immediately after cell disruption. Cells were pelleted by centrifugation for 10 min at 4,500 rpm at 4°C. After resuspension of the pellet in 10% glycerol plus 25 mM Na phosphate buffer (pH 7.2) containing 2 mg/ml lysozyme, the cells were lysed by 3 rounds of bead beating for 20 s at speed 6 in a Precellys 24 beater from Bertin Technologies. The soluble fraction was isolated after another round of centrifugation for 5 min at 8,000 rpm at 4°C. Measurements were taken spectrophotometrically at 30°C and pH 7.2 for the level of NADH at 340 nm (extinction coefficient for NADH [ε] = 6.22 ⋅ 103 ⋅ M−1 ⋅ cm−1). One unit of activity is defined as the amount of enzyme required to produce 1 μmol of product per minute (lactate and NAD+ are formed at an equimolar ratio). The reaction mixture contained Tris-HCl buffer (100 mM; pH 7.2), NADH (0.3 mM), fructose 1,6-bisphosphate (3 mM), MgCl2 (2.5 mM), and sodium pyruvate (30 mM), which was added to initiate the reaction. The reaction was followed by recording the absorption changes at 340 nm for about 40 min.
The activity assay of the methylglyoxal synthase is performed in a similar way, with methylglyoxal as the substrate. It was adapted from the method of Hopper and Cooper (23). The assay was performed after cell disruption, and measurements were taken spectrophotometrically at 37°C and pH 7.0 for the level of S-lactoylglutathione at 240 nm (ε = 3.37 ⋅ 103 ⋅ M−1 ⋅ cm−1). The reaction mixture contained imidazole buffer (40 mM; pH 7.0), dihydroxyacetone phosphate (0.75 mM), reduced glutathione (1.65 mM), glyoxylase enzyme (sufficient to convert 0.8 mM methylglyoxal/min), and cell extract, which was used to initiate the reaction. The progress of the reaction was then followed by recording the absorption at 240 nm for about 15 min.
Calculation of specific ATP synthesis rates and H+/e− ratios.
The total flux of ATP (qATPtotal) can be calculated by addition of the ATP generated by substrate-level phosphorylation (qATPSLP) and the ATP generated by oxidative phosphorylation in the electron transfer chain (qATPETC). For reference, an anaerobically grown glucose-limited chemostat culture was used. Under those conditions, all routes for product formation are defined, and therefore, there are no uncertainties concerning the calculated qATPtotal. For complete oxidation of glucose to CO2, the rate of substrate-level-derived ATP synthesis (qATPSLP) is stoichiometrically coupled to the rate of CO2 production as follows: 1 Glc + 6 O2 + 4 ADP + 4 Pi→6 CO2 + 4 ATP + 6 H2O. Hence, qATPSLP is equal to 2/3 ⋅ qCO2, where qCO2 is the rate of CO2 synthesis. In the generally accepted view, under aerobic conditions, pyruvate-formate lyase is not active (1) and acetate is produced via the conversion of pyruvate by pyruvate dehydrogenase, phosphotransacetylase, and acetate kinase, yielding 1 CO2 and 1 extra ATP per acetate. For strains or conditions that involve acetate (and/or ethanol) production, the additional amount of CO2 produced stoichiometrically with acetate and/or ethanol was taken into account. In addition, if glucose is only partially mineralized, e.g., to lactate by an active lactate dehydrogenase, this yields ATP with a stoichiometry of 1 (mol ATP/mol lactate). Hence, qATPSLP is equal to 2 ⋅ qAce + qEtOH + qLac + (2/3 ⋅ [qCO2 − qAce − qEtOH]), or
| (1) |
where qAce, qEtOH, qLac, and qCO2 are the rates of synthesis of acetate, ethanol, lactate, and CO2, respectively, expressed in mmol ⋅ g (dry weight)−1 ⋅ h−1.
The E. coli genome encodes two additional dissimilatory pathways that use dihydroxyacetone-phosphate (DHAP) or pyruvate as the substrate. Elevated levels of the former substrate may result in a significant flux toward methylglyoxal via the low-affinity methylglyoxal synthase and subsequent conversion of methylglyoxal to lactate (38, 43). As a result, the energy contained in the phosphoanhydride is lost, and hence, lactate formation via the MGOP results in a net loss of 1 mol ATP/mol lactate formed. Pyruvate may also be oxidized by the pyruvate oxidase system (PoxB) (13, 20) to acetate, resulting in the loss of substrate-level phosphorylation during acetate formation. To include these alternative pathways, equation 1 must be modified to equation 2:
| (2) |
The rate of ATP synthesis due to oxidative phosphorylation (qATPETP) can be calculated from the rate of oxygen reduction, the number of protons translocated per electron transferred to oxygen, and the stoichiometry of the number of ATP molecules synthesized per proton flowing back through the ATP synthase. Assuming that this stoichiometry is 4 in E. coli (41) and taking into account the fact that four electrons are needed to reduce molecular oxygen to water, it follows that
| (3) |
where qO2 is the rate of consumption of O2. Combining equations 1 and 2 results in equation 4:
| (4) |
where qATPtotal is the total rate of ATP synthesis. This formula was used for calculating the qATPtotal of strains PS27, PS28, and PS29. To determine the H+/e− ratio of the cytochrome bd-II oxidase in MB37, it was rewritten as follows:
| (5) |
RESULTS
Theoretical consideration of the calculated in vivo contribution of cytochrome bd-II to proton translocation.
The recently published biochemical assessment of the electrogenic properties of cytochrome bd-II oxidase (10) is in striking contrast to previous findings (7), where it was reported that this oxidase in vivo did not contribute to the buildup of a proton motive force (i.e., H+/e− = 0). The conclusion was based on the assumption that lactic acid is produced by E. coli exclusively via pyruvate reduction by lactate dehydrogenase (29) and acetic acid via pyruvate dehydrogenase plus acetate kinase, yielding 1 and 2 ATP, respectively, per molecule of lactic acid and acetic acid formed. As both products were formed at significant rates by the strains under the given conditions, their formation contributes significantly to the total rate of ATP synthesis and hence affects the quantification of the apparent H+/e− stoichiometry of cytochrome bd-II oxidase to a large extent. However, alternative pathways that lead to the formation of these two products have been described in E. coli (2, 12–14, 20, 24, 28, 29, 43), and since they have a lower ATP stoichiometry (mol ATP synthesized per mol product formed), their activity may cause the discrepancy described above. Acetate can also be formed in E. coli by pyruvate oxidase (2), whereas formation of lactate may occur via the methylglyoxal pathway. The route from glucose to acetate via PoxB is associated with the synthesis of 1 ATP/acetate formed (rather than 2 for the acetate kinase route), whereas lactate formation via the methylglyoxal pathway has a net consumption of 1 ATP/lactate (as opposed to the production of 1 ATP/lactate for lactate dehydrogenase).
In order to verify whether these routes could have played a role under the previously used growth conditions (7), two strategies were followed. First, the contribution of PoxB to acetic acid formation was estimated by a theoretical analysis of the previously published data (see Table 3) (7) and by removal of poxB from MB30, MB34, and MB37, resulting in strains PS28, PS27, and PS29, respectively. Second, the potential contribution of the methylglyoxal pathway to lactate production was tested by analysis of the activity of the key enzyme involved.
Table 3.
qATPtotal valuesa
| Strain | Rate (mmol ⋅ g [dry wt]−1 ⋅ h−1) |
|||
|---|---|---|---|---|
| qATPtotal (Ldh + AcK) | qATPtotal (Ldh + PoxB) | qATPtotal (MGOP + AcK) | qATPtotal (MGOP + PoxB) | |
| PS29 | 22.9 ± 0.5 | ND | 19.2 ± 1.0 | ND |
| MB37 | 26.1 ± 0.7 | 20.0 ± 1.0 | 24.8 ± 2.4 | 18.7 ± 2.7 |
| PS27 | 19.0 ± 1.7 | ND | 18.7 ± 1.6 | ND |
| PS28 | 19.5 ± 2.4 | ND | 19.5 ± 2.4 | ND |
Calculated assuming that lactate and acetate are formed by lactate dehydrogenase and acetate kinase (column 2), lactate dehydrogenase and pyruvate oxidase (column 3), methylglyoxal bypass and acetate kinase (column 4), or methylglyoxal bypass and pyruvate oxidase (column 5). For PS27, an H+/e− ratio of 2 was assumed; for all other strains, an H+/e− ratio of 1 was assumed. ND, not detectable.
Contribution of the methylglyoxal pathway and PoxB to the total lactic acid and acetic acid flux, respectively.
A catabolic flux analysis of strains PS27, PS28, and PS29 was carried out in a glucose-limited chemostat, as described previously (7). Table 2 presents all the relevant specific product formation and oxygen consumption rates that were subsequently used to calculate the specific ATP synthesis rates (qATPtotal). Table 3 shows a comparative analysis with regard to ATP synthesis rates that was done for 4 scenarios regarding the conversion of glucose to fermentation products: (i) all fluxes proceed through the LDH pathway and acetate kinase, (ii) all fluxes proceed through the LDH pathway and pyruvate oxidase, (iii) all fluxes proceed through the methylglyoxal pathway and acetate kinase, or (iv) all fluxes proceed completely through the methylglyoxal pathway and PoxB.
Table 2.
Specific product formation and oxygen consumption ratesa
| Strain | Rate (mmol ⋅ g [dry wt]−1 ⋅ h−1) |
|||
|---|---|---|---|---|
| qAce | qLac | qCO2 | qO2 | |
| PS29 | 7.2 ± 0.7 | 1.7 ± 0.7 | 7.0 ± 0.7 | 6.7 ± 0.3 |
| BW25113 | 0.0 ± 0.0 | 0.0 ± 0.0 | 8.3 ± 0.3 | 4.8 ± 1.9 |
| MB37 | 6.2 ± 0.8 | 1.1 ± 1.2 | 11.7 ± 2.1 | 9.6 ± 2.7 |
| PS27 | 1.4 ± 0.4 | 0.1 ± 0.2 | 5.8 ± 1.2 | 6.2 ± 1.3 |
| PS28 | 3.7 ± 1.7 | NDb | 10.0 ± 0.9 | 10.8 ± 1.2 |
Strains (7) were grown in glucose-limited chemostat cultures. See Materials and Methods for growth conditions.
ND, not detectable.
For the amount of ATP required to synthesize E. coli biomass, qATPtotal, a value of 18.8 ± 1.0 mmol ⋅ g (dry weight)−1 ⋅ h−1 was used, derived from an experiment with a fully fermentative anaerobic glucose-limited chemostat culture. Under anaerobic conditions, only acetate and ethanol are produced via the well-established coupled pathways, and therefore, the qATP can be unambiguously determined under such conditions. H+/e− stoichiometries were assumed to be as published previously (36, 37) for the cytochrome bo oxidase (i.e., 2) and cytochrome bd-I oxidase (i.e., 1) and as recently determined biochemically by Borisov et al. (10) for cytochrome bd-II (i.e., 1).
Quantitative analysis of the data for strain MB37 (7), which now includes a contribution of fluxes through the alternative pathways, shows that indeed one has to assume a flux through PoxB in order to arrive at a specific ATP synthesis rate that matches that of the reference value (Table 3). Similarly, analysis of strain PS29 (MB37ΔpoxB) revealed that acetic acid was still synthesized (Table 2). However, the calculated qATPtotal was much lower than for MB37, confirming that in strain MB37, acetate formation is largely catalyzed by PoxB. Furthermore, assuming that scenario 1 holds leads to a qATPtotal value for PS29 that is still higher than the reference value (Table 3). In contrast, an analysis according to scenario 3 results in a specific ATP synthesis rate close to the reference value. This strongly suggests that under the conditions employed, uncoupling of substrate-level phosphorylation takes place in this strain via a high flux through the methylglyoxal pathway. As PS27 and PS28 produce virtually no lactate, assuming that either lactate dehydrogenase or the methylglyoxal pathway contributes to ATP synthesis is not relevant. It is noteworthy that for both strains, a qATPtotal value close to the reference value was calculated (Table 3).
A different calculation can be made to illustrate the sensitivity of the analysis. In Table 3, all qATPtotal values are set to the reference rate (18.8 mmol ⋅ g [dry weight])−1 ⋅ h−1) in order to calculate the resulting H+/e− stoichiometry for cytochrome bd-II oxidase. It can be seen that with the respective flux distributions through the methylglyoxal pathway and PoxB, deviations from the predicted stoichiometries are negligibly small. In conclusion, assuming flux distributions through these alternative pathways results in H+/e− stoichiometries that match the biochemical data.
With respect to the assumed contribution of the methylglyoxal pathway in PS29, enzyme activities were measured in cell extracts (see Materials and Methods) of the various strains grown under identical glucose-limited conditions at a dilution rate of 0.15 h−1. Table 4 shows the activities in comparison with lactate dehydrogenase. Clearly, PS29 showed significantly higher activity. Thus, this finding supports the hypothesis that the methylglyoxal pathway may contribute to lactate formation in the strain and hence lowers the ATP yield in substrate-level phosphorylation in PS29. Contribution to glucose catabolism via methylglyoxal is further substantiated by the fact that under the conditions tested, PS29 produces only l-lactate (see Fig. S1 in the supplemental material), the end product of the conversion of dihydroxyacetone phosphate by methylglyoxal synthase and subsequently glyoxylase, whereas lactate dehydrogenase produces l- and d-lactate in approximately a 1:1 ratio (see Fig. S1 in the supplemental material).
Table 4.
Enzyme activities of cell extracts of strains cultured in a glucose-limited chemostata
| Strain | Activity (mmol ⋅ g protein−1 ⋅ h−1) |
|
|---|---|---|
| Methylglyoxal synthase | Lactate dehydrogenase | |
| BW25113 | 4.1 ± 1.2 | 0.5 ± 0.3 |
| MB37 | 3.1 ± 1.1 | 3.8 ± 0.7 |
| PS27 | 10.9 ± 1.8 | 6.9 ± 0.9 |
| PS28 | 7.7 ± 4.3 | 6.2 ± 2.3 |
| PS29 | 28.2 ± 6.5 | 12.2 ± 3.9 |
See Materials and Methods.
DISCUSSION
In this paper, we have addressed an apparent discrepancy between the physiological (7) and biochemical (10) quantifications of the electrogenic nature of cytochrome bd-II oxidase. Our previous flux analysis, designed to calculate specific ATP synthesis rates, lacked two alternative catabolic pathways and therefore overestimated the ATP synthesis rate by substrate-level phosphorylation. The analysis led to the conclusion that the high Vmax of cytochrome bd-II oxidase was non-proton pumping. In the new analysis presented here, we show that the activities of two uncoupling reactions (i.e., catabolic pathways that have lower net ATP yields than the pathway to the same end product with the maximal yield) explain the observed differences. Our analyses allow us to conclude that under the conditions tested (aerobic, glucose-limited chemostat cultures), a significant flux through these alternative, low-efficiency pathways occurs, although the activity of methylglyoxal synthase in PS29 grown under glucose-limited continuous conditions could be assessed only indirectly via an assay of the enzymatic activity of the key enzyme of the methylglyoxal pathway and of LDH. Unfortunately, the additional removal of genes from strain PS29 (e.g., all lactate dehydrogenases) has proven not to be technically feasible due to an inability to perform the necessary transformation with the gene knockout plasmids. Nevertheless, the observation that the methylglyoxal synthase activity in strain PS29 is 3-fold higher than that of wild-type cells (in which the activity is in agreement with previously published data [10.4 mmol ⋅ g protein−1 ⋅ h−1 [24]) shows that in this strain, the MGO pathway is upregulated and contributes to lactate production. Further confirmation is seen in the fact that only the l-lactate isomer is produced.
Our analysis demonstrates that E. coli invokes loss of catabolic efficiency under energy-limited growth conditions at the level of cytoplasmic ATP synthesis. This may seem rather surprising, but it should be noted that we have previously shown that under these conditions, the electron transport chain employs the non-proton-translocating NDH II (11). The phenomenon of catabolic uncoupling by bypassing substrate-level phosphorylation is seen most clearly in strains that contain cytochrome bd-II oxidase as the sole catalyst transferring electrons to oxygen. It may be that the strongly reduced ubiquinone pool (7) in these strains affects NADH dehydrogenase activity, thereby allowing increased activity of the PoxB pathway, or, alternatively, influences steady-state pools of intermediates (e.g., an increase in intracellular pyruvate by inhibition of pyruvate dehydrogenase), which in turn may increase enzyme activity or gene expression. Similarly, the high flux via the methylglyoxal pathway could be the result of limited activity of the glyceraldehyde 3-phosphate dehydrogenase complex due to a high NADH/NAD+ ratio and, hence, possible buildup of triose phosphate in these cells (46).
The mechanisms and signals that govern the relative contributions of such unexpected pathways as demonstrated here clearly remain to be elucidated. Nevertheless, our analysis touches upon both fundamental and applied issues. First, our results exemplify the complexity of the physiological strategies employed by microorganisms when it comes to energy conservation; they further complicate the questions of whether and when cells induce catabolic networks optimized for either a high growth yield or a high growth rate. The former is related to efficient use of the energy source, and the latter is related to high catabolic rates and responsiveness (11, 21, 27, 34, 44, 47, 48). Second, our findings emphasize the need for extensive knowledge of metabolic pathways when a rational design is formulated in the applied field of metabolic engineering. Pathways that are considered to be exceptional and to be specific for certain conditions only may be paramount for the cell's functioning and hence be more important than assumed. These findings emphasize that thorough quantitative physiological and biochemical understanding is of high importance for effective metabolic engineering.
Supplementary Material
ACKNOWLEDGMENTS
We thank A. Vreugdenhil for her assistance in genetic manipulation of the E. coli strains described in this report.
This work was supported by the SysMo-SUMO2 project and the Erasmus Mundus External Cooperation Window (EMECW).
Footnotes
Published ahead of print 27 July 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Abbe K, Takahashi S, Yamada T. 1982. Involvement of oxygen-sensitive pyruvate formate-lyase in mixed-acid fermentation by Streptococcus mutans under strictly anaerobic conditions. J. Bacteriol. 152: 175– 182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abdel-Hamid AM, Attwood MM, Guest JR. 2001. Pyruvate oxidase contributes to the aerobic growth efficiency of Escherichia coli. Microbiology 147: 1483– 1498 [DOI] [PubMed] [Google Scholar]
- 3. Alexeeva S, de Kort B, Sawers G, Hellingwerf KJ, Teixeira de Mattos MJ. 2000. Effects of limited aeration and of the ArcAB system on intermediary pyruvate catabolism in Escherichia coli. J. Bacteriol. 182: 4934– 4940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Alexeeva S, Hellingwerf KJ, Teixeira de Mattos MJ. 2002. Quantitative assessment of oxygen availability: perceived aerobiosis and its effect on flux distribution in the respiratory chain of Escherichia coli. J. Bacteriol. 184: 1402– 1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Alexeeva S, Hellingwerf KJ, Teixeira de Mattos MJ. 2003. Requirement of ArcA for redox regulation in Escherichia coli under microaerobic but not anaerobic or aerobic conditions. J. Bacteriol. 185: 204– 209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baba T, et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2: 2006 0008 doi:10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bekker M, de Vries S, Ter Beek A, Hellingwerf KJ, Teixeira de Mattos MJ. 2009. Respiration of Escherichia coli can be fully uncoupled via the nonelectrogenic terminal cytochrome bd-II oxidase. J. Bacteriol. 191: 5510– 5517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bogachev AV, Murtazina RA, Skulachev VP. 1996. H+/e− stoichiometry for NADH dehydrogenase I and dimethyl sulfoxide reductase in anaerobically grown Escherichia coli cells. J. Bacteriol. 178: 6233– 6237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Borisov VB, Gennis RB, Hemp J, Verkhovsky MI. 2011. The cytochrome bd respiratory oxygen reductases. Biochim. Biophys. Acta 1807: 1398– 1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Borisov VB, et al. 2011. Aerobic respiratory chain of Escherichia coli is not allowed to work in fully uncoupled mode. Proc. Natl. Acad. Sci. U. S. A. 108: 17320– 17324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Calhoun MW, Oden KL, Gennis RB, Teixeira de Mattos MJ, Neijssel OM. 1993. Energetic efficiency of Escherichia coli: effects of mutations in components of the aerobic respiratory chain. J. Bacteriol. 175: 3020– 3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carter K, Gennis RB. 1985. Reconstitution of the ubiquinone-dependent pyruvate oxidase system of Escherichia coli with the cytochrome o terminal oxidase complex. J. Biol. Chem. 260: 10986– 10990 [PubMed] [Google Scholar]
- 13. Chang YY, Cronan JE., Jr 1983. Genetic and biochemical analyses of Escherichia coli strains having a mutation in the structural gene (poxB) for pyruvate oxidase. J. Bacteriol. 154: 756– 762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cooper RA. 1984. Metabolism of methylglyoxal in microorganisms. Annu. Rev. Microbiol. 38: 49– 68 [DOI] [PubMed] [Google Scholar]
- 15. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97: 6640– 6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Evans CGT, Herbert D, Tempest DW. 1970. The continuous culture of microorganisms, vol 2 Construction of a chemostat. Academic Press, London, United Kingdom [Google Scholar]
- 17. Galkin A, Drose S, Brandt U. 2006. The proton pumping stoichiometry of purified mitochondrial complex I reconstituted into proteoliposomes. Biochim. Biophys. Acta 1757: 1575– 1581 [DOI] [PubMed] [Google Scholar]
- 18. Galkin AS, Grivennikova VG, Vinogradov AD. 1999. →H+/2e- stoichiometry in NADH-quinone reductase reactions catalyzed by bovine heart submitochondrial particles. FEBS Lett. 451: 157– 161 [DOI] [PubMed] [Google Scholar]
- 19. Garrigues C, Loubiere P, Lindley ND, Cocaign-Bousquet M. 1997. Control of the shift from homolactic acid to mixed-acid fermentation in Lactococcus lactis: predominant role of the NADH/NAD+ ratio. J. Bacteriol. 179: 5282– 5287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Grabau C, Cronan JE., Jr 1984. Molecular cloning of the gene (poxB) encoding the pyruvate oxidase of Escherichia coli, a lipid-activated enzyme. J. Bacteriol. 160: 1088– 1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Griffin AS, West SA, Buckling A. 2004. Cooperation and competition in pathogenic bacteria. Nature 430: 1024– 1027 [DOI] [PubMed] [Google Scholar]
- 22. Hinkle PC, Kumar MA, Resetar A, Harris DL. 1991. Mechanistic stoichiometry of mitochondrial oxidative phosphorylation. Biochemistry 30: 3576– 3582 [DOI] [PubMed] [Google Scholar]
- 23. Hopper DJ, Cooper RA. 1972. The purification and properties of Escherichia coli methylglyoxal synthase. Biochem. J. 128: 321– 329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hopper DJ, Cooper RA. 1971. The regulation of Escherichia coli methylglyoxal synthase; a new control site in glycolysis? FEBS Lett. 13: 213– 216 [DOI] [PubMed] [Google Scholar]
- 25. Inoue Y, Kimura A. 1995. Methylglyoxal and regulation of its metabolism in microorganisms. Adv. Microb. Physiol. 37: 177– 227 [DOI] [PubMed] [Google Scholar]
- 26. Kayser A, Weber J, Hecht V, Rinas U. 2005. Metabolic flux analysis of Escherichia coli in glucose-limited continuous culture. I. Growth-rate-dependent metabolic efficiency at steady state. Microbiology 151: 693– 706 [DOI] [PubMed] [Google Scholar]
- 27. Kreft JU, Bonhoeffer S. 2005. The evolution of groups of cooperating bacteria and the growth rate versus yield trade-off. Microbiology 151: 637– 641 [DOI] [PubMed] [Google Scholar]
- 28. Mather MW, Gennis RB. 1985. Kinetic studies of the lipid-activated pyruvate oxidase flavoprotein of Escherichia coli. J. Biol. Chem. 260: 16148– 16155 [PubMed] [Google Scholar]
- 29. Mat-Jan F, Alam KY, Clark DP. 1989. Mutants of Escherichia coli deficient in the fermentative lactate dehydrogenase. J. Bacteriol. 171: 342– 348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mattos MJ, Streekstra H, Tempest DW. 1984. The methylglyoxal bypass as a metabolic uncoupling device in anaerobic glucose-limited chemostat cultures of Klebsiella aerogenes. Arch. Microbiol. 139: 260– 264 [Google Scholar]
- 31. Neves AR, et al. 2002. Effect of different NADH oxidase levels on glucose metabolism by Lactococcus lactis: kinetics of intracellular metabolite pools determined by in vivo nuclear magnetic resonance. Appl. Environ. Microbiol. 68: 6332– 6342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ohnishi T, et al. 1994. Biophysical and biochemical studies of bacterial NADH:quinone oxidoreductase (NDH-1). Biochem. Soc. Trans. 22: 70S. [DOI] [PubMed] [Google Scholar]
- 33. Patridge EV, Ferry JG. 2006. WrbA from Escherichia coli and Archaeoglobus fulgidus is an NAD(P)H:quinone oxidoreductase. J. Bacteriol. 188: 3498– 3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pfeiffer T, Schuster S, Bonhoeffer S. 2001. Cooperation and competition in the evolution of ATP-producing pathways. Science 292: 504– 507 [DOI] [PubMed] [Google Scholar]
- 35. Poole RK, Cook GM. 2000. Redundancy of aerobic respiratory chains in bacteria? Routes, reasons and regulation. Adv. Microb. Physiol. 43: 165– 224 [DOI] [PubMed] [Google Scholar]
- 36. Puustinen A, Finel M, Haltia T, Gennis RB, Wikström M. 1991. Properties of the two terminal oxidases of Escherichia coli. Biochemistry 30: 3936– 3942 [DOI] [PubMed] [Google Scholar]
- 37. Puustinen A, Finel M, Virkki M, Wikström M. 1989. Cytochrome o (bo) is a proton pump in Paracoccus denitrificans and Escherichia coli. FEBS Lett. 249: 163– 167 [DOI] [PubMed] [Google Scholar]
- 38. Saikusa T, Watanabe RHK, Murate K, Kimura A. 1987. Metabolism of 2-oxoaldehydes in bacteria—purification and characterization of methylglyoxal reductase from Escherichia coli. Agric. Biol. Chem. 51: 1893– 1899 [Google Scholar]
- 39. Sharma P, Teixeira de Mattos MJ, Hellingwerf KJ, Bekker M. 2012. On the function of the various quinone species in Escherichia coli. FEBS J. [Epub ahead of print.] doi:10.1111/j.1742–4658.2012.08608.x [DOI] [PubMed] [Google Scholar]
- 40. Sled VD, et al. 1993. Bacterial NADH-quinone oxidoreductases: iron-sulfur clusters and related problems. J. Bioenerg. Biomembr. 25: 347– 356 [DOI] [PubMed] [Google Scholar]
- 41. Steigmiller S, Turina P, Graber P. 2008. The thermodynamic H+/ATP ratios of the H+-ATP synthases from chloroplasts and Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 105: 3745– 3750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sturr MG, Krulwich TA, Hicks DB. 1996. Purification of a cytochrome bd terminal oxidase encoded by the Escherichia coli app locus from a Δcyo Δcyd strain complemented by genes from Bacillus firmus OF4. J. Bacteriol. 176: 1742– 1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tempest DW, Neijssel OM, Teixeira de Mattos MJ. 1983. Regulation of metabolite overproduction in Klebsiella aerogenes. Riv. Biol. 76: 263– 274 [PubMed] [Google Scholar]
- 44. Travisano M, Velicer GJ. 2004. Strategies of microbial cheater control. Trends Microbiol. 12: 72– 78 [DOI] [PubMed] [Google Scholar]
- 45. Unden G, Bongaerts J. 1997. Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim. Biophys. Acta 1320: 217– 234 [DOI] [PubMed] [Google Scholar]
- 46. Weber J, Kayser A, Rinas U. 2005. Metabolic flux analysis of Escherichia coli in glucose-limited continuous culture. II. Dynamic response to famine and feast, activation of the methylglyoxal pathway and oscillatory behaviour. Microbiology 151: 707– 716 [DOI] [PubMed] [Google Scholar]
- 47. Westerhoff HV, Hellingwerf KJ, van Dam K. 1983. Thermodynamic efficiency of microbial growth is low but optimal for maximal growth rate. Proc. Natl. Acad. Sci. U. S. A. 80: 305– 309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Westerhoff HV, Lolkema JS, Otto R, Hellingwerf KJ. 1982. Thermodynamics of growth. Non-equilibrium thermodynamics of bacterial growth. The phenomenological and the mosaic approach. Biochim. Biophys. Acta 683: 181– 220 [DOI] [PubMed] [Google Scholar]
- 49. Wikstrom M. 1984. Two protons are pumped from the mitochondrial matrix per electron transferred between NADH and ubiquinone. FEBS Lett. 169: 300– 304 [DOI] [PubMed] [Google Scholar]
- 50. Wikstrom M, Hummer G. 2012. Stoichiometry of proton translocation by respiratory complex I and its mechanistic implications. Proc. Natl. Acad. Sci. U. S. A. 109: 4431– 4436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Young IG, Jaworowski A, Poulis MI. 1978. Amplification of the respiratory NADH dehydrogenase of Escherichia coli by gene cloning. Gene 4: 25– 36 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.