Abstract
Tumor cell-derived factors, such as interleukin 10 (IL-10), polarize macrophages toward a regulatory M2 phenotype, characterized by the expression of anti-inflammatory cytokines and protumorigenic mediators. Here we explored molecular mechanisms allowing IL-10 to upregulate the protumorigenic protein NGAL in primary human macrophages. Reporter assays of full-length or deletion constructs of the NGAL promoter provided evidence that NGAL production is STAT3 dependent, activated downstream of the IL-10–Janus kinase (Jak) axis, as well as being C/EBPβ dependent. The involvement of STAT3 and C/EBPβ was shown by chromatin immunoprecipitation (ChIP) and ChIP-Western analysis, as well as decoy oligonucleotides scavenging both STAT3 and C/EBPβ in human macrophages. Furthermore, the production of NGAL in macrophages in response to IL-10 induces cellular growth and proliferation of MCF-7 breast cancer cells. We conclude that both STAT3 and C/EBPβ are needed to elicit IL-10-mediated NGAL expression in primary human macrophages. Macrophage-secreted NGAL shapes the protumorigenic macrophage phenotype to promote growth of MCF-7 breast cancer cells. Our data point to a macrophage-dependent IL-10–STAT3–NGAL axis that might contribute to tumor progression.
INTRODUCTION
Macrophages exhibit a great heterogeneity and functional plasticity in response to microenvironmental signals (42). This is exemplified during inflammation, where they contribute to both the initiating and resolution phases. Macrophage heterogeneity has been considered crucial for the outcome of injury and highlights their pivotal role in maintaining tissue integrity. During innate immune responses, macrophages phagocytose and eliminate invading pathogens and even dead cells. Later during inflammation, macrophages contribute to healing and tissue reorganization. Macrophage phenotypes are also associated with malignancies if their activation is not properly controlled during, e.g., chronic inflammatory diseases or if they continue to support tissue vascularization and thus foster tumor growth (23, 42).
It became apparent that not only tumor cell intrinsic genetic and epigenetic changes but also the tumor microenvironment promotes tumor initiation and progression. Tumor cells produce and secrete a number of factors to create a tumor-supportive microenvironment, which contributes to growth, angiogenesis, and metastasis (25). The tumor itself thereby evades the naturally occurring immune surveillance, which protects transformed cells from the attack of their own immune defense system. In many tumors, invading macrophages are found in high numbers (31). These tumor-associated macrophages (TAM) are “educated” by cancer cells to support growth rather than to eradicate tumor cells. The presence of TAM often correlates with a poor patient prognosis, as shown for, e.g., breast, prostate, ovarian and cervical cancers (3). The functional TAM phenotype is at least in part a response to tumor-released components, such as interleukin 10 (IL-10), transforming growth factor β (TGF-β), prostanglandin E2 (PGE2), other chemokines, and tumor hypoxia (19). Signaling pathways that are not fully elucidated control the differentiation and polarization of infiltrating monocytes to TAM, which resembles an alternatively activated M2 macrophage phenotype (25). M2-polarized macrophages support proliferation and regeneration of damaged tissues, which is achieved mainly through phagocytosis of apoptotic cells and the subsequent production of anti-inflammatory chemokines and cytokines. Within the tumor, TAM promote tumor growth and metastasis by secreting growth-promoting mediators, among others IL-10, vascular endothelial growth factor (VEGF), IL-8, PGE2, or TGF-α (51).
IL-10 is an established anti-inflammatory and immunosuppressive cytokine known to promote macrophage polarization toward a tumor-supportive phenotype (reviewed in reference 38). Using a cell-based therapy approach, we previously showed that IL-10 overexpression in primary macrophages enhanced their proresolution activity in complex, inflammation-associated pathologies (15). Interestingly, IL-10-expressing macrophages also promoted the induction of the neutrophil gelatinase-associated lipocalin (NGAL). NGAL is a 25-kDa protein of the lipocalin superfamily and exerts bacteriostatic effects by capturing and depleting siderophores (14). Recent evidence suggests that NGAL acts as a growth and differentiation factor in different cell types (36). Exogenous NGAL has been shown to cause expression of genetic markers reflecting early epithelial progenitors and to support proliferation of epithelial cells (29). Conversely, NGAL induces cell death in neutrophils and lymphocytes, probably to limit inflammation, whereas nonhematopoietic cells and macrophages are resistant (7). Furthermore, we previously showed that apoptotic tumor cells activate the production and secretion of NGAL in macrophages, with the subsequent polarization of these macrophages toward the M2 phenotype (39). Blocking NGAL production in macrophages reduced protective effects achieved with IL-10-overexpressing macrophages in a kidney ischemia/reperfusion injury model, substantiating NGAL-associated proproliferative and anti-inflammatory properties (15).
Taking into account that NGAL conveys proproliferative, proregenerative, and anti-inflammatory properties, we hypothesized that growth and differentiation of tumor cells as well as the tumor-supportive microenvironment rely at least in part on the presence of NGAL in TAM. We aimed at elucidating molecular mechanisms of NGAL production in human macrophages and exploring effects of macrophage-derived NGAL on tumor development and progression.
MATERIALS AND METHODS
Materials.
The Janus kinase (Jak) inhibitor was purchased from Calbiochem (Darmstadt, Germany). SB203580 and LY294002 were ordered from Alexis Biochemical (Lörrach, Germany). STA-21 was delivered by Biomol (Hamburg, Germany). Inhibitors were preincubated for 30 min prior to stimulating macrophages with IL-10 for 3 h. Cell culture supplements and fetal calf serum (FCS) were ordered from PAA Laboratories (Cölbe, Germany). Primers and C/EBPβ and signal transducer and activator of transcription 3 (STAT3) decoy oligonucleotides were bought from Biomers (Ulm, Germany). Restriction enzymes BglII and KpnI were purchased from New England BioLabs (Frankfurt am Main, Germany). Anti-C/EBPβ came from Santa Cruz Biotechnology (Santa Cruz, California), anti-NGAL and anti-IgG were purchased from R&D Systems (Wiesbaden-Nordenstadt, Germany), and anti-STAT3 antibodies were bought from Cell Signaling (Danvers, MA). All chemicals were of the highest grade of purity and commercially available. A loss of cell viability was excluded for cell treatments based on trypan blue staining (Biochrom AG, Berlin, Germany).
Primary macrophage generation.
Human monocytes were isolated from buffy coats (DRK-Blutspendedienst Baden-Württemberg-Hessen, Frankfurt, Germany) using Ficoll-Hypaque gradients (PAA Laboratories). Peripheral blood mononuclear cells were washed twice with phosphate-buffered saline (PBS) containing 2 mM EDTA and subsequently incubated for 1 h at 37°C in RPMI 1640 medium supplemented with 100 U/ml penicillin and 100 μg/ml streptomycin to allow their adherence to culture dishes (Sarstedt, Nümbrecht, Germany). Nonadherent cells were removed. Monocytes were then differentiated into macrophages with RPMI 1640 containing 5% AB-positive human serum (DRK-Blutspendedienst Baden-Württemberg-Hessen, Frankfurt, Germany) for 10 days and achieved approximately 80% confluence.
Preparation of macrophage conditioned medium.
For the generation of conditioned medium, cells were serum starved for 24 h. Primary human macrophages were then stimulated with 20 ng/ml recombinant human IL-10 for 3 h. After removal of the supernatant, macrophages were washed twice with PBS and incubated for another 3-h period with RPMI 1640 medium without serum. The resultant supernatant was termed macrophage conditioned medium (MCM) and subsequently used in further experiments in a 1:1 dilution with regular, FCS-containing growth medium. MCM was harvested by centrifugation (13,000 × g, 10 min) and filtered through 0.2-μm-pore-size filters (Millipore, Schwalbach, Germany) to remove large particles, such as apoptotic bodies. MCM from unstimulated macrophages was used as control medium.
Cancer cells.
The human breast cancer cell lines MCF-7, T47D, and MDA-MB-231, human lung carcinoma cell line A549, and human hepatoblastoma cell line HuH7 were cultured in Dulbecco's modified Eagle medium (DMEM) with high glucose, 15 mM HEPES, and l-glutamine, supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% FCS. Cells were kept in a humidified atmosphere of 5% CO2 in air at 37°C and were passaged every 2 days.
Determination of cell growth.
MCF-7 and MDA-MB-231 breast cancer cells were seeded at a density of 1 × 105 cells in 12-well plates. One day prior to the experiment, cells were serum starved for 16 h. Cells were counted at day 0 and subsequently every 24 h up 72 h poststimulation. Cells were stimulated at day 0 either with recombinant human NGAL (80 ng/ml) diluted in medium obtained from unstimulated macrophages or MCM diluted 1:1 in regular (FCS-containing) growth medium. MCM from unstimulated macrophages served as a control medium and was diluted 1:1 in regular growth medium as indicated for MCM from IL-10-stimulated macrophages.
RNA extraction and quantitative real-time PCR (qRT-PCR).
RNA from primary human macrophages was extracted using the peqGold RNAPure reagent (Peqlab Biotechnology, Erlangen, Germany). Total RNA (1 μg) was transcribed using the Maxima first-strand cDNA synthesis kit (Fermentas, St. Leon-Rot, Germany). Quantitative real-time PCR was performed using the MyIQ real-time PCR system (Bio-Rad Laboratories) and Absolute Blue qPCR SYBR green fluorescein mix (Thermo Scientific, Karlsruhe, Germany). Sequences of specific primers are listed in Table 1. Real-time PCR results were quantified using the Gene Expression Macro (version 1.1) software program from Bio-Rad (Munich, Germany), with 18S mRNA expression as an internal housekeeping gene control.
Table 1.
Sequences of qRT-PCR primers and decoy oligonucleotides
| Oligonucleotide | Sequence (5–3′) or sourcea |
|---|---|
| mRNA expression primers (gene) | |
| Hs_18S | GTA-ACC-CGT-TGA-ACC-CCA-TT |
| CCA-TCC-AAT-CGG-TAG-TAG-CG | |
| Hs_PCNA | AAC-TCC-CAG-AAA-AGC-AAC-AAG-CA |
| CGA-GGA-GGA-ATG-AGA-AGA-AGA-CG | |
| Hs_CD14 | AGA-ATC-CTT-CCT-ACG-GTC-CC |
| TGA-TCA-CCT-CCC-CAC-CTC-T | |
| Hs_NGAL | QuantiTect primer assay (Qiagen) |
| ChIP primers (promoter) | |
| NGAL (STAT3 binding site) | AAG-GAA-GGC-ACA-GAG-GGA-GT |
| GGG-ATC-TAG-GGT-GGG-TTG-AT | |
| NGAL (C/EBPβ binding site) | AGA-GTC-AGG-GGG-ATG-GTC-TC |
| GTG-AGT-TGG-GCT-GAT-GTG-TG | |
| SOCS3 | GTT-CCA-GGA-ATC-GGG-GGG-CGG-G |
| CGG-CTG-GCT-GCG-TGC-GGG-GC | |
| IL-6 | ACG-ACC-TAA-GCT-GCA-CTT-TTC-CC |
| ATC-TTT-GTT-GGA-GGG-TGA-GGG-TGG | |
| CD14 | TCC-TGC-TTG-TTG-CTG-CTG-CT |
| AGT-TGC-AGA-CGC-AGC-GGA-AA | |
| β-Actin | ACC-ATG-GAT-GAT-GAT-ATC-GCC |
| GCC-TTG-CAC-ATG-CCG-G | |
| Decoy oligonucleotides | |
| sc_STAT3 | CGT-ATC-TGA-TCT-GAT-CTA-CGT-CGA |
| TCG-ACG-TAG-ATC-AGA-TCA-GAT-ACG | |
| de_STAT3 | GAT-CCT-TCT-GGG-AAT-TCC-TAG-ATC |
| GAT-CTA-GGA-ATT-CCC-AGA-AGG-ATC | |
| mt_STAT3 | GAT-CCT-TCT-GGG-CCG-TCC-TAG-ATC |
| GAT-CTA-GGA-CGG-CCC-AGA-AGG-ATC | |
| sc_C/EBPβ | GTC-TAG-ATC-AGT-CAG-TCA |
| TGA-CTG-ACT-GAT-CTA-GAC | |
| de_ C/EBPβ | TGC-AGA-TTG-CAC-AAT-CTG |
| CAG-ATT-GTG-CAA-TCT-GCA | |
| mt_ C/EBPβ | TGC-AGA-GAC-TAG-TCT-CTG |
| CAG-AGA-CTA-GTC-TCT-GCA |
Underlining indicates mutation.
ELISA.
Supernatants were collected from IL-10-treated primary human macrophages and clarified by centrifugation. One hundred microliters of each sample was applied to an enzyme-linked immunosorbent assay (ELISA) 96-well-plate and incubated overnight at 4°C. After washing, the plate was blocked with PBS–0.05% Tween–1% BSA for 1 h. Afterwards, an anti-NGAL antibody (R&D, Wiesbaden-Nordenstadt, Germany) was added. Detection was by a biotinylated secondary anti-rat IgG antibody (Dako, Eching, Germany). After additional washing, horseradish peroxidase (HRP)-conjugated avidin (R&D) was incubated for 1 h, and the color reagent (R&D) was added. The total protein amount in the sample was determined by the Lowry method for calculating the NGAL amount per mg of total protein.
ChIP.
Chromatin immunoprecipitation (ChIP) analysis was performed as described previously by Tausendschön et al. (44). Briefly, macrophages were incubated with IL-10 for 3 h. Cells were cross-linked with paraformaldehyde and harvested by centrifugation. Immunoprecipitation was carried out by using anti-STAT3 (Cell Signaling, Danvers, MA), anti-C/EBPβ (Santa Cruz Biotechnology, Heidelberg, Germany), anti-H3 pan, and anti-IgG (Upstate, Billerica, MA) primary antibodies. To pull down immunocomplexes, protein G-agarose beads (Roche, Penzberg, Germany) were used. After reversed cross-linking, analysis of enriched DNA fragments was performed. Five percent preimmunoprecipitation samples were taken as the input control. After the ChIP assay, qRT-PCR was performed for both immunoprecipitated target DNA and input control DNA. Calculation of ChIP results (percentage of input) was performed as described previously (47) by using the delta CT method. Results were calculated by correlation to β-actin and are expressed as fold enrichment. Specific primer sequences can be found in Table 1.
ChIP-Western analysis.
Cells were prepared as previously described for the ChIP method, and immunoprecipitation (IP) was performed for C/EBPβ. ChIP-Western blotting was performed as previously described (16). Briefly, lysis buffer (10 mM Tris-HCl, 6.65 M urea, 10% glycerol, and 1% SDS) was added directly to the beads used for IP of DNA-protein complexes after the washing steps. A 4× concentration of sample buffer was added, and samples were boiled for 10 min at 95°C. After clearing by centrifugation (4,000 rpm, 5 min), supernatants were resolved on 10% polyacrylamide gels and blotted on nitrocellulose membranes. Monoclonal antibody directed against STAT3 was used for complexed protein detection.
Transfection and luciferase reporter assay.
Primary human macrophages were transiently transfected by using the JetPei transfection reagent (Polyplus transfection, Illkirch, France). Cells were transfected with 1 μg of NGAL promoter constructs along with 0.2 μg of the Renilla luciferase control vector pRL-TK (Promega, Mannheim, Germany). After transfection, cells were incubated for 16 h, medium was changed, and cells were incubated for another 24 h; either this was followed by stimulation with IL-10 or the cells remained as controls. Luciferase activities in cell lysates were measured as light emission after addition of luciferase assay buffer (Promega, Mannheim, Germany) with a luminometer (Mithras, Berthold Technologies, Bad Wildbad, Germany). Firefly luciferase activity was normalized to Renilla luciferase activity in the lysate. The background obtained from mock-transfected cells was subtracted from each experimental value.
Plasmids.
The full-length NGAL promoter construct was kindly provided by Tatsushi Muta (Tohoku University, Japan) (26). Progressive deletion constructs were generated by PCR amplification using primers with internal KpnI and BglII restriction sites. After Taq polymerase amplification, direct insertion of PCR products into the pCR2.1-TOPO TA subcloning vector (Invitrogen, Darmstadt, Germany) was performed according to the manual. Positive clones were selected by blue-white screening and subsequently cloned into the pGL3basic vector (Promega, Mannheim, Germany) via KpnI and BglII by ligation using T4 DNA ligase (Fermentas, St. Leon-Rot, Germany). Plasmids were sequenced to confirm correct sequences.
Site-directed mutagenesis.
The online software tool PROMO (10, 28) was used to identify putative binding sites for potential transcription factors in the human NGAL promoter (hpNGAL). The QuikChange II XL site-directed mutagenesis kit (Stratagene, La Jolla, CA) was used to introduce point mutations in the putative STAT3 binding site at position −181 to −170 within the human NGAL promoter. The following primers (Biomers) were used to mutate the sequence from 5′-CAC TCC GGG AAT G-3′ to 5′-CAC TCC GGG CCG G-3′ (mutated sequence underlined). Additionally, point mutations were introduced in the binding site for CCAAT-enhancer binding protein beta (C/EBPβ) at position −757 to −751. The following primers (Biomers) were used to mutate the sequence from 5′-TGC AGA TTG CGC AAT CTG CA-3′ to 5′-TGC AGA GAC TAG TCT CTG CA-3′ (mutated sequence is underlined). Site-directed mutagenesis was confirmed by sequencing.
C/EBPβ and/or STAT3 decoy experiment.
Decoy oligonucleotides were used as described previously (1). Briefly, primary human macrophages were transiently transfected with decoy oligonucleotides containing either the wild-type or mutated C/EBPβ or STAT3 consensus site derived from hpNGAL. Scrambled control oligonucleotides, which do not recognize any known transcription factor binding site, were used as controls. Phosphothiorate-stabilized 5′-terminal fluorescein-labeled oligonucleotides were used. Oligonucleotides (3 μM) were added 24 h prior to cell stimulation. After the medium was changed, macrophages were stimulated with 20 ng/ml IL-10 for 3 h. NGAL mRNA expression was determined by RT-PCR. CD14 expression was determined as a negative control since it was previously described to be an IL-10 target that is regulated independently of STAT3 and C/EBP (34). Sequences for the used oligonucleotides were included in Table 1.
Cell migration assays.
For scratch assays, 2 × 105 MCF-7 cells were seeded in 24-well culture plates. After 24 h, cells were starved for 16 h before the scratch was initiated with a small pipette tip. Detached cells were removed by washing with PBS. Stimulation was with either 80 ng/ml recombinant human NGAL or MCM in the presence or absence of 5 μg of a NGAL-neutralizing antibody (R&D Systems). An isotype-matching IgG control antibody was used as a control (R&D Systems). Figures were acquired using an Axio-Scope microscope (Carl Zeiss Microimaging, Offenbach, Germany).
Immunofluorescence.
Cells were fixed in 4% paraformaldehyde and permeabilized with PBS-Triton X. After washing, cells were blocked with FCS for 1 h and primary antibody was added to detect the proliferation marker, PCNA (Santa Cruz Biotechnology). Sections were mounted with Fluromount-G (Biozol, Eching, Germany). For analyzing cell viability, cells were additionally stained with 4′,6-diamidino-2-phenylindole (DAPI) for 30 min, and cell nuclei were analyzed. Images were obtained with an Axioskop 40 microscope and documented with the AxioVision software program (Zeiss, Göttingen, Germany). Negative controls, lacking primary antibodies, showed no staining. Slides were examined in a blinded manner, and proliferation was quantified by counting the number of PCNA-positive cells in an average of five high-power fields of each section.
Statistical analysis.
Each experiment was performed at least three times. The P values were calculated using one-way analysis of variance (ANOVA), and P values of <0.05 were considered significant.
RESULTS
IL-10 uses a Jak/STAT-dependent pathway for NGAL induction in macrophages.
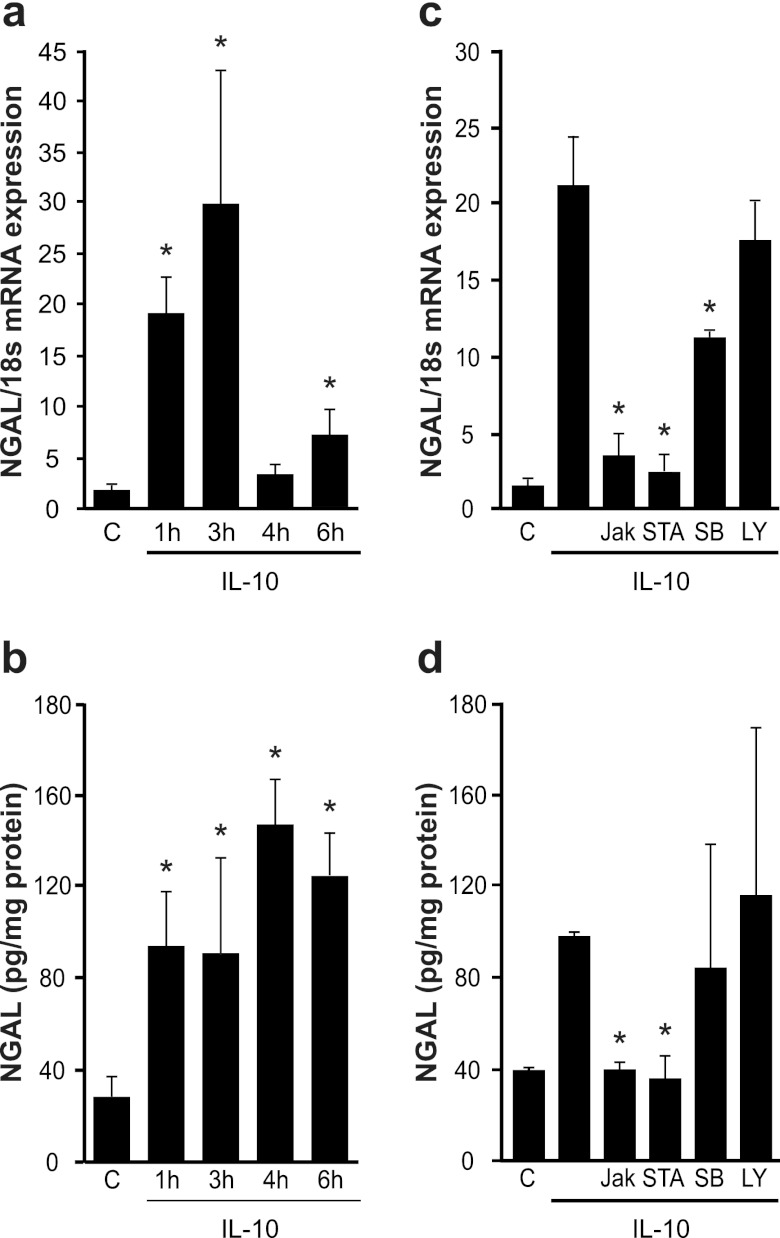
IL-10 induces NGAL production and secretion in primary human macrophages. This was evident at both the mRNA (Fig. 1a) and protein (Fig. 1b) levels and was already observed after 1 h of stimulation. NGAL mRNA expression declined after 4 h and remained low (data not shown). However, protein amounts remained stable for at least 6 h but also decayed with prolonged incubation periods at 24 h (data not shown). We then analyzed signaling pathways involved in IL-10-mediated NGAL expression in primary human macrophages and tested pharmacological inhibitors. Macrophages were pretreated with the indicated inhibitors for 30 min, washed once with PBS, and then stimulated with IL-10 for 3 h. LY29004, an inhibitor of PI3K signaling, had no effect on IL-10-induced NGAL mRNA or protein expression. The p38 kinase inhibitor SB239063 only reduced NGAL mRNA levels but not the protein amount. The small but significant mRNA decrease apparently is not sufficient to reduce the protein amount, which may be explained by the lower half-life of the protein than of the mRNA (Fig. 1c and d). In contrast, significant NGAL mRNA and protein reduction was achieved with the Jak inhibitor and the specific STAT3 inhibitor STA-21 (Fig. 1c and d). This argues for a Jak/STAT3 pathway in IL-10-induced NGAL formation with a minor or no impact of p38 signaling.
Fig 1.
IL-10 signals through a Jak/STAT-dependent pathway to induce NGAL. (a and b) Human primary macrophages remained as controls or were stimulated with 20 ng/ml recombinant human IL-10 for the times indicated. (a) NGAL mRNA expression measured by qRT-PCR. (b) Release of the NGAL protein into macrophage supernatants quantified by ELISA. Data are means ± SD; n = 8; ∗, P < 0.05 versus results for controls. (c and d) Human primary macrophages were pretreated for 30 min with inhibitors prior to stimulation with IL-10 for 3 h. A 10 μM concentration of Jak, STAT3 (STA-21), or phosphatidylinositol 3-kinase (PI3K) (LY294002) and 5 μM p38(SB203580) inhibitors were used. (c) NGAL mRNA expression measured by qRT-PCR. (d) Released NGAL protein quantified by ELISA. Data are means ± SD; n = 8; ∗, P < 0.05 versus IL-10.
IL-10 stimulates NGAL promoter activity.
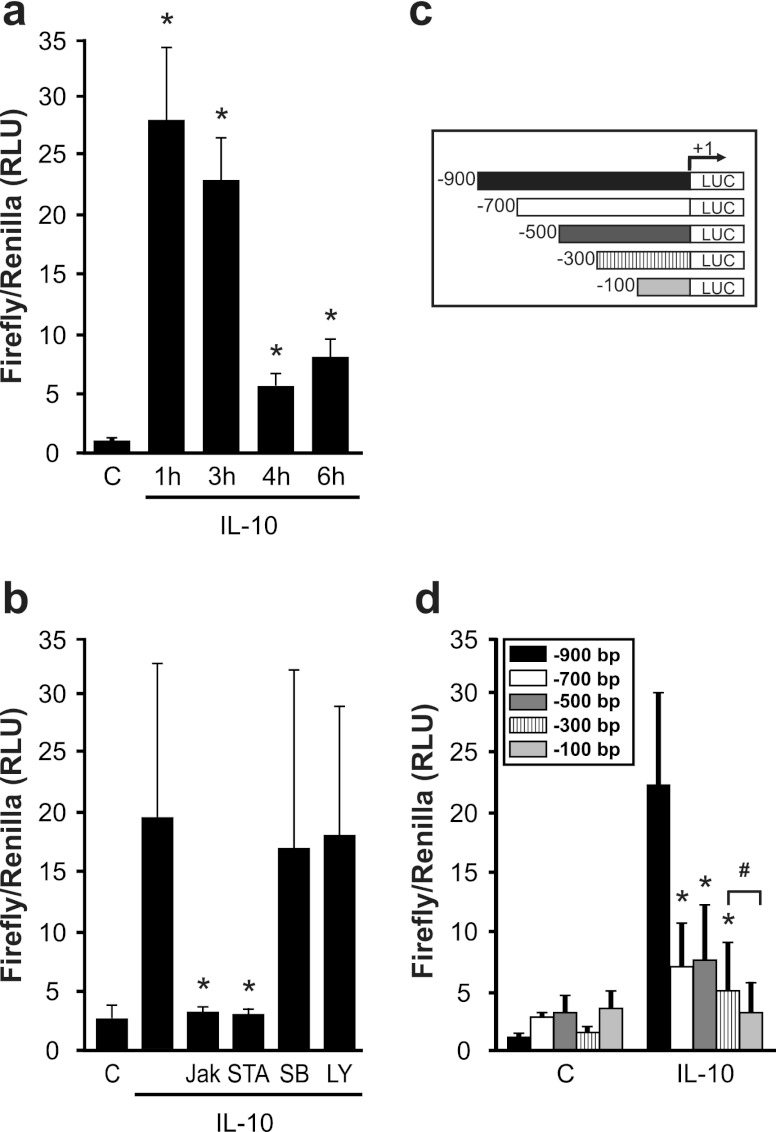
To clarify the molecular mechanism of IL-10-induced NGAL formation, we performed NGAL promoter analysis in macrophages. Transfecting the full-length NGAL promoter construct (pNGAL-900), we noticed its immediate activation in response to IL-10 at 1 h and 3 h, with much lower responses at 4 h and 6 h (Fig. 2a). Furthermore, IL-10-induced NGAL promoter activity was again sensitive to Jak and STAT3 inhibition but was unaffected by LY29004 as well as SB239063 (Fig. 2b).
Fig 2.
IL-10 activates the NGAL promoter. (a and b) Promoter activity was measured by luciferase reporter assay after IL-10 stimulation for the times indicated. Data are means ± SD; n = 8; ∗, P < 0.05 versus results for controls. (b) Macrophages were pretreated for 30 min with inhibitors as indicated in the legend to Fig. 1, and NGAL promoter activity was measured by luciferase reporter assay. Data are means ± SD; n = 8; ∗, P < 0.05 versus results with IL-10. (c) A schematic overview of NGAL promoter deletion constructs. (d) Macrophages were cotransfected with the indicated deletion constructs and a thymidine kinase (TK)-Renilla control construct prior to IL-10 treatment for 3 h. Promoter activity was measured by luciferase reporter assay. Data are means ± SD; n = 8; ∗, P < 0.05 versus results for IL-10 and the bp −900 construct; #, P < 0.05 versus results for IL-10 and the bp −300 construct.
To identify cis-acting elements in the NGAL promoter promoting IL-10-mediated transcriptional activation, we generated a series of 5′ deletion constructs of the full-length promoter, as depicted in Fig. 2c. Analysis of these constructs revealed high promoter activity with the bp −900 fragment and much lower activities with all other constructs (Fig. 1d) after 3 h of IL-10 treatment. Apparently, a distal promoter region spanning bp −700 to −900 contains indispensable putative transcription factor binding sites needed for IL-10-induced NGAL formation. Comparing the bp −300 fragment to the bp −100 construct again revealed a significant loss in activity. Based on promoter deletion analysis, we suggest defining two regions of interest with importance to IL-10-dependent NGAL promoter activation: one region spanning the distance between bp −900 and −700 and another within the region from bp −300 to −100. In silico analysis of the human NGAL promoter done by using the PROMO software program (28) revealed potential binding sites for C/EBPβ at bp −761 and a unique binding site for STAT3 at bp −170. Based on previous reports showing a potential role for C/EBPs in modulating NGAL promoter activity in response to IL-1β (8, 26), we decided to further delineate the role of C/EBPβ. Although we are not aware of any study linking STAT3 and NGAL expression, the well-established role of STAT3 downstream of IL-10 signaling (32) prompted us to hypothesize that STAT3 might be a relevant factor under our conditions.
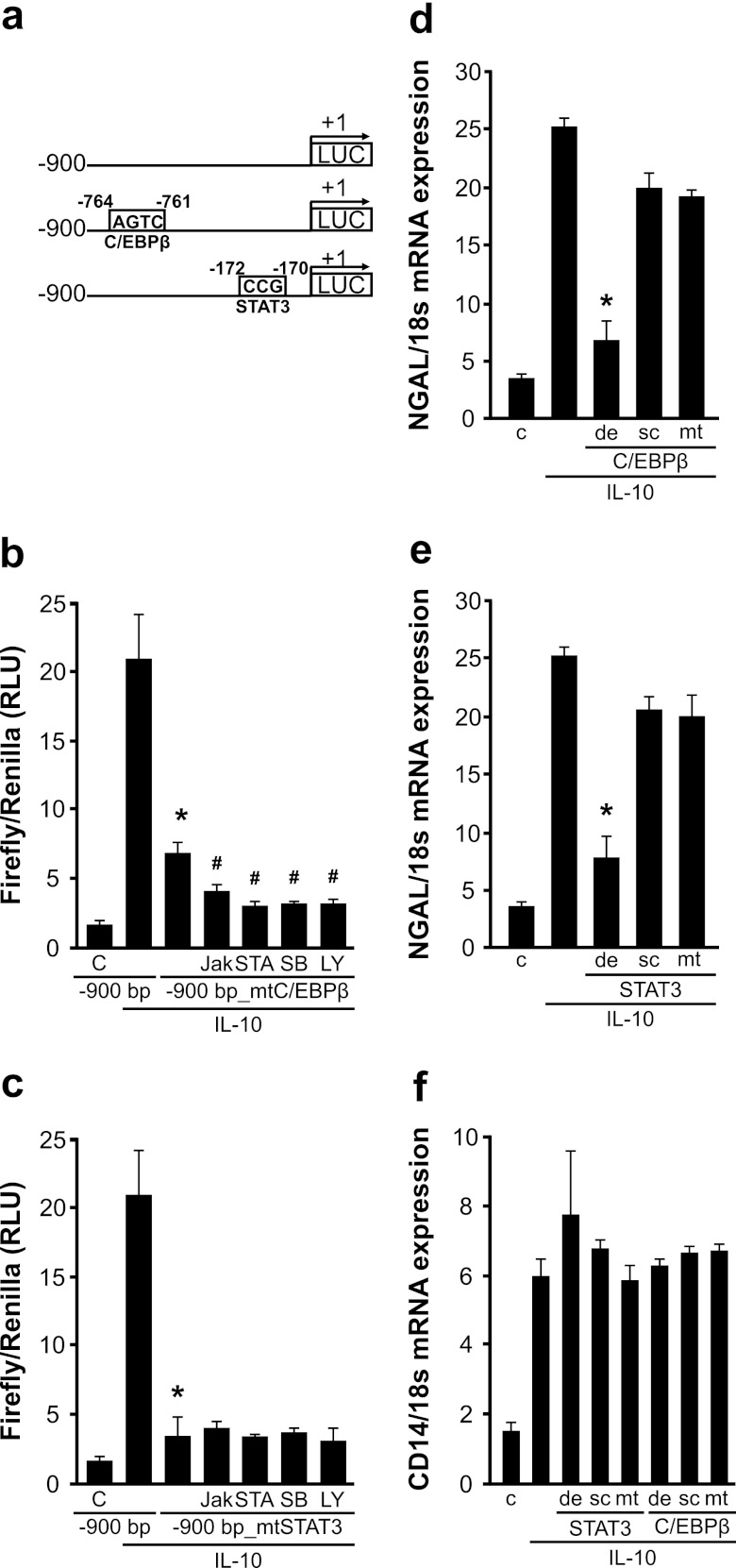
To elucidate the role of C/EBPβ and STAT3, we used the bp −900 promoter construct and introduced point mutations in the putative binding sites for the transcription factor shown in Fig. 3a. Compared to results with the full-length bp −900 promoter construct, mutation of either the C/EBPβ or the STAT3 site significantly reduced IL-10-induced promoter activity (Fig. 3b and c). Since there are more C/EBP-binding sites in the NGAL promoter than the one we focused on, we decided to follow the mutated NGAL promoter activity in the presence of the inhibitors used for analysis of mRNA as well as protein analysis. Results show an additional reduction in NGAL promoter activity in the presence of the inhibitors for the samples transfected with the mutated C/EBPβ reporter construct (Fig. 3b). NGAL promoter activity was significantly impaired using the STAT3-mutated construct, with no further reduction of the promoter activity in the presence of the inhibitors.
Fig 3.
C/EBPβ and STAT3 are crucial for IL-10-mediated NGAL expression. (a) A scheme showing point mutations in the respective putative binding sites of C/EBPβ and STAT3 in the NGAL promoter. (b and c) Luciferase reporter assay for the indicated full-length versus mutated NGAL promoter constructs after IL-10 stimulation for 3 h, where indicated in the presence of 10 μM Jak, STAT3 (STA-21), or PI3K (LY294002) and 5 μM p38(SB203580) inhibitors. Data are means ± SD; n = 8; ∗, P < 0.05 versus results for IL-10 and the bp −900 construct; #, P < 0.05 versus results for IL-10 with the bp −900 construct and C/EBPβ/STAT3. (d, e, and f) Oligonucleotides (3 μM) containing the binding sites for either STAT3 or C/EBPβ (de) or the mutated binding sites for each transcription factor (mt) were transfected separately in primary human macrophages prior to stimulation with IL-10 for 3 h. Scrambled oligonucleotides (sc) were used as controls. NGAL mRNA expression was measured by qRT-PCR. Data are means ± SD; n = 8; ∗, P < 0.05 versus results for IL-10.
To further prove the relevance of STAT3 and C/EBPβ in NGAL expression in response to IL-10, we performed a decoy oligonucleotide approach using primary human macrophages. The method has successfully been used in our lab to attenuate CREB-mediated transcription (1, 45). Oligonucleotides containing wild-type or mutated binding sites for either STAT3 or C/EBPβ were transfected separately into macrophages in order to compete with DNA binding sites for active transcription factors (Fig. 3d and e). Scrambled oligonucleotides were used as controls. Experimentally, macrophages were exposed to 3 μM fluorescence-labeled oligonucleotides for 24 h prior to stimulation with IL-10 for 3 h. Decoy oligonucleotides against the wild-type binding site of both STAT3 and C/EBPβ significantly reduced IL-10-induced NGAL mRNA expression. Neither scrambled nor mutated oligonucleotides altered NGAL mRNA expression. As a specificity control, we screened for expression of CD14, an established IL-10 target known to be regulated independently of STAT3 and C/EBPβ, and observed no changes for any of the oligonucleotides used in this study.
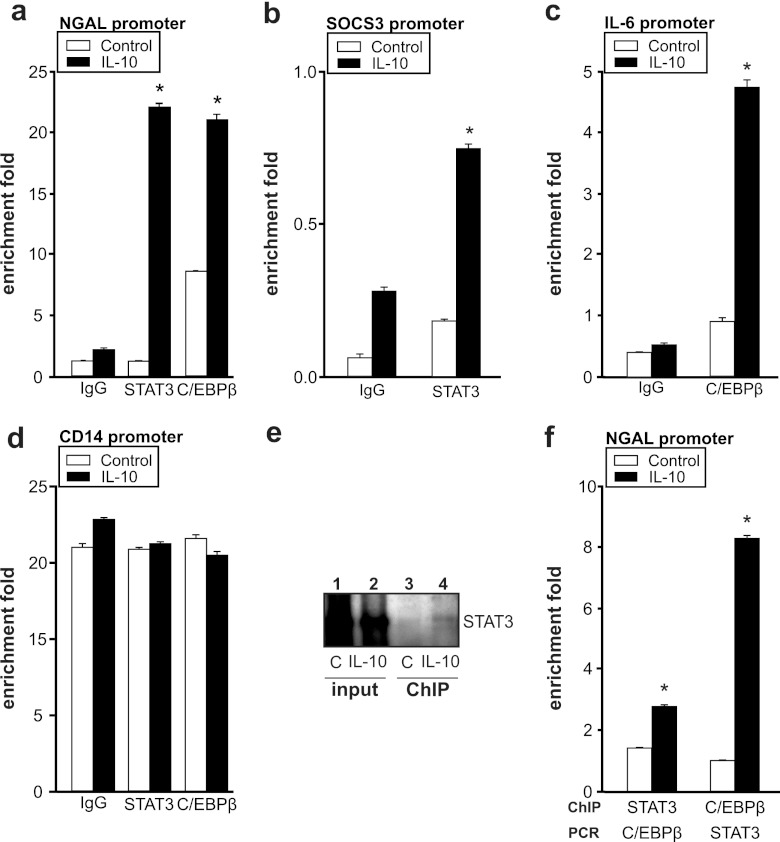
ChIP analysis reveals binding of C/EBPβ and STAT3 to the NGAL promoter.
To explore the recruitment of the transcription factors C/EBPβ and STAT3 to the NGAL promoter, we performed ChIP analysis using antibodies against either STAT3 or C/EBPβ (Fig. 4a) compared to an IgG control antibody. In unstimulated macrophages, neither STAT3 nor C/EBPβ was recruited to its putative binding site in the NGAL promoter. However, stimulation of macrophages with IL-10 for 3 h significantly enhanced association of both STAT3 and C/EBPβ to their predicted binding sequences in the NGAL promoter. In order to evaluate the specificity of the ChIP technique, we included internal positive controls, such as binding of STAT3 to the SOCS3 promoter (Fig. 4b) and binding of C/EBPβ to the IL-6 promoter (Fig. 4c). As a negative control, we ensured the lack of STAT3 and C/EBPβ binding to the CD14 promoter (Fig. 4d). ChIP results are calculated as percentages of input normalized to β-actin and are expressed as fold enrichment.
Fig 4.
ChIP analysis reveals binding of C/EBPβ and STAT3 to the NGAL promoter. ChIP analysis of promoter regions of the NGAL (a), SOCS3 (b), IL-6 (c), or CD14 (d) promoter was performed to determine the recruitment of STAT3 and C/EBPβ by using specific antibodies against STAT3 and C/EBPβ, respectively, after 3 h of IL-10 stimulation. ChIP DNA was amplified by qRT-PCR using primers specific for STAT3 and C/EBPβ binding sites within the NGAL promoter. Data are represented as fold enrichment by normalizing to β-actin. Data are means ± SD; n = 4; ∗, P < 0.05 versus results for controls. (e) ChIP-Western analysis after IL-10 treatment for 3 h. ChIP was performed using a specific antibody against C/EBPβ, followed by immunoblot detection of STAT3 (lanes 3 and 4) compared to unfractionated cross-linked DNA-protein complexes (input) (lanes 1 and 2). A representative picture out of 4 independent experiments is shown. (f) qRT-PCR analysis of either STAT3- or C/EBPβ-specific chromatin-immunoprecipitated DNA. Data are represented as fold enrichment by normalizing to β-actin. Data are means ± SD; n = 4; ∗, P < 0.05 versus results for controls.
Next, we asked whether these transcription factors could interact in order to promote NGAL promoter activity, although the C/EBPβ and STAT3 binding sites were roughly 500 bp apart. For this purpose, we performed a ChIP-Western analysis (Fig. 4b). Chromatin immunoprecipitation was performed with the C/EBPβ antibody, followed by immunoblot detection of STAT3 (Fig. 4e, lanes 3 and 4), compared to the unfractionated cross-linked DNA-protein complexes (input) (Fig. 4e, lanes 1 and 2). Results show a distinct band at 105 kDa, the expected size of STAT3. Moreover, a significant increase in the STAT3 signal was noticed after IL-10 stimulation compared to results for the controls. These data support the assumption that C/EBPβ and STAT3 are both recruited to the NGAL promoter, but further investigation is needed to identify the interaction and/or complex formation of STAT3 and C/EBPβ. To validate these data, we decided to include PCR analysis of different promoter regions after performing ChIP analysis of one of the transcription factors (Fig. 4f). Following chromatin immunoprecipitation of STAT3, we performed RT-PCR analysis for the putative C/EBPβ binding site and vice versa. Accumulation of the STAT3 binding sequence with the promoter region containing the binding site for C/EBPβ and vice versa could be detected. These results corroborate the idea that both STAT3 and C/EBPβ DNA binding sites are needed to facilitate transcriptional activation of NGAL in macrophages after IL-10 stimulation. Further experiments are needed to discriminate the importance of other C/EBPβ binding sites in the NGAL promoter and the speculative assumption that STAT3 and C/EBPβ binding regions might form a loop in order to activate the NGAL promoter in response to IL-10.
Macrophage-derived NGAL promotes tumor growth and proliferation.
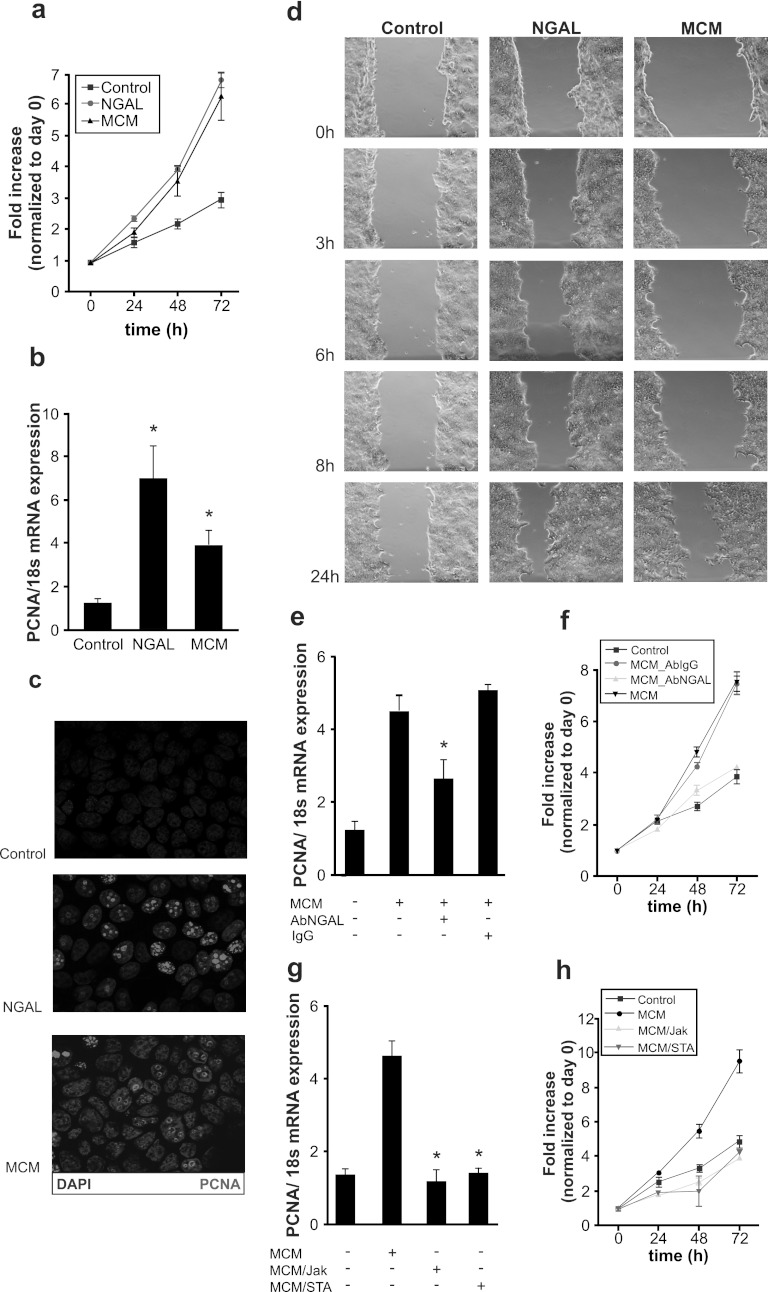
Having proproliferative and regenerative actions of NGAL in mind (15, 39), we looked into a protumorigenic capacity of macrophage-derived NGAL. We generated macrophage-conditioned medium (MCM) from primary human macrophages stimulated with IL-10 for 3 h. In turn, MCF-7 breast cancer cells were then treated either with 80 ng/ml of recombinant human NGAL or MCM. MCM from unstimulated macrophages served as control medium (Fig. 5a). Both exogenous NGAL and MCM significantly enhanced MCF-7 cell growth compared to that of untreated controls cells, which were cultured in MCM from unstimulated macrophages as appropriate. Using the same experimental setup, we determined the expression of the proliferating cell nuclear antigen (PCNA). NGAL and MCM provoked a significant increase of PCNA mRNA after 24 h (Fig. 5b). To confirm the proliferation-promoting action of NGAL on cancer cells, we stained MCF-7 cells for PCNA (red) after their treatment with NGAL or MCM. As seen in Fig. 5c, PCNA protein expression was significantly induced in MCM-exposed cells and reached even higher expression values in NGAL-treated cells. In the following set of experiments, we analyzed the impact of NGAL versus MCM on migration/proliferation (Fig. 5d). Therefore, we performed a wound-scratch assay using MCF-7 cells. Compared to controls, NGAL and MCM provoked scratch closure. This became evident following NGAL addition after 6 h, while MCM lagged behind but clearly provoked scratch closure at 8 to 24 h. The relatively slow repopulation of the scratched area argued for cellular proliferation rather than migration as the underlying mechanism, thus supporting previous findings for NGAL in promoting proliferation and growth. Autocrine effects of NGAL, generated by the cancer cell itself, were excluded because mRNA expression of NGAL in MCF-7 cells treated with either NGAL or MCM remained unaltered compared to results for controls (Fig. 6a).
Fig 5.
Macrophage-derived NGAL promotes tumor growth and proliferation. (a, b, and c) MCF-7 breast cancer cells were treated with either 80 ng/ml recombinant human NGAL or MCM. MCM from untreated macrophages was used as a control. (a) Growth characteristics were analyzed by cell counting every 24 h. Data are expressed as fold increase normalized to results for day 0. (b) qRT-PCR analysis of the proliferation marker PCNA in response to NGAL and MCM treatment for 24 h. Data are means ± SD; n = 8; ∗, P < 0.05 versus results for controls. (c) Immunofluorescent staining for proliferation (PCNA, white) and nuclei (DAPI, gray) after treatment with NGAL and MCM for 24 h. (d) Scratch-wound assay for NGAL- and MCM-treated MCF-7 cells for times indicated. (e and f) MCF-7 cells were treated with MCM in the absence or presence of a neutralizing NGAL antibody (5 μg) versus an IgG control antibody (5 μg) for 24 h. mRNA expression of the proliferation marker PCNA was measured by qRT-PCR (e), and growth characteristics were analyzed by cell counting (f). (g and h) MCF-7 cells were treated with complete MCM or MCM from macrophages pretreated for 30 min with either Jak inhibitor (MCM/Jak) or the STAT3 inhibitor STA-21 (MCM/STA). mRNA expression of the proliferation marker PCNA was measured by qRT-PCR (g), and the growth curve was analyzed by cell counting (h). Data are means ± SD; n = 6 for qRT-PCR measurements; ∗, P < 0.05 versus results with MCM.
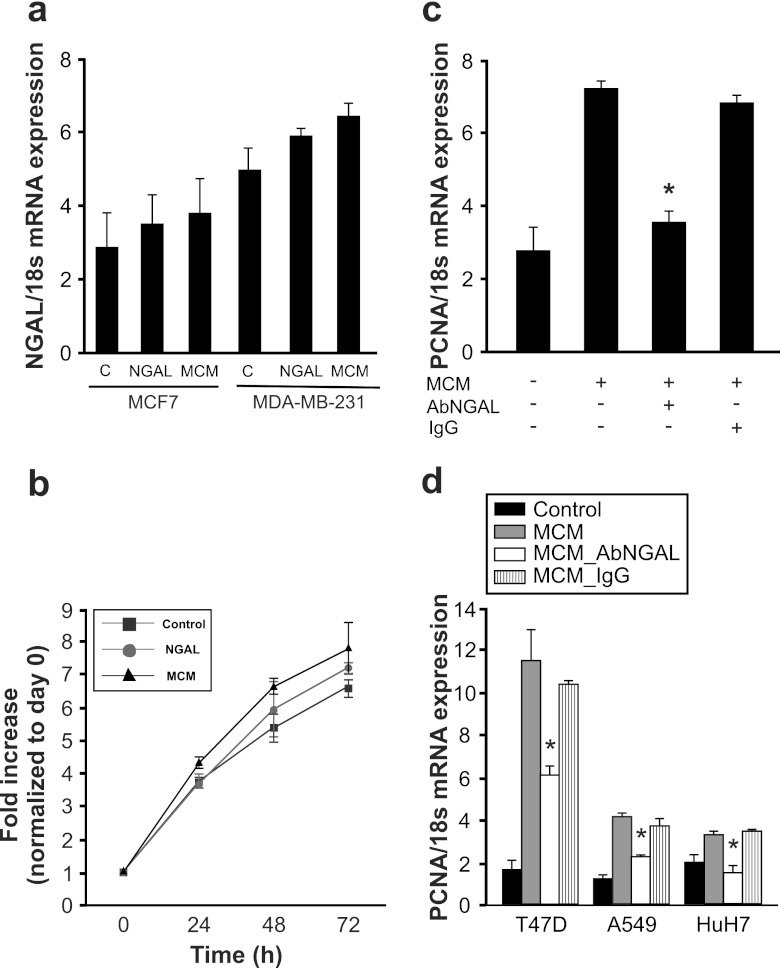
Fig 6.
Growth-promoting characteristics of NGAL on various cancer cells. (a) MCF-7 and MDA-MB-231 breast cancer cells remained as controls or were stimulated with 80 ng/ml recombinant human NGAL or MCM for 24 h. NGAL mRNA expression was measured by qRT-PCR. Data are means ± SD; n = 6; ∗, P < 0.05 versus results for controls. (b and c) Growth-promoting effects of NGAL on MDA-MB-231 cells was measured by growth analysis (b) and qRT-PCR of the proliferation marker PCNA (c). The impact of NGAL in MCM was determined using a neutralizing antibody against NGAL (5 μg) versus the IgG control antibody (5 μg). Data are means ± SD; n = 4; ∗, P < 0.05 versus results with MCM. (d) Human cancer cells remained as controls or were stimulated with MCM in the presence or absence of a neutralizing NGAL antibody (5 μg) versus an IgG control antibody (5 μg) for 24 h. qRT-PCR was performed to measure the proliferation marker PCNA in T47D breast cancer cells, A549 lung carcinoma cells, or HuH7 hepatoblastoma cells. Data are means ± SD; n = 5; ∗, P < 0.05 versus results with MCM.
In order to prove that NGAL, present in MCM from macrophages treated with IL-10, contributes to the proliferative and growth-promoting effects of MCM, we neutralized NGAL and checked PCNA expression (Fig. 5e). MCM elicited a robust mRNA increase in PCNA, an effect that was significantly attenuated by neutralizing NGAL, while addition of the IgG control antibody was without effect. In extending experiments, we followed MCF-7 growth for up to 72 h (Fig. 5f) and confirmed that MCM-stimulated cell growth was reduced to control levels by neutralizing NGAL.
Having established that NGAL expression in human macrophages after IL-10 treatment is regulated by the Jak-STAT3 pathway, we generated macrophage-conditioned medium from macrophages with impaired Jak-STAT3 signaling (Fig. 5g). Compared to control MCM, MCM derived from cultures in the presence of Jak inhibitor and STAT3 inhibitor (STA-21) significantly reduced mRNA expression of PCNA. Long-term cell growth characteristics for up to 72 h again confirmed stimulation of growth by MCM, but impaired growth with MCM was derived from cells with Jak-STAT3 signaling being impaired (Fig. 5h).
Since MCF-7 cells are noninvasive breast cancer cells and have low endogenous NGAL levels (Fig. 6a), possible autocrine effects due to cancer cell-secreted NGAL are small. In order to substantiate the growth-promoting effect of macrophage-generated NGAL on invasive breast cancer cells as well, we tested MDA-MB-231 breast cancer cells, which have higher endogenous NGAL levels (Fig. 6a). Growth characteristics upon stimulation with recombinant NGAL protein (80 ng/ml) diluted in MCM from unstimulated macrophages and MCM from IL-10-treated macrophages show a growth-promoting effect in MDA-MB-231 cells (Fig. 6b). In order to prove that NGAL in MCM from macrophages treated with IL-10 contributes to the proliferative effects of MCM in MDA-MB-231 cells, we neutralized NGAL with a specific antibody and checked PCNA expression (Fig. 6c). MCM induced a significant mRNA increase in PCNA, which was significantly attenuated by neutralizing NGAL, while addition of the IgG control antibody had no effect.
To exclude potential cell-specific effects, we verified PCNA mRNA expression in response to MCM in human T47D breast cancer cells, human A549 lung carcinoma cells, and human HuH7 hepatoblastoma cells (Fig. 6d). In all cell lines tested, MCM provoked PCNA mRNA expression that was significantly reduced by neutralizing NGAL, while the control IgG left the proliferative index unaltered.
DISCUSSION
The present study adds to the emerging concept that NGAL is a critical player during tumor development. The novelty of our study is based on unraveling the functions of NGAL released from tumor-associated macrophages. Here we have provided evidence that macrophages produce and secrete the growth-promoting mediator NGAL in response to their activation by the anti-inflammatory cytokine IL-10, which is known to be present in the tumor microenvironment. In our study, the cell origin of IL-10 was not addressed. However, it was shown that IL-10 could be secreted by MCF-7 cells themselves after stimulation with procathepsin D (13). It could be hypothesized that procathepsin D, which induces a massive production of IL-10 in tumor cells, may originate with the surrounding tumor microenvironment.
Macrophage activation is linked to tumor attack or a tumor-supportive role. These opposing macrophage functions result from numerous microenvironmental signals (24). Blocking macrophage infiltration attenuated tumor development, resulting in no or largely retarded growth (21). It was also shown that tumor size, vascularization, and metastasis correlated with infiltration of macrophages and their ability to secrete growth-promoting mediators, such as VEGF, IL-8, and TGF-α (51). We obtained evidence that NGAL contributes to the portfolio of molecules that determine the anti-inflammatory and growth-supporting M2-polarized macrophage phenotype. In recent years, a number of studies suggested NGAL as a potential cancer marker, especially for breast cancer (2, 4, 41). Moreover, its expression has been postulated to be a poor prognosis marker. Overexpression of NGAL in human breast carcinoma directly increased tumor growth and angiogenesis (18). Our results corroborate these data by showing growth-promoting effects of NGAL on both noninvasive MCF-7 and invasive MDA-MB-231 tumor cells. However, our work extends existing information that NGAL can also be secreted by tumor-infiltrating immune cells, predominantly macrophages, in quantities that recapitulate authentic NGAL growth-promoting effects on tumor cells. Additionally, we are not aware of any other study dealing with the role of NGAL from TAM in promoting tumor growth.
At the molecular level, NGAL was reported to be upregulated by NF-κB in epithelial cells of inflamed lungs (5, 6). Further promoter studies revealed the importance of the NF-κB-binding cofactor IκB-ξ (26). Chromatin immunoprecipitation assays and knockdown experiments demonstrated that both IκB-ξ and NF-κB were essential for the activation of the NGAL promoter after IL-1β stimulation. Studies using an IκBα superrepressor in human thyroid carcinoma cells underscored the importance of NF-κB in NGAL regulation (12). Since an active NF-κB pathway is linked to the pathogenesis of inflammation-associated tumor formation, NGAL might be contributing via this signaling cascade. However, we did not concentrate on the NF-κB pathway, because alternatively activated macrophages are supposed to suppress this pathway (37). The use of diverse inhibitors allowed us to identify the Jak-STAT pathway, with little or no impact of p38 signaling, as being responsible for IL-10-induced NGAL production, while signaling through PI3K could be ruled out. The anti-inflammatory response to IL-10 in macrophages is driven mainly by STAT3. The role of STAT3 is to suppress the transcription of proinflammatory genes (11, 22). Furthermore, STAT3-dependent signaling is widely recognized as crucial to most of the characterized biological functions of IL-10 in macrophages (32, 46, 48). The IL-10–Jak–STAT axis plays a central role in both acute and chronic inflammation (30). STAT3 was previously described as a crucial factor in various tumors (9, 20, 49). Activation of NGAL requires a flanking C/EBP site in its promoter in order to get fully activated (26, 50). NGAL is highly expressed in adipocytes, which is largely dictated by C/EBP-dependent trans activation of the NGAL promoter during 3T3-L1 adipogenesis (50). During early stages of adipogenesis, the Jak-STAT signaling pathway is mainly involved in regulating C/EBPβ transcription (52). Furthermore, upon stimulation of Toll-like receptors in A549 lung adenocarcinoma cells, activation of the NGAL promoter was significantly suppressed in C/EBPβ-deficient cells. IκB-ξ-mediated transcriptional activation of HEK293 cells required the presence of both the NF-κB and C/EBP binding sites in the NGAL promoter (26). Due to their Rel homology domain and the basic leucine zipper domain, it was suggested that NF-κB and C/EBP proteins could form a direct transcription complex in order to fully activate target promoters (17, 27, 40). In our study, we also observe the importance of two transcription factors located at distant sites of the NGAL promoter. Our data suggest that both the STAT3 and C/EBPβ sites are required for IL-10 to elicit a full transcriptional NGAL response in human macrophages. It should be noted that IL-10 signaling involves C/EBP proteins to promote its anti-inflammatory and immunomodulatory actions (35, 43). This is the case for the IL-6 promoter. IL-10 treatment reduced IL-6 cytokine levels by augmenting C/EBPβ DNA binding activity in human intestinal epithelial cells (35). Additionally, C/EBPs are induced by granulocyte colony-stimulating factor (G-CSF) directly downstream of STAT3 during granulopoiesis in the IL-3-dependent cell line 32D clone 3 (33). It is interesting that promoters of genes that exhibit IL-10-dependent induction/repression harbor both STAT3 and C/EBP binding sites. Multiple C/EBP binding sites but a unique STAT3 site are found in the NGAL promoter, which might guarantee the maximum induction under distinct conditions and in diverse cell types. Transcription factors might form transcription complexes by utilizing different C/EBP binding sites in response to different stimuli. We identified the importance of both STAT3 and C/EBPβ by ChIP and ChIP-Western analysis upon stimulation with IL-10 in primary human macrophages. Our hypothesis of a possible complex formation of STAT3 with C/EBPβ remains to be fully answered. The identification and biochemical characterization of a putative complex of STAT3 and C/EBPβ would add valuable knowledge to the molecular mechanisms underlying transcriptional activation of IL-10 target genes, such as NGAL. Our study focused on the importance of the C/EBPβ binding site within the bp −900 to −700 region of the NGAL promoter but did not exclude other C/EBPβ binding sites throughout the NGAL promoter. As a consequence, no direct conclusions on the interaction between STAT3 and C/EBPβ could be drawn from the current results. Further experiments are needed to define the specificity of the recruitment of C/EBPβ to the investigated site, since the C/EBPβ pulldown in the ChIP experiments may also cause a pulldown of other fragments of the NGAL promoter, which may contain C/EBPβ binding sites. Furthermore, false positives can be generated by the pulldown of a small fraction of large-size DNA fragments, which also could contain binding sites for C/EBPβ. Therefore, further experiments will involve the identification of the putative complex of STAT3 and C/EBPβ, as well as the biochemical characterization of the interaction between STAT3 and C/EBPβ. It is likely that both the distance and the nature of the complex between transcription factor binding sites, as well as the chromatin structure of each site, affects transcriptional activation of target promoters.
Understanding molecular and genetic mechanisms that regulate tumor-promoting genes in TAM could offer new perspectives for efficient therapy. Nevertheless, further research is needed to define the diverse biological effects of NGAL within the tumor microenvironment. Regarding its role in cancer progression, we are currently scrutinizing the growth-promoting effects of NGAL from TAM, since our study particularly points to the importance of tumor-supporting macrophages in producing tumor growth-stimulating molecules like NGAL.
ACKNOWLEDGMENTS
We have no conflicting financial interests.
The work was supported by grants from Deutsche Forschungsgemeinschaft (BR999, ECCPS), Sander Foundation (2007.070.2), and Deutsche Krebshilfe (109599). M.J. was supported by a grant from the Fritz-Thyssen-Stiftung and from the Medical Faculty, Goethe University Frankfurt. J.M. was supported by the Deutscher Akademischer Austauschdienst (DAAD).
We thank Esther Imelmann for her support in designing NGAL promoter deletion constructs.
Footnotes
Published ahead of print 30 July 2012
REFERENCES
- 1. Barra V, Kuhn AM, von Knethen A, Weigert A, Brune B. 2011. Apoptotic cell-derived factors induce arginase II expression in murine macrophages by activating ERK5/CREB. Cell. Mol. Life Sci. 68: 1815–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bauer M, et al. 2008. Neutrophil gelatinase-associated lipocalin (NGAL) is a predictor of poor prognosis in human primary breast cancer. Breast Cancer Res. Treat. 108: 389–397 [DOI] [PubMed] [Google Scholar]
- 3. Bingle L, Brown NJ, Lewis CE. 2002. The role of tumour-associated macrophages in tumour progression: implications for new anticancer therapies. J. Pathol. 196: 254–265 [DOI] [PubMed] [Google Scholar]
- 4. Bolignano D, et al. 2010. Neutrophil gelatinase-associated lipocalin (NGAL) in human neoplasias: a new protein enters the scene. Cancer Lett. 288: 10–16 [DOI] [PubMed] [Google Scholar]
- 5. Cowland JB, Muta T, Borregaard N. 2006. IL-1 beta-specific up-regulation of neutrophil gelatinase-associated lipocalin is controlled by I kappa B-xi. J. Immunol. 176: 5559–5566 [DOI] [PubMed] [Google Scholar]
- 6. Cowland JB, Sorensen OE, Sehested M, Borregaard N. 2003. Neutrophil gelatinase-associated lipocalin is up-regulated in human epithelial cells by IL-1 beta, but not by TNF-alpha. J. Immunol. 171: 6630–6639 [DOI] [PubMed] [Google Scholar]
- 7. Devireddy LR, Teodoro JG, Richard FA, Green MR. 2001. Induction of apoptosis by a secreted lipocalin that is transcriptionally regulated by IL-3 deprivation. Science 293: 829–834 [DOI] [PubMed] [Google Scholar]
- 8. Du ZP, Zhuo LV, Wu BL, Xu LY. 2011. Neutrophil gelatinase-associated lipocalin and its receptor: independent prognostic factors of oesophageal squamous cell carcinoma. J. Clin. Pathol. 64: 69–74 [Correction, 64:113.] [DOI] [PubMed] [Google Scholar]
- 9. Ernst M, Putoczki TL. 2012. Stat3: linking inflammation to (gastrointestinal) tumourigenesis. Clin. Exp. Pharmacol. Physiol. 39: 711–718 [DOI] [PubMed] [Google Scholar]
- 10. Farre D, et al. 2003. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 31: 3651–3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fiorentino DF, Zlotnik A, Mosmann TR, Howard M, Ogarra A. 1991. Il-10 inhibits cytokine production by activated macrophages. J. Immunol. 147: 3815–3822 [PubMed] [Google Scholar]
- 12. Fujino RS, et al. 2006. Spermatogonial cell-mediated activation of an I kappa B zeta-independent nuclear factor-kappa B pathway in Sertoli cells induces transcription of the lipocalin-2 gene. Mol. Endocrinol. 20: 904–915 [DOI] [PubMed] [Google Scholar]
- 13. Fusek M, Vetvickova J, Vetvicka V. 2007. Secretion of cytokines in breast cancer cells: the molecular mechanism of procathepsin D proliferative effects. J. Interferon Cytokine Res. 27: 191–199 [DOI] [PubMed] [Google Scholar]
- 14. Goetz DH, et al. 2002. The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol. Cell 10: 1033–1043 [DOI] [PubMed] [Google Scholar]
- 15. Jung M, et al. 2012. Infusion of IL-10-expressing cells protects against renal ischemia through induction of lipocalin-2. Kidney Int. 81: 969–982 [DOI] [PubMed] [Google Scholar]
- 16. Lau KW, Tian YM, Raval RR, Ratcliffe PJ, Pugh CW. 2007. Target gene selectivity of hypoxia-inducible factor-alpha in renal cancer cells is conveyed by post-DNA-binding mechanisms. Br. J. Cancer 96: 1284–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. LeClair KP, Blanar MA, Sharp PA. 1992. The P50 subunit of NF-kappa-B associates with the NF-IL6 transcription factor. Proc. Natl. Acad. Sci. U. S. A. 89: 8145–8149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Leng XH, et al. 2009. Inhibition of lipocalin 2 impairs breast tumorigenesis and metastasis. Cancer Res. 69: 8579–8584 [DOI] [PubMed] [Google Scholar]
- 19. Lewis CE, Pollard JW. 2006. Distinct role of macrophages in different tumor microenvironments. Cancer Res. 66: 605–612 [DOI] [PubMed] [Google Scholar]
- 20. Li S, Wang N, Brodt P. 22 December 2011. Metastatic cells can escape the pro-apoptotic effects of TNF-alpha through increased autocrine IL-6/STAT3 signalling. Cancer Res. doi:10.1158/0008-5472.CAN-11-1357 [DOI] [PubMed] [Google Scholar]
- 21. Lin EY, et al. 2006. Macrophages regulate the angiogenic switch in a mouse model of breast cancer. Cancer Res. 66: 11238–11246 [DOI] [PubMed] [Google Scholar]
- 22. Malefyt RD, Abrams J, Bennett B, Figdor CG, Devries JE. 1991. Interleukin-10 (Il-10) inhibits cytokine synthesis by human monocytes - an autoregulatory role of Il-10 produced by monocytes. J. Exp. Med. 174: 1209–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mantovani A, Sica A, Locati M. 2005. Macrophage polarization comes of age. Immunity 23: 344–346 [DOI] [PubMed] [Google Scholar]
- 24. Mantovani A, et al. 2004. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 25: 677–686 [DOI] [PubMed] [Google Scholar]
- 25. Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. 2002. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 23: 549–555 [DOI] [PubMed] [Google Scholar]
- 26. Matsuo S, Yamazaki S, Takeshige K, Muta T. 2007. Crucial roles of binding sites for NF-kappa B and C/EBPs in I kappa B-zeta-mediated transcriptional activation. Biochem. J. 405: 605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Matsusaka T, et al. 1993. Transcription factors NF-IL6 and NF-kappa-B synergistically activate transcription of the inflammatory cytokines, interleukin-6 and interleukin-8. Proc. Natl. Acad. Sci. U. S. A. 90: 10193–10197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Messeguer X, et al. 2002. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 18: 333–334 [DOI] [PubMed] [Google Scholar]
- 29. Mishra J, et al. 2004. Amelioration of ischemic acute renal injury by neutrophil gelatinase-associated lipocalin. J. Am. Soc. Nephrol. 15: 3073–3082 [DOI] [PubMed] [Google Scholar]
- 30. Moore KW, Malefyt RD, Coffman RL, O'Garra A. 2001. Interleukin-10 and the interleukin-10 receptor. Annu. Rev. Immunol. 19: 683–765 [DOI] [PubMed] [Google Scholar]
- 31. Murdoch C, Giannoudis A, Lewis CE. 2004. Mechanisms regulating the recruitment of macrophages into hypoxic areas of tumors and other ischemic tissues. Blood 104: 2224–2234 [DOI] [PubMed] [Google Scholar]
- 32. Murray PJ. 2006. Understanding and exploiting the endogenous interleukin-10/STAT3-mediated anti-inflammatory response. Curr. Opin. Pharmacol. 6: 379–386 [DOI] [PubMed] [Google Scholar]
- 33. Numata A, et al. 2005. Signal transducers and activators of transcription 3 augments the transcriptional activity of CCAAT/enhancer-binding protein alpha in granulocyte colony-stimulating factor signaling pathway. J. Biol. Chem. 280: 12621–12629 [DOI] [PubMed] [Google Scholar]
- 34. Rahimi AA, Gee K, Mishra S, Lim W, Kumar A. 2005. STAT-1 mediates the stimulatory effect of IL-10 on CD14 expression in human monocytic cells. J. Immunol. 174: 7823–7832 [DOI] [PubMed] [Google Scholar]
- 35. Robb BW, Hershko DD, Paxton JH, Luo GJ, Hasselgren PO. 2002. Interleukin-10 activates the transcription factor C/EBP and the interleukin-6 gene promoter in human intestinal epithelial cells. Surgery 132: 226–231 [DOI] [PubMed] [Google Scholar]
- 36. Schmidt-Ott KM, et al. 2007. Dual action of neutrophil gelatinase-associated lipocalin. J. Am. Soc. Nephrol. 18: 407–413 [DOI] [PubMed] [Google Scholar]
- 37. Schottelius AJG, Mayo MW, Sartor RB, Badwin AS. 1999. Interleukin-10 signaling blocks inhibitor of kappa B kinase activity and nuclear factor kappa B DNA binding. J. Biol. Chem. 274: 31868–31874 [DOI] [PubMed] [Google Scholar]
- 38. Sica A, Allavena P, Mantovani A. 2008. Cancer related inflammation: the macrophage connection. Cancer Lett. 267: 204–215 [DOI] [PubMed] [Google Scholar]
- 39. Sola A, et al. 2011. Sphingosine-1-phosphate signalling induces the production of Lcn-2 by macrophages to promote kidney regeneration. J. Pathol. 225: 597–608 [DOI] [PubMed] [Google Scholar]
- 40. Stein B, Cogswell PC, Baldwin AS. 1993. Functional and physical associations between NF-kappa-B and C/Ebp family members—a Rel domain-Bzip interaction. Mol. Cell. Biol. 13: 3964–3974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stoesz SP, et al. 1998. Heterogeneous expression of the lipocalin NGAL in primary breast cancers. Int. J. Cancer 79: 565–572 [DOI] [PubMed] [Google Scholar]
- 42. Stout RD, et al. 2005. Macrophages sequentially change their functional phenotype in response to changes in microenvironmental influences. J. Immunol. 175: 342–349 [DOI] [PubMed] [Google Scholar]
- 43. Tanaka N, et al. 2005. Interleukin-10 induces inhibitory C/EBP beta through STAT-3 and represses HIV-1 transcription in macrophages. Am. J. Respir. Cell Mol. 33: 406–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tausendschon M, Dehne N, Brune B. 2011. Hypoxia causes epigenetic gene regulation in macrophages by attenuating Jumonji histone demethylase activity. Cytokine 53: 256–262 [DOI] [PubMed] [Google Scholar]
- 45. von Knethen A, Lotero A, Brune B. 1998. Etoposide and cisplatin induced apoptosis in activated RAW264.7 macrophages is attenuated by cAMP-induced gene expression. Oncogene 17: 387–394 [DOI] [PubMed] [Google Scholar]
- 46. Weber-Nordt RM, Gabler A, Janson C, Weber F, Ho AD. 1999. Constitutive STAT3-DNA binding coincides with a single amino acid exchange in the aminoterminal domain of the STAT3 protein in acute meyloid leukemia. Blood 94: 75a [Google Scholar]
- 47. Wen H, Andrejka L, Ashton J, Karess R, Lipsick JS. 2008. Epigenetic regulation of gene expression by Drosophila Myb and E2F2-RBF via the Myb-MuvB/dREAM complex. Genes Dev. 22: 601–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Williams L, Bradley L, Smith A, Foxwell B. 2004. Signal transducer and activator of transcription 3 is the dominant mediator of the anti-inflammatory effects of IL-10 in human macrophages. J. Immunol. 172: 567–576 [DOI] [PubMed] [Google Scholar]
- 49. Xiong H, et al. 2012. Roles of STAT3 and ZEB1 in E-cadherin downregulation and human colorectal cancer epithelial-mesenchymal transition. J. Biol. Chem. 287: 5819–5832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yan QW, et al. 2007. The adipokine lipocalin 2 is regulated by obesity and promotes insulin resistance. Diabetes 56: 2533–2540 [DOI] [PubMed] [Google Scholar]
- 51. Yu JL, Rak JW. 2003. Host microenvironment in breast cancer development—inflammatory and immune cells in tumour angiogenesis and arteriogenesis. Breast Cancer Res. 5: 83–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang KM, Guo W, Yang Y, Wu JR. 2011. JAK2/STAT3 pathway is involved in the early stage of adipogenesis through regulating C/EBP beta transcription. J. Cell Biochem. 112: 488–497 [DOI] [PubMed] [Google Scholar]