Abstract
This study explores microbial community structure in managed aquifer recharge (MAR) systems across both laboratory and field scales. Two field sites, the Taif River (Taif, Saudi Arabia) and South Platte River (Colorado), were selected as geographically distinct MAR systems. Samples derived from unsaturated riverbed, saturated-shallow-infiltration (depth, 1 to 2 cm), and intermediate-infiltration (depth, 10 to 50 cm) zones were collected. Complementary laboratory-scale sediment columns representing low (0.6 mg/liter) and moderate (5 mg/liter) dissolved organic carbon (DOC) concentrations were used to further query the influence of DOC and depth on microbial assemblages. Microbial density was positively correlated with the DOC concentration, while diversity was negatively correlated at both the laboratory and field scales. Microbial communities derived from analogous sampling zones in each river were not phylogenetically significantly different on phylum, class, genus, and species levels, as determined by 16S rRNA gene pyrosequencing, suggesting that geography and season exerted less sway than aqueous geochemical properties. When field-scale communities derived from the Taif and South Platte River sediments were grouped together, principal coordinate analysis revealed distinct clusters with regard to the three sample zones (unsaturated, shallow, and intermediate saturated) and, further, with respect to DOC concentration. An analogous trend as a function of depth and corresponding DOC loss was observed in column studies. Canonical correspondence analysis suggests that microbial classes Betaproteobacteria and Gammaproteobacteria are positively correlated with DOC concentration. Our combined analyses at both the laboratory and field scales suggest that DOC may exert a strong influence on microbial community composition and diversity in MAR saturated zones.
INTRODUCTION
Water utilities and agencies throughout the world are recognizing water reuse as a viable option to augment groundwater supplies with impaired and reclaimed water in response to population growth and water scarcity. Infiltration of these waters through natural subsurface systems can provide sufficient pathogen inactivation and chemical contaminant attenuation. For example, managed aquifer recharge (MAR) systems, such as riverbank filtration, soil-aquifer treatment, and aquifer recharge and recovery, have been used in Europe and North America for decades to augment local supplies with storm water, impaired surface water, and reclaimed water (46).
These multiobjective treatment systems enable contaminant attenuation through processes such as sorption and biotransformation driven by the indigenous microbial communities (5). Organic compounds delivered to these systems originate from the burial of organic detritus or from the transport of dissolved organic carbon (DOC) in water recharging the underlying groundwater. Allochthonous organic matter from terrestrial and often anthropogenic sources, such as municipal, industrial, and agricultural wastewater, contributes significantly to the organic content in freshwater systems (36). While much of the DOC released during human activity, including many emerging contaminants of concern, can be readily mineralized during MAR, certain trace organic chemicals, such as anticonvulsant drugs (e.g., primidone, carbamazepine) or artificial sweeteners (e.g., sucralose), appear to be more recalcitrant to biotransformation (22, 28, 42, 45).
Although microorganisms play an important role in DOC removal during MAR, a comprehensive understanding of community characteristics in MAR systems is still missing. Sediments and soils utilized as the infiltration layer of MAR systems represent one of the most diverse and complex microbial ecosystems, and it has been estimated that a single gram of soil may contain thousands of bacterial species (38, 54). The microbial community structure could be directly linked with the function of the community; thus, the elucidation of the detailed composition could reflect the metabolic potentials of the community. Just as importantly, environmental factors influencing microbial community structure in MAR systems have not yet been systematically studied. The microbial community might be influenced and shaped by different physicochemical factors, including pH, temperature, water content, oxygen concentration, organic carbon content, and nutrient availability. For example, Fierer and Jackson (19) revealed that microbial communities in soil samples exhibit higher diversity under more acidic conditions when contrasted with that in alkaline environments; subsequent studies have reported similar phenomena relating to pH (35, 48). Thus, to provide a basis for improved design and operation of MAR systems, there is a need to comprehensively characterize microbial communities and to elucidate influencing environmental factors that shape microbial community structure in MAR systems.
High-throughput sequencing tools (454; Illumina) enable a detailed understanding of microbial communities by sequencing huge numbers of 16S and 18S small-subunit (SSU) rRNA gene sequences (24). Until now, high-throughput sequencing of 16S rRNA genes has successfully been applied in diverse natural and engineered environments, including lakes, oceans, soils, activated sludge, and human and animal intestines (8, 13, 21, 47, 59). However, to the best of our knowledge, no equivalent study has yet been performed for the infiltration zone of MAR systems. The characteristics of the microbial communities and the role of influencing environmental factors in MAR systems at both the laboratory and field scales were thus elucidated mainly using high-throughput sequencing and statistical analysis in this study. A series of seasonal samples derived from two representative MAR field systems located in geographically distinct regions (Taif, Saudi Arabia, and Colorado) were analyzed. Field analyses were complemented with laboratory-scale sediment columns to further query DOC, infiltration zone depth, and the applicability of these more controlled systems to the actual environment. An enhanced understanding of the microbial community in MAR and the interplay with environmental variables is helpful to guide future design and improve the operational conditions for enhanced water treatment using MAR systems.
MATERIALS AND METHODS
MAR field sites.
Geographically distinct MAR field sites in North America and the Middle East were selected for this study. Native sediment and water samples were collected from four cross sections (CSs) of the Taif River located in Taif, Saudi Arabia, in February, April, and June of 2011. Samples were collected at two upstream cross sections (CS1 and CS2), at a secondary treated municipal wastewater discharge point, and at two cross sections (CS3 and CS4) downstream of the discharge (see Fig. S1a in the supplemental material). For each cross section, samples were collected from the saturated shallow (depth, 1 to 2 cm) and intermediate (depth, 10 to 50 cm) infiltration zone, in addition to adjacent unsaturated riverbed samples (see Fig. S1b in the supplemental material). Samples were collected in an analogous manner from a single cross section of the South Platte River in Brighton, CO, in November 2010, April 2011, and June 2011. This cross-section site, described by Hoppe-Jones et al. (28), is located downstream of a major wastewater discharge. Water samples were also collected during each sampling event at both sites. Bulk water quality parameters, including DOC, ammonia, and nitrate levels, UV absorbance at 254 nm, and other physicochemical parameters, were analyzed according to standard methods (4). Samples were archived at 4°C (water) and −20°C (sediment) prior to respective chemical and microbial analyses.
Laboratory-scale column experiments.
Column experiments were established both at the King Abdullah University of Science and Technology (KAUST) and at the Colorado School of Mines (CSM) using sediments from the two field sites. Small-scale column experiments at KAUST were conducted in duplicate under moderate DOC condition. Each consisted of a series of two glass columns 30 cm by 4 cm (Kimble Chase Kontes) for a total length of 60 cm connected with Viton tubing (Masterflex). Columns were filled with Taif River unsaturated riverbed sediments (sieve fraction, 0.2 to 2 mm). Columns were operated against gravity, fully saturated, and protected from light. They were fed with modified OECD synthetic sewage solution consisting of 1.6 mg peptone (BD Difco), 0.8 mg yeast extract (BD Difco), 7.8 mg humic acid sodium salt (Sigma-Aldrich), 12 mg CaCl2 · 2H2O, 42.5 mg NaCl, 36 mg MgSO4, 1.75 mg K2HPO4, 0.35 mg KH2PO4, 10 mg KNO3, 0.25 mg FeCl3, 11 mg Na2SO4, and trace metal solution described by Dantas et al. (16) per liter of water. An IPC8 peristaltic pump (Ismatec, Switzerland) was utilized to continuously deliver the solution at a loading rate of 1.15 m/day. A hydraulic retention time of 10 to 12 h was confirmed for the column series by using KBr as a conservative tracer (data not shown). Columns were conditioned for 2 weeks to stabilize DOC and nitrate concentrations in the column effluents, and sediments were harvested after 4 weeks under sterile conditions from column depths of 1 cm, 15 cm, 30 cm, 45 cm, and 60 cm.
In addition, at CSM an intermediate-scale column system was employed under low-DOC conditions that consisted of a series of four 1-m Plexiglas columns filled with aquifer material (median particle diameter [d50] = 0.8 mm, free organic carbon [foc] = 0.003%) to achieve longer retention times. The column system was operated in flowthrough mode at a loading rate of 0.09 m/day and a hydraulic retention time of 25 days under saturated flow conditions (45). Low-DOC nanofiltration permeated material (average, 0.6 mg/liter) was introduced as the feed water for the duration of the work described herein. Samples were collected twice a week from column influent and effluent and analyzed for DOC, UV absorbance, pH, and conductivity. After 5 months of applying nanofiltration permeated material, duplicate sediment samples were collected from the top of each column (0-m, 1-m, 2-m, and 3-m depths). Sediments were archived at −20°C prior to DNA extraction.
DNA extraction and pyrosequencing.
For each sample, approximately 5 g sediments was homogenized after removing larger pebbles and plant materials. DNA was extracted in duplicate for each sample using a PowerSoil kit (MO BIO Laboratories, Carlsbad, CA), and the DNA solutions (approximately 100 μl for each extraction) were pooled to reduce sample variability. 16S rRNA was partially amplified (generating product approximately 300 bp in length) from the DNA extracts using a nearly universal bacterial and archaeal primer set (13). The forward primer consisted of the 454 Life Sciences Adaptor A, the universal primer 515F (GTGYCAGCMGCCGCGGTAA) (13), a CA sequence inserted as a linker between the barcode and the rRNA primer, and a unique 12-bp error-correcting Golay barcode used to tag each PCR product (18, 23). The reverse primer contained the 454 Life Sciences Adaptor B, universal primer 806R (GGACTACHVGGGTWTCTAAT) (13), and a 2-base linker sequence (TC). The PCR amplification (denaturation at 94°C for 3 min, then amplification with 35 cycles of 94°C for 45 s, 50°C for 30 s, and 72°C for 90 s, and a final extension of 10 min at 72°C) and gel purification procedures were used without modification (18) and processed by the Genomics Core Laboratory at KAUST for pyrosequencing on a 454 FLX Titanium genome sequencer (Roche).
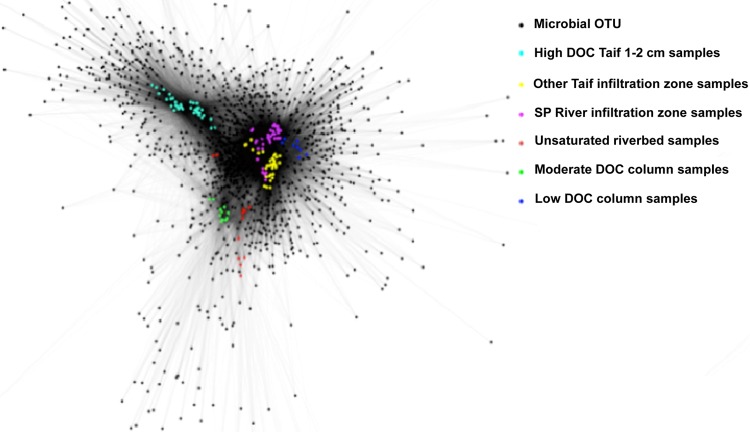
16S rRNA gene pyrosequences were primarily processed using the Quantitative Insights Into Microbial Ecology (QIIME; version 1.3.0) pipeline (12) with default settings combined with RDP's Pyrosequencing Pipeline (http://pyro.cme.msu.edu/). The minimum acceptable length was set to 200 bp. Only those sequences with a quality score of no less than 25, no ambiguous bases, as well as no mismatches in the primer sequence were retained for the following analyses (12). A total of 1,477,291 sequences were obtained from a total of 136 individual samples. In total, 81,925 operational taxonomic units (OTUs) were obtained for all these sequences. The Cytoscape program (version 2.6.0) (52) was utilized to generate Fig. 1, illustrating the OTUs shared by all samples of this study. Only OTUs that harbored more than 200 sequences in all samples were selected for this analysis. The number of sequences assigned to each sample ranged from approximately 2,830 to 59,000. Beta diversity, which compares the species diversity between samples, was evaluated using a weighted UniFrac distance matrix and visualized with principal coordinate analysis (PCoA) in QIIME. The raw pyrosequencing data of this study were deposited in the NCBI Sequence Read Archive with the accession number SRA048577.
Fig 1.
Microbial OTU network generated by Cytoscape (version 2.6.0) (52), depicting OTUs shared among all samples of this study. Only OTUs that harbored in total more than 200 sequences in all samples were selected. Each circle represents an OTU, and each colored circle represents a sample. The lines represent the OTUs present in a particular sample. High-DOC Taif 1–2 cm samples, shallow-infiltration-zone samples from the Taif River exposed to high DOC concentrations (>6 mg/liter); Other Taif infiltration zone samples, all infiltration zone samples from the Taif River except high-DOC Taif 1- to 2-cm samples; SP River infiltration zone samples, all infiltration zone samples from the South Platte River; Unsaturated riverbed samples, unsaturated riverbed samples from the Taif and South Platte Rivers.
qPCR.
A CFX96 Touch real-time PCR system (Bio-Rad) and associated software were used to quantify total Bacteria, Archaea, and fungi. The primers used for the quantification are listed in Table S1 in the supplemental material. The quantitative real-time PCR (qPCR) was performed in a 50-μl final reaction volume according to vendor instructions (iQ SYBR green Supermix; Bio-Rad). PCR amplifications were carried out as follows: for Bacteria and Archaea, 95°C for 3 min, followed by 40 cycles of 95°C for 15 s and 55°C for 30 s with a final melting curve period of 50 to 95°C with a heating increment rate of 0.5°C per 5 s and a continuous fluorescence measurement; for fungi, 95°C for 3 min, followed by 40 cycles of 95°C for 30 s and 55°C for 45 s with the same final melting curve period described above. Published procedures (37) coupled to primers listed in Table S1 in the supplemental material were used to generate standard curves from the 16S rRNA gene fragment of Bacteria and Archaea and the target gene fragment of fungi using serial dilutions (102 to 109 copies per microliter) of the plasmids. All qPCRs were performed in triplicate.
Statistics.
Statistical analyses were performed using the SPSS version 16.0 release (IBM Corporation). Canonical correspondence analysis was performed using the PAST program (25).
RESULTS
Water and sediment properties in field- and laboratory-scale MAR systems.
Water and sediment properties for the Taif and South Platte River cross sections are summarized in Table 1. Sampling locations along the Taif River were selected to represent environmental conditions upstream and downstream of municipal water discharge where DOC concentrations increased 2- to 3-fold as a result of the discharge (Table 1; see Fig. S1a in the supplemental material). To complement the field data, moderate-DOC columns containing Taif sediments were exposed to synthetic feed water. The feed solution provided a DOC concentration of 5.0 mg/liter (approximately 90% biodegradable DOC), with the majority consumed in the first 30 cm of travel (Table 2). In contrast, low-DOC columns were fed with nanofiltration permeated material (Table 2; average, 0.6 mg/liter), where approximately 10% of the DOC was removed during the 4-meter residence (∼25 days). This suggests that the nanofiltration permeated material contained small amounts of biodegradable DOC.
Table 1.
Physicochemical properties of water and sediment samples taken from Taif River, Saudi Arabia, and South Platte River, Colorado, in 2011
| Property | Value for the following sampling time (day/mo/yr), sampling site: |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 23/2/2011,a CS2 | 24/4/2011a |
25/5/2011a |
20/6/2011a |
23/1/2011,b CS | 14/5/2011,b CS | |||||||||
| CS1 | CS2 | Discharge pipe | CS3 | CS4 | CS2 | Discharge pipe | CS4 | Discharge pipe | CS3 | CS4 | ||||
| Water quality parameter | ||||||||||||||
| pH | 8.1 | 8.2 | 8.0 | 7.6 | 8.1 | 7.7 | 6.4 | 7.6 | 5.7 | 8.4 | 8.4 | 8.1 | 7.49 | 7.28 |
| Conductivity (μS/cm) | 1,329 | 964 | 1,096 | 813 | 1,220 | 914 | 1,296 | 1,305 | 1,044 | 1,348 | 2,120 | 1,227 | 875 | 746 |
| Temp (°C) | 26.3 | 24.6 | 26.7 | 27.8 | 27.9 | 29.5 | 32.0 | 29.0 | 27.0 | 27.5 | 27.4 | 26.4 | 8.9 | 16.0 |
| NO3− as N concn (mg/liter) | 11.0 | 12.9 | 1.0 | 9.6 | 2.6 | 12.8 | 11.2 | 2.8 | 9.3 | 6.4 | 8.0 | 4.8 | ||
| UV at 254 nm (cm−1) | 0.04 | 0.05 | 0.1 | 0.08 | 0.1 | 0.08 | 0.1 | 0.2 | 0.1 | 0.2 | 0.2 | 0.14 | 0.12 | |
| DOC concn (mg/liter) | 0.9 | 1.8 | 2.1 | 8.2 | 3.9 | 6.0 | 2.8 | 5.5 | 5.7 | 4.7 | 6.2 | 6.3 | 8.78 | 6.63 |
| SUVAc (liters mg−1 m−1) | 2.2 | 2.4 | 1.8 | 2.1 | 2.0 | 2.8 | 1.9 | 2.9 | 2.6 | 3.0 | 2.4 | 1.57 | 1.78 | |
| Sediment parameter, pH sediment (1:2.5 DI water) | 9.5 | 9.4 | 8.8 | 8.1 | ||||||||||
Samples from the Taif River, Saudi Arabia.
Samples from the South Platte River, Colorado.
SUVA, specific UV absorbance, which is UV absorption at 254 nm (UV254; measured in m−1) divided by the dissolved organic carbon (measured in mg/liter).
Table 2.
Physicochemical properties of water samples taken from moderate- and low-DOC columns in Saudi Arabia and Colorado, respectively
| Sampling site | Concn (mg/liter) |
|
|---|---|---|
| NO3− as N | DOC | |
| Moderate-DOC column | ||
| Feed | 2.7 ± 0.2 | 5.0 ± 0.1 |
| 30 cm | 0.9 ± 0.1 | |
| 60 cm | 3.0 ± 0.2 | 0.6 ± 0.1 |
| Δa | 0.3 | −4.4 |
| Low-DOC column | ||
| Feed | 2.7 ± 0.0 | 0.6 ± 0.1 |
| 1 m | 3.0 ± 0.1 | 0.5 ± 0.1 |
| 4 m | 3.0 ± 0.1 | 0.5 ± 0.1 |
| Δb | 0.3 | −0.1 |
The variance of the values between influent and effluent (60 cm).
The variance of the values between influent and effluent (4 m).
Microbial biomass in field- and laboratory-scale MAR systems.
The 16S rRNA gene copy numbers of Bacteria and Archaea as well as the target gene copy number of fungi are listed in Tables 3 and 4. Generally, the 16S rRNA gene copy numbers of Bacteria observed in samples from the shallow infiltration zone (average, 2.1E+09 per gram) were higher than those observed in samples from the intermediate infiltration zone (average, 2.7E+08 per gram). Moreover, sediments exposed to higher DOC concentrations proximal to and downstream of wastewater discharge (CS3 and CS4) contained significantly (P < 0.01) higher 16S rRNA gene copy numbers of Bacteria than those from the upstream sites (CS1 and CS2) (Table 3). A similar spatial trend where Bacteria gene copy number decreased with depth was observed in the laboratory-scale moderate-DOC-sediment columns (Table 4). In contrast, the low-DOC-sediment column system did not exhibit a significant change in Bacteria copy number with depth. Analogous trends for both Archaea and fungi were also observed in sediment column samples (Table 4). Hence, it appears that for both the field- and laboratory-scale MAR systems described, total bacterial, archaeal, and fungal copy numbers can be positively correlated to DOC concentration.
Table 3.
Range of 16S rRNA gene (total Bacteria and Archaea) or target gene (fungi) copy numbers determined by qPCR and Shannon diversity indexes in samples from the two rivers
| Sampling site | DOCa concn (mg/liter) | 16S rRNA gene copy no./g of sediment |
Target gene copy no./g of sediment for fungi | Shannon diversity index | |
|---|---|---|---|---|---|
| Bacteria | Archaea | ||||
| Taif River | |||||
| Infiltration zone, 1–2 cm | |||||
| CS1 | 1.8 | 3.7E+08–5.1E+08 | 8.6E+07–1.4E+08 | 8.3E+06–8.6E+06 | 8.2–9.3 |
| CS2 | 0.9–2.8 | 9.9E+07–5.0E+08 | 3.4E+07–1.0E+08 | 8.6E+05–1.4E+07 | 8.9–9.5 |
| WPb | 4.7–8.2 | 2.9E+08–1.2E+10 | 5.3E+06–2.6E+07 | 2.3E+05–1.2E+07 | 6.5–8.3 |
| CS3 | 3.9–6.2 | 2.0E+09–2.3E+09 | 1.8E+06–2.0E+06 | 7.3E+06–2.2E+07 | 7.7–9.0 |
| CS4 | 5.7–6.3 | 7.2E+08–2.1E+09 | 1.2E+05–8.3E+05 | 2.8E+05–6.7E+06 | 7.3–8.2 |
| Infiltration zone, 10–50 cm | |||||
| CS1 | 6.2E+07–1.7E+08 | 1.5E+07–3.4E+07 | 1.0E+05–2.3E+05 | 9.3–9.7 | |
| CS4 | 5.8E+08 | 8.1E+07 | 1.1E+07 | 9.1–9.3 | |
| Unsaturated | |||||
| CS1 | 3.5E+08–5.9E+08 | 9.9E+06–5.0E+07 | 2.7E+06–5.4E+06 | 7.0–9.1 | |
| CS2 | 3.9E+07 | NAc | 3.8E+05 | 7.2–7.5 | |
| CS3 | 4.4E+05–3.4E+10 | 1.7E+08–2.8E+08 | NA–4.6E+08 | 6.4–7.8 | |
| CS4 | 1.4E+08 | 1.3E+07 | 9.3E+05 | 8.6 | |
| South Platte River | |||||
| Infiltration zone, 1–2 cm | 5.2–6.7 | 2.9E+08–5.7E+08 | 4.8E+06–5.1E+06 | 3.6E+06–8.5E+06 | 8.8–8.9 |
| Infiltration zone, 10–50 cm | 7.9E+06–2.1E+08 | 2.2E+05–2.9E+07 | 3.9E+04–1.3E+07 | 8.5–9.9 | |
| Unsaturated | 7.0E+08–1.6E+10 | 3.6E+07–1.6E+09 | 1.6E+07–5.1E+08 | 8.4–9.2 | |
The DOC concentrations for water samples listed in Table 1 correspond to shallow-infiltration-zone (1- to 2-cm) sediments. DOC concentrations for proximal intermediate-infiltration-zone (10- to 50-cm) and unsaturated samples were not determined in this study.
WP, wastewater discharge pipeline.
NA, not available.
Table 4.
16S rRNA gene (total Bacteria and Archaea) and target gene (fungi) copy numbers determined by qPCR and Shannon diversity indexes in two saturated sediment columns
| Sampling site | 16S rRNA gene copy no./g of sediment |
Target gene copy no./g of sediment for fungi | Shannon diversity index | |
|---|---|---|---|---|
| Bacteria | Archaea | |||
| Moderate-DOC column | ||||
| 1 cm | 5.0E+08–1.30E+09 | 2.5E+06–5.30E+06 | 2.9E+07–1.50E+08 | 5.9 |
| 15 cm | 3.8E+07–4.50E+07 | 4.80E+05 | 2.2E+06–6.60E+06 | 7.7 |
| 30 cm | 1.4E+07–2.00E+07 | 1.60E+05 | 5.9E+05–6.10E+05 | 7.9 |
| 45 cm | 9.6E+06–1.70E+07 | 1.30E+05 | 1.6E+05–3.30E+05 | 8.1 |
| 60 cm | 4.6E+06–5.70E+07 | 1.80E+05 | 1.6E+05–1.30E+06 | 8.2 |
| Low-DOC column | ||||
| 0 m | 1.10E+09 | 3.50E+06 | 2.60E+07 | 7.6 |
| 1 m | 7.90E+08 | 3.90E+06 | 7.00E+07 | 7.8 |
| 2 m | 2.50E+08 | 2.90E+06 | 3.60E+06 | 7.9 |
| 3 m | 3.50E+08 | 1.40E+06 | 4.40E+06 | 7.7 |
Bacterial community structure in field-scale MAR systems.
There was a strong similarity between samples collected across and within the field-scale MAR sites. With respect to specific sampling locations, no significant differences in phylum-level distribution existed in the shallow infiltration zone for the five sampling locations along the Taif River (Kendall's coefficient of concordance [W] matched-pair test, P = 0.36) or for the cross section of the South Platte River (P = 0.40). Furthermore, the relative abundance of bacterial phyla between samples collected at different times of the year at the same location was not distinctly different (Kendall's W matched-pair test, P > 0.36 for all comparisons). Strikingly, no significant differences in relative bacterial abundance at the phylum level were observed between samples derived from the Taif versus South Platte Rivers in either the shallow infiltration zone (Kendall's W matched-pair test, P = 0.45) or intermediate infiltration zone (P = 0.16). This trend extended to the class level for the shallow infiltration zone (P = 0.25) but not the intermediate infiltration zone. As a result of these trends, further interpretation focuses on samples grouped with respect to location, time, and region.
The relative abundance of sequences belonging to each bacterial phylum in the three conserved environments, (i) shallow infiltration zone, (ii) intermediate infiltration zone, and (iii) unsaturated riverbeds, of the two rivers are summarized in Table 5. For shallow-infiltration-zone samples, the phylum Proteobacteria was the most abundant. It was followed by Bacteroidetes mainly, including the classes Sphingobacteria (7.6%) and Flavobacteria (2.5%), Acidobacteria, Actinobacteria, Firmicutes, including Clostridia (1.6%), Planctomycetes, Chloroflexi, and Cyanobacteria.
Table 5.
Relative abundance of major bacterial groups in field samples combined from Taif and South Platte River riverbeds and two laboratory column systemsa
| Bacterial group | Relative abundance (%) |
||||||
|---|---|---|---|---|---|---|---|
| Infiltration zone |
Unsaturated | Low-DOC column | Moderate-DOC column |
||||
| 1–2 cm | 10–50 cm | 1 cm | 15 cm | 30–60 cm | |||
| Proteobacteria | 50.9 | 40.8 | 22.0 | 36.5 | 75.7 | 33.7 | 33.9 |
| Alphaproteobacteria | 12.1 | 11.2 | 9.7 | 7.9 | 7.8 | 7.5 | 10.2 |
| Betaproteobacteria | 18.5 | 14.5 | 3.9 | 8.5 | 59.6 | 15.7 | 13.8 |
| Gammaproteobacteria | 7.8 | 8.3 | 5.1 | 7.5 | 6.6 | 3.7 | 2.8 |
| Deltaproteobacteria | 7.8 | 3.5 | 2.1 | 4.0 | 0.2 | 6.3 | 6.1 |
| Other | 3.8 | 3.3 | 1.1 | 8.5 | 1.6 | 0.5 | 1.0 |
| Acidobacteria | 3.2 | 6.9 | 3.6 | 9.2 | 0.1 | 3.2 | 4.2 |
| Actinobacteria | 2.5 | 6.3 | 17.4 | 1.5 | 5.7 | 4.1 | 5.3 |
| Bacteroidetes | 18.8 | 10.8 | 9.6 | 5.2 | 12.4 | 12.6 | 11.5 |
| Chloroflexi | 4.1 | 1.4 | 0.7 | 0.2 | 0 | 0.3 | 0.4 |
| Cyanobacteria | 1.5 | 0.4 | 0.2 | 1.5 | 0 | 0 | 0 |
| Firmicutes | 2.2 | 4.6 | 27.4 | 2.9 | 2.8 | 26.8 | 20.1 |
| Gemmatimonadetes | 0.3 | 0.7 | 0.8 | 0.5 | 0.2 | 1.1 | 2.3 |
| Nitrospira | 0.7 | 2.9 | 0.6 | 6.5 | 0 | 0.5 | 1.0 |
| Planctomycetes | 2.7 | 4.1 | 1.4 | 3.5 | 0.1 | 0.6 | 2.1 |
| Verrucomicrobia | 1.2 | 2.1 | 1.4 | 1.3 | 0.2 | 5.5 | 4.1 |
| Other | 11.0 | 16.7 | 10.1 | 26.1 | 2.7 | 11.3 | 14.6 |
The two laboratory column systems included low-DOC-sediment columns regardless of depth and moderate-DOC-sediment columns at a 1-cm depth, a 15-cm depth, and collectively binned for 30- to 60-cm depths.
In the intermediate infiltration zone (Table 5), the phylum Proteobacteria was still the most abundant group. The other dominant phyla included Bacteroidetes, mainly consisting of Sphingobacteria (7.2%), Acidobacteria, Actinobacteria, Firmicutes consisting of Bacilli (3.0%) and Clostridia (1.4%), Planctomycetes, and Nitrospira.
For proximal unsaturated riverbed samples (Table 5), Firmicutes shifted to the most abundant phylum, mainly including Bacilli (25.4% in average of both rivers), with a corresponding increase in the abundance of Actinobacteria, including Actinobacteria (17.4%), while Proteobacteria decreased to 22.0% of the total abundance. The other abundant bacterial groups included Bacteroidetes, Acidobacteria, and Planctomycetes.
The relative abundance of bacterial phyla was not significantly different between the shallow and intermediate infiltration zones (Wilcoxon matched-pair test, P = 0.5). However, the relative abundance of bacteria in the unsaturated riverbed samples differed significantly from that of bacteria derived from the intermediate-infiltration-zone samples on the phylum level (Wilcoxon matched-pair test, P = 0.03) and from both the shallow- and intermediate-infiltration-zone samples on the class level (Wilcoxon matched-pair test, P = 0.05 and 0.001, respectively).
When comparing bacterial community structure at the genus level, differences in relative abundance between samples became much more significant (Kendall's W matched-pair test, P < 0.05). However, the same types of samples still exhibited similar microbial community structure on the basis of the relative abundance of genera. Importantly, similarities were observed between the intermediate infiltration zones in the geographically distanced rivers and between those from the shallow infiltration zones with similar DOC concentrations, such as Taif River CS1 and CS2, Taif River CS3 and CS4, as well as the cross section of the South Platte River (P > 0.1 for Kendall's W matched-pair test for most comparisons).
Overall, the shallow infiltration zones of CS1, CS2, CS3, and CS4 of the Taif River as well as the cross section of the South Platte mainly included (in order of total abundance) the bacterial genera Thauera (3.4%), Terrimonas (1.4%), Pseudoxanthomonas (1.4%), Nocardioides (1.0%), Sphingomonas (0.9%), Nitrospira (0.9%), Porphyrobacter (0.8%), and Rhodobacter (0.7%). Most of these bacterial genera are typical aerobic residents in sediments, soil, and freshwater environments and are known for their ability to degrade chemical pollutants (31, 51).
The shallow infiltration zones under comparably higher DOC conditions at the wastewater discharge point of the Taif River also contained many bacterial genera typical of sediment and soil environments, such as Pseudomonas (1.6%), Thauera (1.5%), Geobacter (1.5%), Thiobacillus (1.3%), Cloacibacterium (0.9%), Aquabacterium (0.8%), and Rhodobacter (0.7%). Furthermore, they also harbored abundant diverse anaerobic sulfate-reducing bacteria, Desulfuromonas (2.5%), Desulfobulbus (1.0%), Desulfobacter (0.9%), Desulfococcus (0.4%), and Desulfocapsa (0.4%), and the sulfur-oxidizing genera Sulfurimonas (0.4%), and Sulfuricurvum (0.3%). The results suggest an increased proportion of anaerobic conditions in these environments with comparably higher DOC conditions, which is likely caused by the mineralization of DOC by indigenous bacteria.
For intermediate infiltration zones of both rivers, the major bacterial genera included Nitrospira (2.9%), Hyphomicrobium (0.9%), Sphingomonas (0.6%), Terrimonas (0.5%), Flavobacterium (0.4%), Methylibium (0.4%), Gemmatimonas (0.4%), and Pseudomonas (0.2%). Similar to the shallow infiltration zones, most of these genera are prevalent in sediment and soil environments.
Finally, bacterial genera abundant in the unsaturated riverbed samples were composed of Arthrobacter (0.9%), Kocuria (0.9%), Halomonas (0.7%), Truepera (0.5%), Gemmatimonas (0.5%), Salinimicrobium (0.4%), and Bacillus (0.3%). Several genera among these are recognized to have notable survival or extremophile-type characteristics consistent with a desiccating environment. Halomonas spp. and Salinimicrobium spp. are adapted to halophilic environments, Truepera spp. are resistant to radiation (3), and Bacillus sp. strains are well-known sporeformers.
Bacterial community structure in laboratory-scale MAR systems.
Further insight into the bacterial community structure as a function of DOC and sampling depth as well as across lab and field scales was explored using sediment column systems simulating MAR infiltration zones. There was no significant difference in the relative abundance of bacterial phyla between sediments from different depths in low-DOC columns (Kendall's W matched-pair test, P = 0.40). The phylum Proteobacteria was the most abundant (Table 5). Furthermore, no significantly different relative abundance of bacterial genera was observed among the different depths of low-DOC columns (Kendall's W matched-pair test, P = 0.487). The major bacterial genera were Nitrospira (6.5%), Meiothermus (0.6%), Aquicella (0.6%), Mycobacterium (0.5%), and Gemmatimonas (0.5%). As described in the previous section, most of these genera were also observed in intermediate infiltration zones of both rivers, strengthening the validity of laboratory-scale simulations of these environmental systems.
In contrast to these low-DOC columns, the relative abundance of bacterial phyla was significantly different among sediment samples taken from certain depths for the moderate-DOC sediment columns (Kendall's W matched-pair test, P < 0.001). This was not an artifact of sampling technique, as harvests from the same depth of duplicate sampling columns contained similar distributions (Kendall's W matched-pair test, P > 0.15). Specifically, the sediment samples taken from a depth of 1 cm contained a significantly different bacterial phylum distribution when contrasted with those derived from 15 cm (Kendall's W matched-pair test, P = 0.001); these were in turn significantly different from those taken from 30 cm (Kendall's W matched-pair test, P = 0.017). At subsequent depths (30 cm, 45 cm, and 60 cm), the bacterial phylum distributions could be grouped because they were not distinctly different from one another (Kendall's W matched-pair test, P = 0.517).
As illustrated in Table 5, samples collected from the moderate-DOC columns at a depth of 1 cm contained 75.7% Proteobacteria, with the Betaproteobacteria class contributing to 59.6% of the total sequences. At 15 cm, the relative abundance of Proteobacteria decreased to 33.7%, with the Betaproteobacteria contributing 15.7%. At the same time, the relative abundance of Firmicutes rose to 26.8%. For samples taken from the depth of 30 to 60 cm, the average percentage of Proteobacteria was 33.9% (13.8% Betaproteobacteria) and that of Firmicutes was 20.1%.
Shared OTUs among field- and laboratory-scale MAR system samples.
The sharing of OTUs among field and column samples appeared to be determined by sampling region and DOC, as illustrated in Fig. 1. OTUs shared by environmentally derived shallow-infiltration-zone samples exposed to high DOC concentrations (>6 mg/liter; samples mainly taken from the wastewater discharge point on the Taif River) were distinct from the OTUs in other samples. Notably, they were distinguished from the low-DOC-sediment-column samples as well as Taif and South Platte River infiltration zone samples exposed to lower DOC concentrations (<6 mg/liter). This result was generally accordant with the bacterial genera present in shallow- and intermediate-infiltration-zone samples of both rivers. On the other hand, OTUs in the unsaturated riverbed samples from both the Taif and South Platte Rivers were distributed in a dispersed manner, indicating that these samples did not share many OTUs compared to other clustered samples. The sharing of OTUs with the moderate-DOC-sediment column samples is most likely explained by the fact that the source material from the unsaturated riverbed was used to establish the moderate-DOC-sediment-column experiments.
Microbial community diversity in field- and laboratory-scale MAR systems.
Microbial community diversity was assessed for all samples in this study by computing the Shannon diversity index (Tables 3 and 4). For shallow-infiltration-zone samples, the diversity index generally decreased from 8.9 ± 0.7 in upstream samples (CS1 and CS2) to 8.2 ± 0.8 in downstream samples (CS3 and CS4) and 7.4 ± 0.9 at the wastewater discharge. This indicates a clear negative correlation between diversity and DOC concentration (Table 3). With respect to specific sampling locations, microbial community diversity clearly increased from the shallow-infiltration-zone to the intermediate-infiltration-zone samples, also suggesting a negative correlation to both DOC concentration and total microbial 16S rRNA gene copy number.
This phenomenon was further confirmed in sediment column samples. The diversity index increased significantly from 5.9 at a depth of 1 cm to 8.2 at a depth of 60 cm for moderate-DOC-sediment-column samples (Table 4), while DOC concentration and microbial density decreased along the same flow path. Samples from the low-DOC-column system exhibited little to no increase in microbial diversity, with indexes from 7.6 at 0 m to 7.7 at 3 m. These index values were similar to those of the more DOC-depleted field environments described above.
Correlation between microbial community structure and environmental factors.
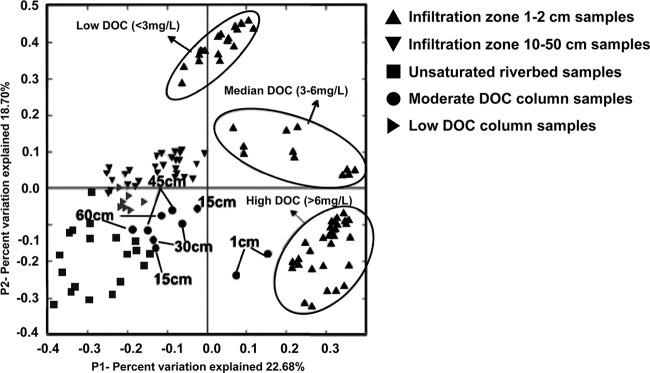
PCoA analysis was adopted to analyze relationships between microbial communities from sediments derived from field- and laboratory-scale systems (Fig. 2). General dissimilarities were represented by the scattered pattern of data points. The analysis revealed different microbial assemblages, represented by distinct clusters correlating to the environmentally derived shallow and intermediate infiltration zones and the unsaturated riverbed samples. Shallow-infiltration-zone samples from the Taif and South Platte Rivers could be further grouped into three clusters corresponding to different DOC concentration ranges (low [<3 mg/liter], moderate [3 to 6 mg/liter], and high [>6 mg/liter]).
Fig 2.
PCoA (P1 versus P2) of microbial communities in all field- and laboratory-scale samples elucidated in this study. Microbial communities can be grouped into clusters corresponding to different DOC concentration ranges for field materials (circled) and by depth in laboratory systems.
Consideration of laboratory-scale column sediment samples in the same analysis revealed that microbial communities in moderate-DOC columns could be classified according to depth and corresponding declining DOC concentration (Table 2 and Fig. 2). Microbial communities in samples derived from the top centimeter were closely related to those in high-DOC samples from the shallow infiltration zones of both rivers. In contrast, microbial communities in samples from deeper locations (15, 30, 45, and 60 cm) in the laboratory columns were closely related to those in the intermediate infiltration zones of environmentally derived river samples. This trend further extended to sediments derived from the low-DOC laboratory columns, where the microbial communities in this cluster were closely related to those in the intermediate-infiltration-zone samples and deeper samples from the moderate-DOC columns (Fig. 2).
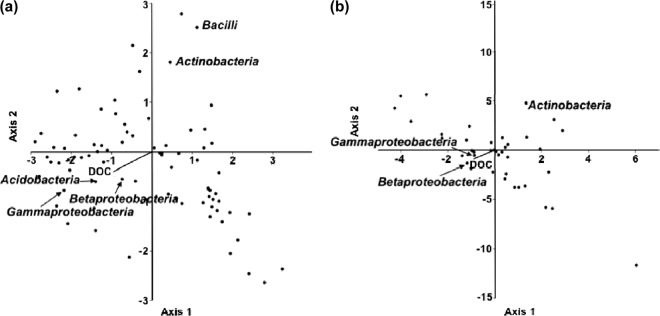
It is noteworthy that the distribution trend relating to declining DOC concentration for column samples was similar to that for shallow-infiltration-zone samples from field sites. This suggests that DOC concentration exerts a similar influence on the microbial community in both field- and laboratory-scale MAR systems. Canonical correspondence analysis was performed to further elucidate this association (Fig. 3). This analysis revealed that the abundance of Betaproteobacteria and Gammaproteobacteria was positively associated with DOC concentration.
Fig 3.
Canonical correspondence analysis (CCA) of microbial groups reveals that Betaproteobacteria and Gammaproteobacteria positively correlate with DOC concentration in field samples derived from the Taif River (a) and South Platte River (b).
DISCUSSION
Both field (Taif and South Platte River) and laboratory (low- and moderate-DOC-column) MAR sediments were selected to investigate the microbial community of the infiltration zone in MAR systems. The results of this study suggest that the behavior of the microbial community in sediment column samples from simulated MAR systems under different DOC concentrations was highly consistent with that of communities derived from field settings of the Taif and South Platte Rivers, confirming that controlled sediment columns can be used to reproduce and further explore the microbial characteristics and perhaps functions representative of field-scale MAR infiltration zones. Our results strengthen a prior and intuitive observation that the total biomass in MAR systems is positively correlated to the DOC concentration (44) and that the majority of DOC and nutrients in MAR systems is removed during initial percolation through shallow sediments or soil, where concentrations decline sharply with depth (2, 11, 33). Accordingly, microbial biomass can be modeled as a power function of depth, where the top layer (0 to 20 cm) of the sediment typically harbors the highest density of microorganisms (27, 44). The more oligotrophic vadose zone, which can range from a few meters to several hundreds of meters, represents a different regime where recalcitrant organic compounds become more prominent (27, 49).
Parallels can be drawn to analogous studies that characterize microbial abundance, community structure, metabolic status, and functional microbial groups and their response to toxic chemicals in the shallow infiltration zone of sediments (1, 9, 20, 26, 58). However, prior to the current study, little was known regarding seasonal or spatial variations in microbial community structure or physiochemical parameters shaping the microbial community specifically in MAR systems. Janssen (29) reported that worldwide soil and sediment microbial communities are dominated by nine bacterial phyla: Proteobacteria (mainly the Alpha-, Beta-, and Deltaproteobacteria subdivisions), Actinobacteria, Acidobacteria, Chloroflexi, Bacteroidetes, Firmicutes, Planctomycetes, Verrucomicrobia, and Gemmatimonadetes. In accordance, our study supports this generalization, and Proteobacteria, Bacteroidetes, Actinobacteria, and Firmicutes appear to be the dominant phyla in samples collected from both field- and laboratory-scale MAR systems. While previous research has suggested that biogeographical differences exist for microbial communities and taxon-area relationships have been observed for microorganisms (39), our results demonstrate that microbial communities exhibit a remarkably stable structure on the phylum level in analogous sediment samples derived from two distinct regions. This suggests that analogous biogeochemical ecosystems can harbor similar phylogenetic communities, despite spatial distance; notably dominant OTUs, representative of species-level comparisons, were typically shared by the same types of samples rather than by region (Fig. 1 and 2). Thus, it appears that in the case of MAR and quite possibly other riparian systems, dominant OTUs present in sediment ecosystems might be globally widespread.
Furthermore, no significant changes in the microbial community structure in the same sampling location of both the Taif and South Platte Rivers were observed in different seasons (winter, spring and summer), indicating that surface temperature or other seasonal perturbations did not play an important role in shaping microbial community structure in shallow sediments. Analogous findings concerning seasonal effects in lake sediment ecosystems have recently been reported (55). Although season did not appear to exert a strong pressure, a variety of other physicochemical conditions, such as pH, temperature, water content, oxygen concentration, DOC concentration, nutrient availability, and depth, can still influence the microbial community structure in soil and sediment ecosystems (10, 60). Our specific findings for MAR indicate that water content (saturated and unsaturated) and DOC concentration might be two major determinants of microbial community structure. Since microorganisms in soil or sediment are generally adsorbed to solid matrices or form biofilms (6, 17), water content could directly affect the diffusion and availability of substrates as well as influence the osmotic status of microbial cells (30, 32, 43).
Canonical correlation analysis is a way of measuring the linear relationship between two sets of multidimensional variables. It seeks a pair of linear transformations, one for each of the sets of variables, such that when the set of variables is transformed, the corresponding coordinates are maximally correlated. In this study, the relative abundance of Betaproteobacteria and Gammaproteobacteria was closely correlated to DOC concentration by canonical correlation analysis; thus, they are the major groups whose abundance was positively linked to DOC concentration in saturated samples (Fig. 3). This result is consistent with previous reports in analogous environmental systems. Wakelin et al. (56) reported that Gammaproteobacteria were correlated with DOC concentrations in stream sediments. Langenheder and Prosser (34) also demonstrated that Betaproteobacteria and Gammaproteobacteria were the major degraders of organic pollutants in sediments, and these classes are well-known for their ability to degrade diverse organic compounds that include some persistent organic chemicals (40, 57). Environments with proximal unsaturated conditions exhibited the emergence of Bacilli and Actinobacteria. These Gram-positive and spore-forming organisms are more likely to survive in dry and desiccating environments (7) and have also been shown to have the potential to degrade recalcitrant organic pollutants problematic to water reuse, such as nitrosamines (50).
In concert with selection for Betaproteobacteria and Gammaproteobacteria, overall microbial community diversity was negatively correlated to DOC concentration (Tables 3 and 4). A possible explanation can be grounded in the different compositions and availability of organics. The laboratory- and field-scale systems with higher DOC concentrations that we studied contained biodegradable organic carbon that was more easily assimilated by microorganisms, as evidenced by percent removal (Tables 1 and 2). As a consequence, these systems favor microbial groups that dominate the community due to their high metabolism and assimilation rates. After the exhaustion of this more bioavailable organic carbon, DOC concentrations drop, while the portion of refractory and less bioavailable DOC increases (5). From here, it stands to reason that low DOC concentrations in the same ecosystems allow the inhabitance of a more diverse microbial community in which numerous microbial species degrade the more recalcitrant and varied refractory organic substrates. It is interesting that the diversity index for low-DOC sediment columns was comparable to that of the tail end of the moderate-DOC column system, where DOC concentration and recalcitrance were analogous. While not yet confirmed in the field, this suggests that a shift toward increased microbial diversity will be observed in the more oligotrophic depths of full-scale MAR systems.
In summary, this study helps to elucidate the microbial community composition and diversity found in the infiltration zone of MAR systems across a variety of spatial and temporal scales. Conservation of microbial communities suggests that similar bioattenuation attributes may be achieved regardless of geographical location by manipulating the concentration of DOC in feed water. It should be noted that this conclusion should be restricted to waters that are buffered by a carbonate/bicarbonate buffer system and generally dominated by readily degradable organic compounds, as tested in this study. Anyway, these natural water treatment systems play an important role in contaminant attenuation and demonstrate particular promise for low energy and economic requirements for the remediation of recalcitrant trace organic chemicals (28, 50). Our findings provide a better understanding of microbiological profiles in MAR systems and establish a foundation for understanding, predicting, and optimizing performance variables at the laboratory scale.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by discretionary investigator funds (to J.E.D. and P.E.S.) and user facilities (to S.A. and M.A.) at King Abdullah University of Science and Technology (KAUST). The material presented is also based in part upon work supported by the National Science Foundation under grant no. CBET-1055396 (to J.O.S.) and Cooperative Agreement EEC-1028968 (to J.E.D. and J.O.S.).
Footnotes
Published ahead of print 13 July 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Aelion CM, Swindoll CM, Pfaender FK. 1987. Adaptation to and biodegradation of xenobiotic compounds by microbial communities from a pristine aquifer. Appl. Environ. Microbiol. 53:2212–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ajwa HA, Rice CW, Sotomayor D. 1998. Carbon and nitrogen mineralization in tallgrass prairie and agricultural soil profiles. Soil Sci. Soc. Am. J. 62:942–951 [Google Scholar]
- 3. Albuquerque L, et al. 2005. Truepera radiovictrix gen. nov., sp. nov., a new radiation resistant species and the proposal of Trueperaceae fam. nov. FEMS Microbiol. Lett. 247:161–169 [DOI] [PubMed] [Google Scholar]
- 4.American Public Health Association 1998. Standard methods for the examination of water and wastewater. American Public Health Association, Washington, DC [Google Scholar]
- 5. Amy G, Drewes JE. 2007. Soil aquifer treatment (SAT) as a natural and sustainable wastewater reclamation/reuse technology: fate of wastewater effluent organic matter (EfOM) and trace organic compounds. Environ. Monit. Assess. 129:19–26 [DOI] [PubMed] [Google Scholar]
- 6. Andrews JS, Rolfe SA, Huang WE, Scholes JD, Banwart SA. 2010. Biofilm formation in environmental bacteria is influenced by different macromolecules depending on genus and species. Environ. Microbiol. 12:2496–2507 [DOI] [PubMed] [Google Scholar]
- 7. Angert ER. 2005. Alternatives to binary fission in bacteria. Nat. Rev. Microbiol. 3:214–224 [DOI] [PubMed] [Google Scholar]
- 8. Benson AK, et al. 2010. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc. Natl. Acad. Sci. U. S. A. 107:18933–18938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Blume E, et al. 2002. Surface and subsurface microbial biomass, community structure and metabolic activity as a function of soil depth and season. Appl. Soil Ecol. 20:171–181 [Google Scholar]
- 10. Bossio DA, Scow KM. 1995. Impact of carbon and flooding on the metabolic diversity of microbial communities in soils. Appl. Environ. Microbiol. 61:4043–4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bundt M, Widmer F, Pesaro M, Zeyer J, Blaser P. 2001. Preferential flow paths: biological ‘hot spots' in soils. Soil Biol. Biochem. 33:729–738 [Google Scholar]
- 12. Caporaso JG, et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Caporaso JG, et al. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U. S. A. 108:4516–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reference deleted.
- 15.Reference deleted.
- 16. Dantas G, Sommer MOA, Oluwasegun RD, Church GM. 2008. Bacteria subsisting on antibiotics. Science 320:100–103 [DOI] [PubMed] [Google Scholar]
- 17. Elliott DR, et al. 2010. Dynamic changes in microbial community structure and function in phenol degrading microcosms inoculated with cells from a contaminated aquifer. FEMS Microbiol. Ecol. 71:247–259 [DOI] [PubMed] [Google Scholar]
- 18. Fierer N, Hamady M, Lauber CL, Knight R. 2008. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc. Natl. Acad. Sci. U. S. A. 105:17994–17999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fierer N, Jackson RB. 2006. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. U. S. A. 103:626–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Foulquier A, Mermillod-Blondin F, Malard F, Gibert J. 2011. Response of sediment biofilm to increased dissolved organic carbon supply in groundwater artificially recharged with stormwater. J. Soils Sediments 11:382–393 [Google Scholar]
- 21. Galand PE, Casamayor EO, Kirchman DL, Lovejoy C. 2009. Ecology of the rare microbial biosphere of the Arctic Ocean. Proc. Natl. Acad. Sci. U. S. A. 106:22427–22432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Grunheid S, Amy G, Jekel M. 2005. Removal of bulk dissolved organic carbon (DOC) and trace organic compounds by bank filtration and artificial recharge. Water Res. 39:3219–3228 [DOI] [PubMed] [Google Scholar]
- 23. Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. 2008. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat. Methods 5:235–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hamady M, Knight R. 2009. Microbial community profiling for human microbiome projects: tools, techniques, and challenges. Genome Res. 19:1141–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hammer Ø Harper DAT, Ryan PD. 2001. Past: paleontological statistics software package for education and data analysis. Palaeont. Elec. 4:e1–e9 [Google Scholar]
- 26. Hansel CM, Fendorf S, Jardine PM, Francis CA. 2008. Changes in bacterial and archaeal community structure and functional diversity along a geochemically variable soil profile. Appl. Environ. Microbiol. 74:1620–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Holden PA, Fierer N. 2005. Microbial processes in the vadose zone. Vadose Zone J. 4:1–21 [Google Scholar]
- 28. Hoppe-Jones C, Oldham G, Drewes JE. 2010. Attenuation of total organic carbon and unregulated trace organic chemicals in U.S. riverbank filtration systems. Water Res. 44:4643–4659 [DOI] [PubMed] [Google Scholar]
- 29. Janssen PH. 2006. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 72:1719–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kieft TL, et al. 1993. Microbial abundance and activities in relation to water potential in the vadose zones of arid and semiarid sites. Microb. Ecol. 26:59–78 [DOI] [PubMed] [Google Scholar]
- 31. Kim JM, et al. 2008. Influence of soil components on the biodegradation of benzene, toluene, ethylbenzene, and o-, m-, and p-xylenes by the newly isolated bacterium Pseudoxanthomonas spadix BD-a59. Appl. Environ. Microbiol. 74:7313–7320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Konings WN, Albers S-V, Koning S, Driessen AJM. 2002. The cell membrane plays a crucial role in survival of bacteria and archaea in extreme environments. Antonie Van Leeuwenhoek 81:61–72 [DOI] [PubMed] [Google Scholar]
- 33. Konopka A, Turco R. 1991. Biodegradation of organic compounds in vadose zone and aquifer sediments. Appl. Environ. Microbiol. 57:2260–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Langenheder S, Prosser JI. 2008. Resource availability influences the diversity of a functional group of heterotrophic soil bacteria. Environ. Microbiol. 10:2245–2256 [DOI] [PubMed] [Google Scholar]
- 35. Lauber CL, Hamady M, Knight R, Fierer N. 2009. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 75:5111–5120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lennon JT, Pfaff LE. 2005. Source and supply of terrestrial organic matter affects aquatic microbial metabolism. Aquat. Microb. Ecol. 39:107–119 [Google Scholar]
- 37. Li D, et al. 2010. Characterization of bacterial community structure in a drinking water distribution system during an occurrence of red water. Appl. Environ. Microbiol. 76:7171–7180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lozupone CA, Knight R. 2007. Global patterns in bacterial diversity. Proc. Natl. Acad. Sci. U. S. A. 104:11436–11440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Martiny JB, et al. 2006. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4:102–112 [DOI] [PubMed] [Google Scholar]
- 40. Moore ERB, et al. 2006. Nonmedical: Pseudomonas, p 646–703. In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E. (ed), The prokaryotes, 3rd ed. Springer-Verlag, New York, NY [Google Scholar]
- 41.Reference deleted.
- 42. Oppenheimer J, Eaton A, Badruzzaman M, Haghani AW, Jacangelo JG. 2011. Occurrence and suitability of sucralose as an indicator compound of wastewater loading to surface waters in urbanized regions. Water Res. 45:4019–4027 [DOI] [PubMed] [Google Scholar]
- 43. Potts M. 1994. Desiccation tolerance of prokaryotes. Microbiol. Mol. Biol. Rev. 58:755–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rauch-Williams T, Drewes JE. 2006. Using soil biomass as an indicator for the biological removal of effluent-derived organic carbon during soil infiltration. Water Res. 40:961–968 [DOI] [PubMed] [Google Scholar]
- 45. Rauch-Williams T, Hoppe-Jones C, Drewes JE. 2010. The role of organic matter in the removal of emerging trace organic contaminants during managed aquifer recharge. Water Res. 44:449–460 [DOI] [PubMed] [Google Scholar]
- 46. Ray C, et al. 2008. Riverbank filtration for drinking water supply. In Encyclopedia of hydrological sciences. John Wiley & Sons, Inc., New York, NY [Google Scholar]
- 47. Roesch LFW, et al. 2007. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 1:283–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rousk J, et al. 2010. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 4:1340–1351 [DOI] [PubMed] [Google Scholar]
- 49. Schütz K, Nagel P, Vetter W, Kandeler E, Ruess L. 2009. Flooding forested groundwater recharge areas modifies microbial communities from top soil to groundwater table. FEMS Microbiol. Ecol. 67:171–182 [DOI] [PubMed] [Google Scholar]
- 50. Sharp JO, Sales C, Alvarez-Cohen L. 2010. Functional insights towards propane-enhanced n-nitrosodimethylamine removal by two actinomycetes. Biotechnol. Bioeng. 107:924–932 [DOI] [PubMed] [Google Scholar]
- 51. Shinoda Y, et al. 2004. Aerobic and anaerobic toluene degradation by a newly isolated denitrifying bacterium, Thauera sp. strain DNT-1. Appl. Environ. Microbiol. 70:1385–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Smoot M, Ono K, Ruscheinski J, Wang P-L, Ideker T. 2011. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27:431–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reference deleted.
- 54. Torsvik V, Goksoyr J, Daae FL. 1990. High diversity in DNA of soil bacteria. Appl. Environ. Microbiol. 56:782–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tšertova N, Kisand A, Tammert H, Kisand V. 2011. Low seasonal variability in community composition of sediment bacteria in large and shallow lake. Environ. Microbiol. Rep. 3:270–277 [DOI] [PubMed] [Google Scholar]
- 56. Wakelin SA, Colloff MJ, Kookana RS. 2008. Effect of wastewater treatment plant effluent on microbial function and community structure in the sediment of a freshwater stream with variable seasonal flow. Appl. Environ. Microbiol. 74:2659–2668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Willems A, Vos PD. 2006. Comamonas, p 723–736. In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E. (ed), The prokaryotes, 3rd ed. Springer-Verlag, New York, NY [Google Scholar]
- 58. Yagi JM, Neuhauser EF, Ripp JA, Mauro DM, Madsen EL. 2010. Subsurface ecosystem resilience: long-term attenuation of subsurface contaminants supports a dynamic microbial community. ISME J. 4:131–143 [DOI] [PubMed] [Google Scholar]
- 59. Zhang H, Ziv-El M, Rittmann BE, Krajmalnik-Brown R. 2010. Effect of dechlorination and sulfate reduction on the microbial community structure in denitrifying membrane-biofilm reactors. Environ. Sci. Technol. 44:5159–5164 [DOI] [PubMed] [Google Scholar]
- 60. Zhou J, et al. 2002. Spatial and resource factors influencing high microbial diversity in soil. Appl. Environ. Microbiol. 68:326–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.