Abstract
Hepcidin is a liver-derived peptide hormone and the master regulator of systemic iron homeostasis. Decreased hepcidin expression is a common feature in alcoholic liver disease (ALD) and in mouse models of ethanol loading. Dysregulation of hepcidin signaling in ALD leads to liver iron deposition, which is a major contributing factor to liver injury. The mechanism by which hepcidin is regulated following ethanol treatment is unclear. An increase in liver hypoxia was observed in an acute ethanol-induced liver injury model. The hypoxic response is controlled by a family of hypoxia-inducible transcription factors (HIFs), which are composed of an oxygen-regulated alpha subunit (HIFα) and a constitutively present beta subunit, aryl hydrocarbon receptor nuclear translocator (HIFβ/Arnt). Disruption of liver HIF function reversed the repression of hepcidin following ethanol loading. Mouse models of liver HIF overexpression demonstrated that both HIF-1α and HIF-2α contribute to hepcidin repression in vivo. Ethanol treatment led to a decrease in CCAAT-enhancer-binding protein alpha (C/EBPα) protein expression in a HIF-dependent manner. Importantly, adenoviral rescue of C/EBPα in vivo ablated the hepcidin repression in response to ethanol treatment or HIF overexpression. These data provide novel insight into the regulation of hepcidin by hypoxia and indicate that targeting HIFs in the liver could be therapeutic in ALD.
INTRODUCTION
Patients with alcoholic liver disease (ALD) accumulate iron in the liver (23). Free iron enhances reactive oxygen species (ROS) production in the liver, leading to oxidative stress, which is a major contributing factor to alcohol-induced liver injury (53). The development of liver fibrosis positively correlates with liver iron staining in ALD, and the presence of iron deposits in the liver of patients with alcoholic cirrhosis is predictive of death (10). There is compelling evidence that iron-mediated oxidative stress may be an important pathological mechanism for the increased incidence of hepatocellular carcinoma in individuals with hepatic iron overload who consume alcohol (2). Lastly, hepatic iron overload increases the risk of insulin resistance and diabetes due to hepatic inflammation (25).
Hepcidin is a small antimicrobial peptide produced in the liver and secreted into the bloodstream which regulates systemic iron homeostasis (24, 33). Hepcidin functions by binding to the only known mammalian iron exporter, ferroportin (FPN), which leads to its internalization and degradation (28). FPN is primarily expressed on macrophages of the reticuloendothelial system and absorptive enterocytes in the small intestine (6). Thus, hepcidin acts to restrict intestinal iron absorption and prevent release of iron from stores. Conversely, hepcidin deficiency leads to increased iron absorption and mobilization of iron stores, which can cause iron overload (6, 29). Previous publications have shown that in rodents and humans, hepcidin is downregulated in response to ethanol treatment (5, 11, 14, 16, 17, 19, 20, 31). Moreover, it has been shown that the increase in liver iron following alcohol consumption in ALD patients is directly due to low hepcidin expression (5, 11, 14, 16, 17, 19, 20, 31). However, the mechanism(s) by which hepcidin is dysregulated following ethanol exposure remains unclear.
The present study demonstrated that alcohol treatment in mice led to a robust hypoxic response compared to that seen with vehicle-treated mice and that this hypoxia was associated with a strong inhibition of hepcidin expression. Changes in gene expression during hypoxia are primarily controlled by a family of hypoxia-inducible transcription factors (HIFs) (43). HIFs consist of an oxygen-regulated alpha subunit (HIFα) and a constitutively expressed beta subunit, aryl hydrocarbon receptor nuclear translocator (HIFβ/Arnt). Under conditions of normal oxygen tension, the HIFα subunit is hydroxylated by prolyl hydroxylase enzymes (PHDs) at specific proline residues. This modification is recognized by the protein coded by the Von Hippel-Lindau tumor suppressor gene (Vhl), which is part of an E3 ubiquitin ligase complex. VHL facilitates ubiquitination of the HIFα subunit, which leads to its subsequent degradation by the proteasome. Genetic disruption of Vhl results in the presence of constitutively active HIF in vivo and is a well-characterized model to study HIF function (13, 21). Under conditions of low oxygen, PHD activity is decreased and the HIFα subunit is stabilized, which allows it to accumulate in the cytoplasm and then translocate into the nucleus and heterodimerize with Arnt. The HIF complex binds to promoters of many genes involved in the hypoxic response and activates their transcription (42).
Hypoxia is known to repress hepcidin, and several mechanisms for the hypoxic repression have been proposed (7, 30, 35, 49). However, the precise mechanism is still unclear. Moreover, the role of hypoxia in the repression of hepcidin in ALD has not been examined. The present report clearly demonstrates that alcohol-induced hepcidin repression can be blocked by a conditional disruption of Arnt in the hepatocytes, which leads to total loss of HIF transcriptional activity. Furthermore, we identified a novel signaling cascade that involved HIF-mediated degradation of CCAAT-enhancer-binding protein alpha (C/EBPα). C/EBPα overexpression rescued the repression of hepcidin observed with ethanol loading and partially reversed the repression caused by HIF overexpression. These data demonstrate a novel connection between hypoxia and hepcidin expression in vivo and could lead to development of therapies targeting HIFs or C/EBPα in the liver of ALD patients.
MATERIALS AND METHODS
Animal experiments.
VhlF/F, VhlF/F Hif-1αF/F, VhlF/F Hif-2αF/F, and VhlF/F ArntF/F mice were previously described (46) and are in the same genetic background (129S6/SvEv). Temporal liver-specific knockouts constructed using serum albumin enhancer-driven tamoxifen-inducible estrogen receptor-conjugated Cre (SA-Cre-ERT2) were achieved as previously described (38). In these animals, tamoxifen treatment causes the estrogen receptor Cre fusion protein to translocate to the nucleus, which leads to Cre-mediated excision of floxed genes. The hypoxia reporter mouse (oxygen-dependent-degradation domain–luc [ODD-luc]) animal model was obtained from Jackson Laboratories (Bar Harbor, ME) and has been described previously (40). For viral expression, animals were administered 1 × 109 PFU of purified virus in 200 μl of normal saline solution by tail vein injection. Blood was collected by submandibular bleeding. For ex vivo imaging experiments, animals were administered d-luciferin (Promega Corp., Madison, WI) (50 mg/kg of body weight) by intraperitoneal injection, and tissues were imaged after 15 min using an IVIS 200 imaging system (Caliper Life Sciences, Hopkinton, MA). For tamoxifen experiments, tamoxifen (Sigma-Aldrich, St. Louis, MO) (2 mg in corn oil) was injected intraperitoneally once daily for two consecutive days and mice were sacrificed 5 days or 2 weeks later. For ethanol experiments, mice were gavaged every 12 h with increasing doses of 25% ethanol starting at 5 mg/kg and increasing by 0.5-mg/kg increments each treatment. Animals were euthanized 12 h after the last gavage of 7.5 mg/kg. The mice were housed in a light- and temperature-controlled room and were given water and chow ad libitum. Tissues were harvested and used fresh or were flash-frozen in liquid nitrogen and stored at −80°C for future use. All animal studies were carried out in accordance with Institute of Laboratory Animal Resources guidelines and approved by the University Committee on the Use and Care of Animals at the University of Michigan.
Cell culture.
Huh7 and HEK293A cells were maintained at 37°C in 5% CO2 and 21% O2. Cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and antibiotics-antimycotics. Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer, and Western blot analysis was performed. For transfection experiments, HEK293A cells were transfected using FuGENE 6 transfection reagent (Promega Corporation, Madison, WI) 24 h before hypoxia treatment. For adenoviral infection experiments, cells were treated at a multiplicity of infection (MOI) of virus of 100 24 h before sample collection. Viruses were purified by the University of Michigan Vector Core. MG132 (Cayman Chemical, Ann Arbor, MI) was used at a concentration of 10 μM for 24 h. For hypoxia experiments, cells were incubated in 1% O2 and 5% CO2 with balance N2 at 37°C for 24 h.
Western blotting.
Tissues were homogenized and lysed in RIPA buffer for whole-cell extracts, and nuclear proteins were isolated using an NE-PER nuclear extraction kit (Pierce, Rockford, IL). Proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes using standard methods. Membranes were incubated with antibodies against C/EBPα (sc-61), histone H1 (sc-8030), HIF-1α (sc-10790), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (sc-25778; Santa Cruz Biotech, Santa Cruz, CA). Histone H3 (catalog no. 4499) and pSmad1, -5, and -8 (pSmad1/5/8; catalog no. 9511) antibodies were from Cell Signaling Technology (Danvers, MA). HIF-2α (NB100-122) antibody was from Novus (Novus Biologicals, Littleton, CO). All antibodies were used at a 1:1,000 dilution.
Real-time quantitative PCR.
RNA from fresh or frozen tissue and cells was isolated using Isol-RNA lysis reagent (Prime, Gaithersburg, MD) and subjected to reverse transcription using Moloney murine leukemia virus (MMLV) reverse transcriptase (RT; Fisher Scientific, Waltham, MA). cDNA was quantified using SYBR green dye and run on a 7900HT Fast real-time RT-PCR system (primers are listed in Table S1 in the supplemental material). Threshold cycle (CT) values were normalized to β-actin and expressed as fold differences from control values.
Site-directed mutagenesis.
Site-directed mutagenesis was performed using a QuikChange II XL site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA) (primers are listed in Table S1 in the supplemental material).
Measurement of serum EPO.
Serum erythropoietin (EPO) was measured using a mouse enzyme-linked immunosorbent assay (ELISA) kit per the manufacturer's recommendation (Abcam, Cambridge, MA).
RESULTS
Repression of hepcidin during ethanol loading is independent of the Bmp/Smad signaling pathway.
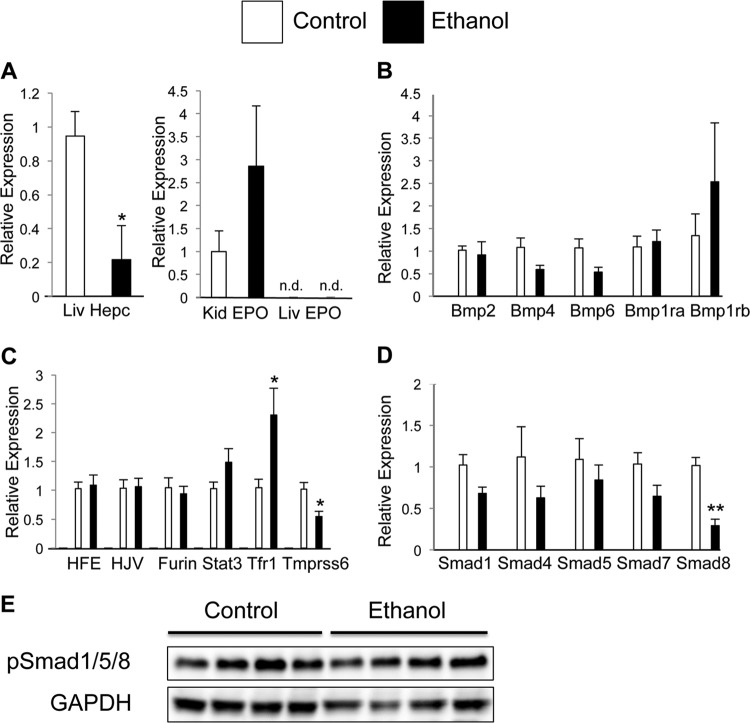
Mice were treated with increasing concentrations of ethanol by oral gavage every 12 h for a total of 5 doses. This model parallels the binge drinking observed in a significant proportion of ALD patients. Livers collected from control and ethanol-loaded mice were examined for gene expression. Hepcidin expression was strongly repressed in response to ethanol treatment (Fig. 1A), whereas EPO expression in the kidney was not statistically significant (P = 0.23) (Fig. 1A). In order to determine the mechanism by which hepcidin is repressed, a number of known hepcidin regulatory genes were examined by real-time quantitative PCR (qPCR). No changes in bone morphogenetic protein (Bmp) ligands or their respective receptors were observed following ethanol treatment (Fig. 1B). There was an increase in transferrin receptor 1 (Tfr1) expression (Fig. 1C). Moreover, ethanol treatment significantly decreased expression of Tmprss6, which is a critical regulator of hepcidin expression (Fig. 1C) (45). However, these changes could not account for the repression of hepcidin expression seen following ethanol treatment, as Tmprss6 is a negative regulator of hepcidin expression and an increase in Tfr1 expression should increase liver iron uptake and thus increase hepcidin expression. There was a significant repression of mothers against decapentaplegic homolog 9 (Smad8) (Fig. 1D). The Smad family of transcription factors is well characterized in their regulation of hepcidin expression (22). However, when phosphorylated proteins Smad1, -5, and -8 were examined by Western blot analysis, no significant changes were found (Fig. 1E), demonstrating that an alternate mechanism is responsible for the repression of hepcidin.
Fig 1.
Repression of hepcidin during ethanol treatment is independent of Bmp/Smad and EPO signaling. Following ethanol treatment, gene expression was determined by qPCR analysis of (A) kidney (Kid) EPO and liver (Liv) EPO and hepcidin (Hepc) expression, (B) liver expression of Bmp ligands and receptors, (C) hepcidin regulatory genes, and (D) Smads. mRNA expression was normalized to β-actin. (E) Western blot analysis of pSmad1/5/8 protein from whole-cell extracts of liver following ethanol treatment. Loading was normalized to GAPDH. n.d., not detected. n = 6 to 8 animals per group. Each bar represents the mean ± standard error (SE). *, P < 0.05 as determined by Student's t test; **, P < 0.01.
Hypoxic signaling is critical in ethanol-mediated hepcidin repression.
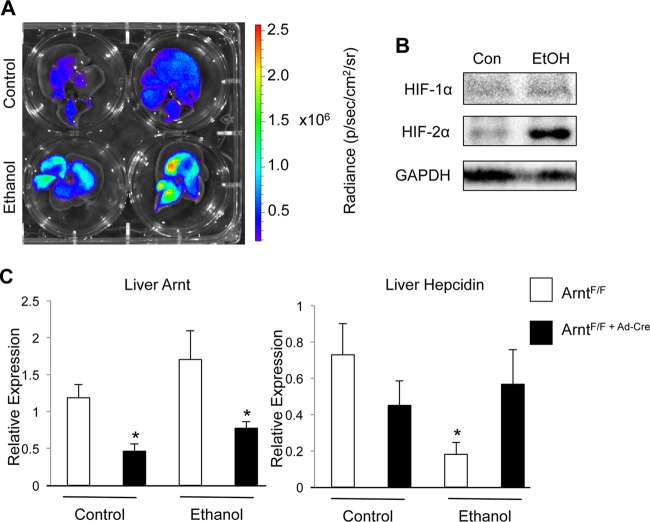
In order to test the possibility that hypoxia is involved in the repression of hepcidin during ethanol loading, the hypoxia reporter mouse (ODD-luc) was utilized. This mouse model expresses a fusion protein consisting of the oxygen-dependent-degradation domain (ODD) from HIF-1α fused to luciferase. This protein is constitutively expressed in all tissues but is stable only under conditions where endogenous HIF would also be stabilized. Thus, this model allows detection of hypoxia in live mice or specific detection of hypoxia in a tissue ex vivo (40). ODD-luc mice treated with ethanol showed a strong induction of hypoxia in the liver, as seen by ex vivo imaging (Fig. 2A). In addition, Western blot analysis was performed on liver nuclear extracts from control and ethanol-treated mice. HIF-2α expression was strongly increased in response to ethanol treatment (Fig. 2B). HIF-1α was not detected, although it is possible that HIF-1α is stabilized acutely following ethanol treatment and is degraded more rapidly than HIF-2α. To assess if liver hypoxia is critical for hepcidin repression following ethanol treatment, intravenous administration of adenoviral Cre, which traffics specifically to the liver (44), was used to induce recombination in mice containing loxP recombination sites flanking exon 6 of Arnt, referred to as ArntF/F mice (47). Arnt is the obligate heterodimer partner for HIF1α, HIF2α, and HIF3α; thus, disruption results in complete inactivation of HIF signaling. This model of Arnt knockdown is preferable to an albumin-Cre-mediated recombination, as it eliminates the possibility of developmental effects on hepcidin expression (51). Arnt knockdown was validated by qPCR (Fig. 2C). Control mice with intact Arnt expression in the liver had decreased hepcidin expression following ethanol treatment. Disruption of Arnt in the liver completely blocked repression of hepcidin in response to ethanol loading (Fig. 2C). These data suggest that HIF signaling is essential in hepcidin repression by ethanol treatment.
Fig 2.
HIF signaling in the liver is required for repression of hepcidin expression by ethanol treatment. ODD-luc mice were treated with ethanol and imaged using an IVIS 200 imaging system. (A) Representative livers from two control and two ethanol-treated ODD-luc mice. (B and C) Western blot analysis of HIF-1α, HIF-2α, and histone 3 in liver nuclear extracts from control (Con) and ethanol-treated (EtOH) mice. Liver-specific Arnt disruption was induced by tail vein injection of adenovirus expressing Cre recombinase (Ad-Cre) 3 days before ethanol treatment, and (B) hepcidin and (C) Arnt gene expression was analyzed in control mice (ArntF/F) and Arnt knockout mice (ArntF/F+Ad-Cre). Gene expression was normalized to β-actin. n = 6 to 8 animals per group. Each bar represents the mean ± SE. Asterisks denote statistical significance versus control results at P < 0.05 as determined by Student's t test.
Repression of hepcidin expression by HIFs does not require erythropoiesis.
Hypoxia is a well-known but poorly understood repressor of hepcidin expression. An initial connection between hypoxia and hepcidin expression was made in which direct HIF binding to the hepcidin promoter led to repression (35). However, this finding was not uniformly observed (49), thereby leaving the relationship between HIF and hepcidin unclear. A recent study demonstrated that overexpression of HIFs in the liver caused a decrease in hepcidin expression that required erythropoiesis (26). However, reports of studies that examined erythropoietic hepcidin repression make no mention of the repression of hepcidin in alcohol loading (3, 9, 48). In addition, a study of VhlR200W homozygote humans, who have activated HIF signaling, demonstrated reduced hepcidin levels that did not correlate with an increase in serum EPO, suggesting that HIFs could repress hepcidin independently of erythropoiesis (12).
To understand the role of HIFs in hepcidin repression and to avoid the confounding developmental defects of Vhl deletion, a model of inducible liver gene knockout was used (38). Tissues that lack Vhl are unable to degrade the HIFα subunit under normal oxygen conditions, leading to constitutive HIF stability and activity. VhlF/F mice were crossed with SA-Cre-ERT2 transgenic mice to generate a temporal and conditional disruption of Vhl (VhlLivKO) using the tamoxifen-inducible cre fusion protein (41). In these mice, Cre-mediated recombination specifically in the liver is induced by tamoxifen treatment, allowing temporal control over gene expression. Moreover, to assess the influence of HIF-dependent pathways on iron-regulatory gene expression in the liver, mice with a double disruption of Vhl and Hif-1α or Hif-2α (Vhl/Hif-1αLivKO and Vhl/Hif-2αLivKO) were also assessed. These compound knockouts remove the possibility that the effects of Vhl ablation on hepcidin expression are the result of a HIF-independent mechanism. By the use of compound conditional liver knockouts, mice with loss of Vhl alone or in conjunction with HIF-1α demonstrated a robust decrease in hepcidin levels, high EPO expression, and a large increase in circulating EPO 2 weeks after induction of Cre by tamoxifen treatment (Table 1). Double disruption of Arnt and Vhl (Vhl/ArntLivKO) completely inhibits all HIFα function, since it is an obligate heterodimer partner for both HIF-1α and HIF-2α. The livers of the Vhl/ArntLivKO mice showed a loss of hepcidin repression, demonstrating a HIF-dependent mechanism. Surprisingly, compound deletion of Vhl and HIF-2α still led to strong hepcidin repression despite the absence of any increase in circulating EPO levels (Table 1). These data suggest that there is a HIF-dependent mechanism of hepcidin repression that does not require enhanced erythropoiesis.
Table 1.
HIF represses hepcidin through EPO-dependent and -independent mechanisms
| Parameter and genotype | Value | SD | n |
|---|---|---|---|
| Liver hepcidin | |||
| VhlF/F | 1.000 CT fold differencea | 0.568 | 5 |
| VhlLivKO | 0.004 CT fold differenceb | 0.004 | 5 |
| VhlLivKO Hif-1αLivKO | 0.024 CT fold differencec | 0.047 | 4 |
| VhlLivKO Hif-2αLivKO | 0.092 CT fold differencec | 0.034 | 4 |
| VhlLivKO ArntLivKO | 0.872 CT fold difference | 0.326 | 4 |
| Liver EPO | |||
| VhlF/F | 1.000 CT fold difference | 0.797 | 5 |
| VhlLivKO | 1,237.208 CT fold differenceb | 201.080 | 5 |
| VhlLivKO Hif-1αLivKO | 1,629.958 CT fold differenceb | 617.123 | 4 |
| VhlLivKO Hif-2αLivKO | 38.632 CT fold differenceb | 15.223 | 4 |
| VhlLivKO ArntLivKO | 8.997 CT fold differenceb | 3.594 | 4 |
| Circulating EPO | |||
| VhlF/F | 145 ng/ml | 113 | 5 |
| VhlLivKO | 940 ng/mlb | 336 | 5 |
| VhlLivKO Hif-1αLivKO | 985 ng/mlb | 277 | 4 |
| VhlLivKO Hif-2αLivKO | 206 ng/ml | 59 | 4 |
CT fold difference data represent mRNA normalized to β-actin.
Data denote significance at P < 0.01 versus control results by Student's t test.
Data denote significance at P < 0.05 versus control results by Student's t test.
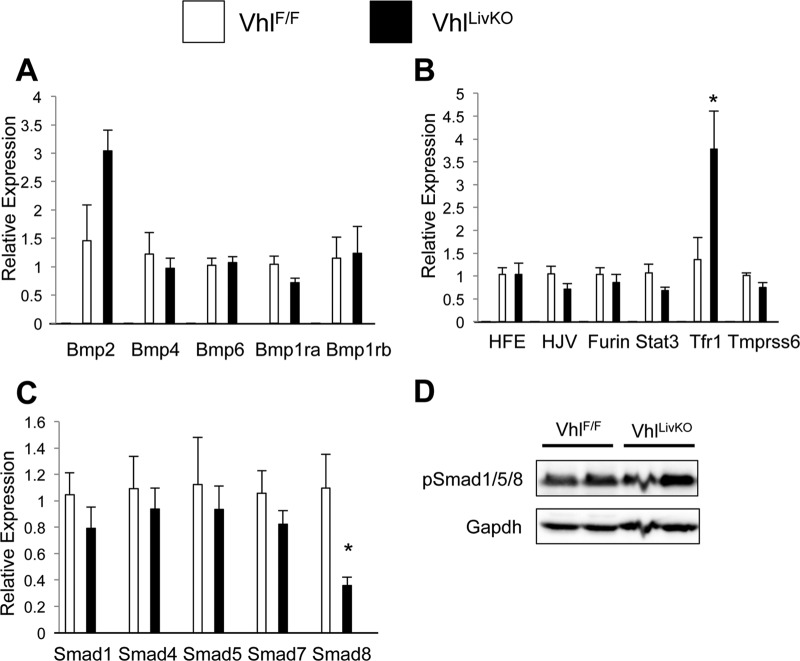
Examination of gene expression in the liver Vhl knockout mice after 5 days demonstrated no change in Bmp ligand or receptor expression (Fig. 3A). Consistent with ethanol treatment, Tfr1 expression was increased and Smad8 mRNA expression was significantly decreased following liver Vhl knockout (Fig. 3B and C). However, there was no detectable decrease in the levels of phosphorylated Smad proteins (Fig. 3D). These data establish that there is a HIF-dependent but Bmp/Smad- and erythropoietin-independent pathway for hepcidin repression.
Fig 3.
Hepcidin repression by HIFs is independent of Bmp/Smad signaling. (A, B, and C) VhlF/F and VhlF/F;AlbERcre mice were administered tamoxifen (TM), and then mRNAs from livers of the mice were examined 5 days later by qPCR analysis of (A) Bmp ligand and receptor expression, (B) hepcidin regulatory genes, and (C) Smads. mRNA expression was normalized to β-actin. (D) Western blot analysis was performed on whole-cell extracts of livers from VhlF/F and VhlF/F;AlbERcre mice treated with tamoxifen (TM) for 5 days and examined for pSmad1/5/8 expression. Loading was normalized to GAPDH. n = 6 to 8 animals per group. Each bar represents the mean ± SE. Asterisks denote statistical significance versus control results at P < 0.05 as determined by Student's t test.
A decrease in liver C/EBPα protein expression is responsible for hepcidin repression by HIFs.
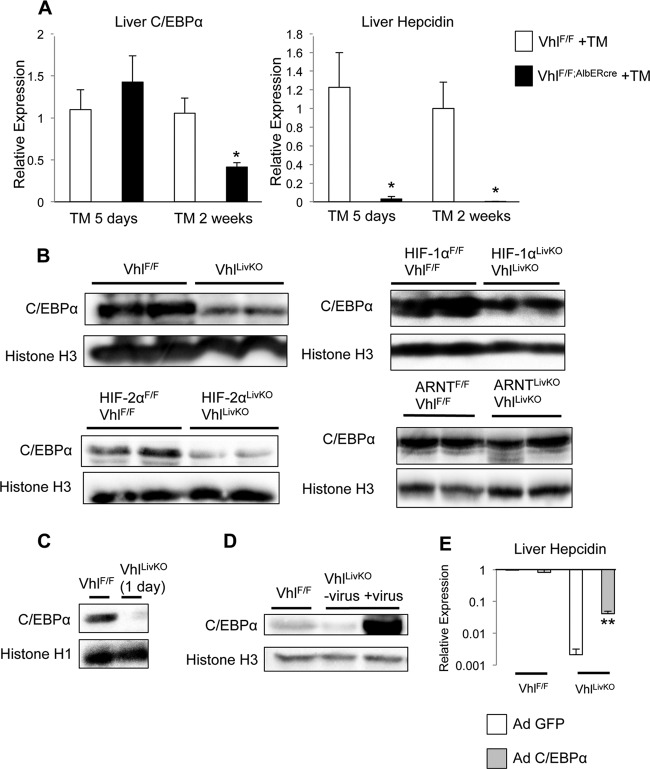
C/EBPα is a transcription factor that is also critical in maintaining basal hepcidin expression (8). In order to determine whether C/EBPα is involved in the repression of hepcidin expression by HIFs, conditional deletion of Vhl was performed for different amounts of time. Five days following induction of Cre recombination by tamoxifen treatment, hepcidin expression was strongly repressed without a decrease in C/EBPα mRNA levels (Fig. 4A). Two weeks after recombination, hepcidin mRNA and C/EBPα mRNA levels were both decreased (Fig. 4A). Examination of C/EBPα protein expression by Western blot analysis showed that liver Vhl knockout alone or in conjunction with HIF-1α led to a significant decrease in the protein expression (Fig. 4B). EPO has been shown to decrease C/EBPα expression (36). To completely rule out the role of EPO, C/EBPα protein was examined in the mice with the compound Vhl and HIF-2α knockout. These mice had no increase in EPO or erythropoiesis. The Vhl and HIF-2α double-knockout mice demonstrated lower C/EBPα protein expression (Fig. 4B), whereas the Vhl and Arnt double-knockout mice showed no change in C/EBPα protein expression (Fig. 4B). C/EBPα protein expression was decreased as early as 1 day after induction of Cre by tamoxifen, demonstrating that the decrease in C/EBPα expression is an early event (Fig. 4C). To test whether C/EBPα could rescue hepcidin expression in liver Vhl knockout mice, animals were infected with an adenoviral construct expressing C/EBPα. Successful adenoviral overexpression was confirmed by Western blot analysis (Fig. 4D). Infection of Vhl knockout animals with C/EBPα led to a substantial derepression of hepcidin expression (Fig. 4E). To confirm that the green fluorescent protein (GFP)-expressing adenovirus infection did not affect hepcidin levels, mice infected with adenovirus expressing GFP demonstrated levels of expression of hepcidin similar to those seen with uninfected mice (see Fig. S1 in the supplemental material). These data demonstrate that the repression of hepcidin expression by HIFs is mediated by a decrease in C/EBPα protein expression.
Fig 4.
Hepcidin repression by HIF requires a decrease in C/EBPα protein expression. (A) VhlF/F and VhlF/F;AlbERcre mice were treated with tamoxifen (TM) and sacrificed either 5 days or 2 weeks following Cre recombination, and liver expression of C/EBPα and hepcidin was determined by qPCR and normalized to β-actin. (B) Western blot analysis was performed on nuclear extracts to detect C/EBPα in VhlF/F and VhlF/F;AlbERcre mice or in the Vhl compound-disrupted mice following 2 weeks of tamoxifen (TM) treatment. (C) C/EBPα protein expression in the livers of VhlF/F and VhlF/F;AlbERcre mice was examined by Western blot analysis 1 day following tamoxifen (TM) treatment. (D) Mice were infected with recombinant C/EBPα adenovirus (Ad-C/EBPα) and 3 days later examined for C/EBPα protein expression by Western blot analysis. Loading was normalized to histone H1 or histone H3. (E) VhlF/F and VhlF/F;AlbERcre mice were infected with adenovirus for 3 days and then treated with tamoxifen (TM) for an additional 5 days, and hepcidin expression was assessed by qPCR normalized to β-actin. n = 6 to 8 animals per group. Each bar represents the mean ± SE. Single asterisks denote statistical significance versus control results at P < 0.05 as determined by Student's t test. **, P < 0.01.
Hypoxia promotes proteasomal degradation of C/EBPα.
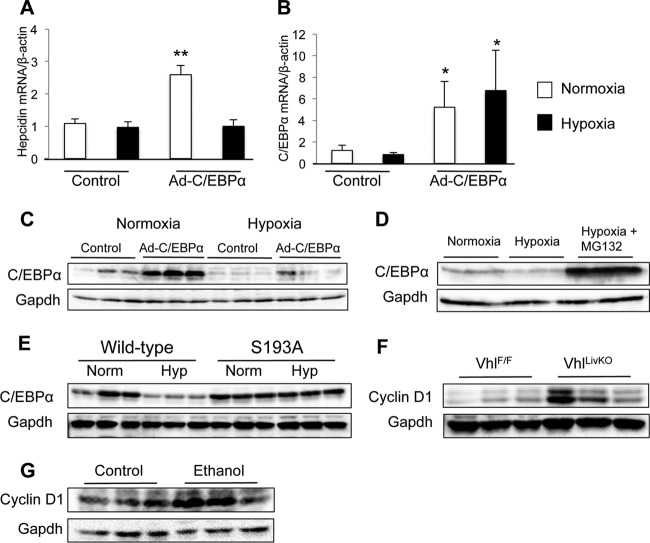
Since the decrease in C/EBPα protein expression was observed independently of a change in mRNA, the possibility that hypoxia affects C/EBPα protein stability was tested. Huh7 cells, representing a human hepatoma cell line, have been used in multiple studies to examine regulatory mechanisms of hepcidin expression (4, 7, 27, 39). Treatment of Huh7 cells with hypoxia caused no change in endogenous hepcidin expression (Fig. 5A). However, adenoviral overexpression of C/EBPα increased hepcidin expression, and this increase was abolished by hypoxia treatment (Fig. 5A). In addition, hypoxia treatment did not decrease endogenous or exogenous C/EBPα mRNA expression (Fig. 5B). Western blot analysis of Huh7 protein extracts showed that hypoxia led to a decrease in exogenous levels of C/EBPα (Fig. 5C). There was no substantial decrease in the presence of endogenous C/EBPα, which is expressed at very low levels in this cell line. This could explain why hypoxia did not decrease endogenous hepcidin expression. Studies have shown that C/EBPα is degraded through the ubiquitin-proteasome pathway (50). To determine if this process is responsible for hypoxia-dependent C/EBPα destabilization, cultured Huh7 cells were treated with the proteasome inhibitor MG132 under hypoxia conditions. Proteasome inhibition drastically increased C/EBPα protein stability under hypoxia conditions (Fig. 5D). A previous publication showed that phosphorylation at serine 193 was critical for proteasomal degradation of C/EBPα (50). To determine if phosphorylation at this site was important for HIF-mediated C/EBPα degradation, we transfected HEK293A cells with either a wild-type or an S193A-mutated C/EBPα expression construct and subjected the cells to normoxia and hypoxia. HEK293A cells were used, since these cells are easily transfected with plasmid DNA. Similar to Huh7 cells, wild-type C/EBPα is degraded under hypoxia conditions in HEK293A cells (Fig. 5E). However, the S193A mutation abolished the protein degradation, indicating that phosphorylation at this site is essential for HIF-mediated C/EBPα degradation. Cyclin-dependent kinase 4 has been shown to phosphorylate C/EBPα at S193 and is regulated by cyclin D. To determine whether HIFs could cause an increase in CDK activity, Western blot analysis was performed on liver whole-cell extracts from liver Vhl knockout mice. These animals have an increase in liver cyclin D1 protein (Fig. 5F). Moreover, an increase in cyclin D1 protein was also detected in the livers of ethanol-treated mice (Fig. 5G). In conjunction with the in vivo data, these experiments demonstrate that HIF-dependent hepcidin repression is mediated by destabilization of C/EBPα protein.
Fig 5.
Proteasomal degradation of C/EBPα is necessary for HIF-dependent hepcidin repression. (A, B, and C) Huh7 cells were infected with Ad-C/EBPα at an MOI of 100 1 day before exposure to hypoxia (1% O2) for 24 h. Gene expression was examined by qPCR for (A) hepcidin and (B) C/EBPα, and (C) Western blot analysis was performed on whole-cell extracts to detect C/EBPα. (D) Western blot analysis for C/EBPα in Huh7 cells infected with Ad-C/EBPα at an MOI of 100 1 day before exposure to hypoxia (Hyp) (1% O2) and 10 μM MG132 for 24 h. (E) HEK293A cells were transfected with wild-type or S193A mutant C/EBPα expression constructs and exposed to normoxia (Norm) or hypoxia (Hyp) for 24 h, and C/EBPα expression was examined by Western blot analysis. (F) Liver Vhl knockout mice were analyzed for liver cyclin D1 expression by Western blot analysis in whole-cell extracts 2 weeks after Cre recombination. (G) Control and ethanol-treated animals were examined for liver cyclin D1 expression by Western blot analysis in whole-cell extracts. Loading was normalized to GAPDH. n = 3 samples per group; the experiments were repeated at lest three times. Each bar represents the mean ± SE. Single asterisks denote statistical significance versus control results at P < 0.05 as determined by Student's t test. **, P < 0.01.
A HIF-dependent decrease in C/EBPα protein stability is responsible for hepcidin repression in ethanol loading.
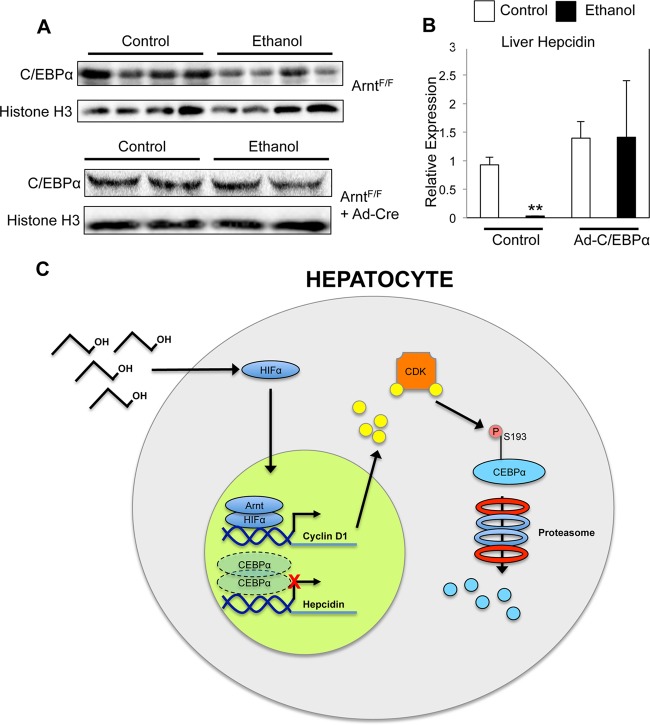
To determine whether the HIF-mediated C/EBPα protein decrease was relevant to hepcidin repression following ethanol treatment, Western blot analysis for C/EBPα was performed on livers from control and ethanol-treated mice. Ethanol treatment caused a decrease in C/EBPα protein expression in the liver (Fig. 6A). However, mice deficient for Arnt expression in the liver showed no decrease in C/EBPα protein expression following ethanol treatment (Fig. 6A). To test if C/EBPα overexpression could rescue the ethanol-induced repression of hepcidin, mice were infected with an adenoviral vector expressing C/EBPα and then treated with ethanol. C/EBPα overexpression prevented the decrease in hepcidin expression following ethanol treatment (Fig. 6B).
Fig 6.
C/EBPα overexpression in vivo blocks repression of hepcidin by ethanol treatment. (A) Liver-specific Arnt disruption was induced by tail vein injection of adenovirus expressing Cre recombinase (Ad-Cre) 3 days before ethanol treatment, and Western blot analysis was performed to detect C/EBPα in control and ethanol-treated livers. Protein loading was normalized to histone H3. (B) Mice were infected with adenovirus expressing C/EBPα (Ad-C/EBPα) 3 days before ethanol treatment, and hepcidin expression was analyzed by qPCR. Expression was normalized to β-actin. (C) A schematic diagram illustrating the mechanism by which alcohol-induced activation of HIF controls hepcidin repression through Cdk-dependent C/EBPα degradation. n = 6 to 8 animals per group. Each bar represents the mean ± SE. **, P < 0.01 (Student's t test).
DISCUSSION
Iron overload in ALD contributes to oxidative stress and tissue damage (23). Investigating the mechanisms behind ethanol-mediated hepcidin repression could lead to new therapies to combat iron overload in ALD. This report provides novel insight not only into the mechanisms of hepcidin repression following ethanol loading but also into the role of HIFs in controlling hypoxic hepcidin repression. Mechanisms of hepcidin repression that involve hypoxia, such as high-altitude exposure, hemolytic anemia, and phlebotomy, are all complicated by increased erythropoiesis (1, 3, 37). This study took advantage of a temporal liver-specific double disruption of Vhl and HIF-2α, which strongly represses hepcidin expression without an increase in the presence of circulating erythropoietin. This model allowed delineation of hypoxic and erythropoietic pathways for hepcidin expression. Also, the ethanol-loading model described in this report represents the first demonstration of the physiological consequence of liver hypoxia in the absence of increased erythropoiesis.
The present report demonstrates that ethanol-mediated repression of hepcidin is accomplished through a HIF-dependent decrease in C/EBPα protein stabilization. The proposed mechanism is that ethanol treatment stabilizes HIF in hepatocytes, leading to C/EBPα degradation by the proteasome and thus preventing it from activating hepcidin (Fig. 6C). The HIF-dependent changes that lead to this destabilization remain unclear. Previous studies have shown that C/EBPα can be phosphorylated by cyclin-dependent kinases (Cdk) at serine 193, subsequently leading to its ubiquitination and degradation through the ubiquitin-proteasome pathway (50). Mutation of serine 193 to alanine blocks hypoxia-induced degradation, suggesting that the HIF and Cdk pathways may interact to control C/EBPα stability. Notably, cyclin D1, an important cofactor for Cdk activity, is upregulated in a HIF-dependent manner (18, 54).
Prior studies have shown that ethanol treatment can lead to a decrease in C/EBPα mRNA and DNA-binding activity (5, 15, 17). Moreover, EPO has been shown to decrease levels of C/EBPα mRNA. In the present study, 5 days after induction of HIF overexpression, in experiments utilizing the conditionally and temporally disrupted Vhl mice, hepcidin expression was potently repressed and C/EBPα mRNA expression was unchanged (Fig. 4A). Two weeks after HIF overexpression, both C/EBPα expression and hepcidin mRNA expression were strongly repressed (Fig. 4A). At that later time point, the decrease in C/EBPα mRNA expression was likely mediated by EPO signaling through its receptor, as a previous publication demonstrated that an EPO receptor antibody blocked erythropoietic C/EBPα mRNA repression (36). The rapid destabilization of C/EBPα protein by HIFs represents a novel mechanism for HIF-C/EBPα interactions. The data presented herein demonstrate that HIF-mediated C/EBPα destabilization plays an important role in the liver hypoxic response in vivo. This raises the question of whether the mechanism for C/EBPα degradation during hypoxia is intact in other tissues as well. For example, mutations in C/EBPα have been identified in multiple cases of acute myeloid leukemia (AML) (32) and C/EBPα is dysregulated in more than half of patients with the disease (34). In addition, it has been shown that HIF inhibition can selectively kill human AML cancer stem cells (52). Further exploration of this pathway in additional cell types could lead to a better understanding of the pathogenesis of C/EBPα-related diseases, which could improve therapy.
HIF repression of hepcidin is a well-documented observation, and several putative mechanisms have been proposed (7, 30, 35, 49). Through an in vivo mechanistic assessment, the data in the present paper identify a novel erythropoietin-independent mechanism by which HIF represses hepcidin expression. It is possible that low levels of hepcidin allow increased activity of iron-dependent enzymes and processes, which could be beneficial, but the physiological role of this repression is completely unclear. No publications to date have shown the importance of HIF repression of hepcidin in maintaining systemic iron homeostasis. Although its normal physiological role is not apparent, the present report definitively demonstrates that alcohol induces pathological hypoxia and that HIF activity is essential in alcohol-mediated hepcidin repression. This finding may be critical in identifying therapies to limit iron-induced injury in ALD patients.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a Rackham predoctoral fellowship (E.R.A.), NIH grants CA148828 and DK095201 and the University of Michigan Gastrointestinal Peptide Center (Y.M.S.), NIH grant DK47918 (M.B.O.), and institutional NIH P30 grant DK34933.
We declare no conflicts of interest related to this work.
Footnotes
Published ahead of print 6 August 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Anderson ER, Xue X, Shah YM. 2011. Intestinal hypoxia-inducible factor-2alpha (HIF-2alpha) is critical for efficient erythropoiesis. J. Biol. Chem. 286:19533–19540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Asare GA, Bronz M, Naidoo V, Kew MC. 2008. Synergistic interaction between excess hepatic iron and alcohol ingestion in hepatic mutagenesis. Toxicology 254:11–18 [DOI] [PubMed] [Google Scholar]
- 3. Bondi A, et al. 2005. Hepatic expression of hemochromatosis genes in two mouse strains after phlebotomy and iron overload. Haematologica 90:1161–1167 [PubMed] [Google Scholar]
- 4. Braliou GG, et al. 2008. 2-Oxoglutarate-dependent oxygenases control hepcidin gene expression. J. Hepatol. 48:801–810 [DOI] [PubMed] [Google Scholar]
- 5. Bridle K, et al. 2006. Hepcidin is down-regulated in alcoholic liver injury: implications for the pathogenesis of alcoholic liver disease. Alcohol. Clin. Exp. Res. 30:106–112 [DOI] [PubMed] [Google Scholar]
- 6. Chaston T, et al. 2008. Evidence for differential effects of hepcidin in macrophages and intestinal epithelial cells. Gut 57:374–382 [DOI] [PubMed] [Google Scholar]
- 7. Chaston TB, et al. 2011. Hypoxia inhibits hepcidin expression in HuH7 hepatoma cells via decreased SMAD4 signaling. Am. J. Physiol. Cell Physiol. 300:C888–C895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Courselaud B, et al. 2002. C/EBPalpha regulates hepatic transcription of hepcidin, an antimicrobial peptide and regulator of iron metabolism. Cross-talk between C/EBP pathway and iron metabolism. J. Biol. Chem. 277:41163–41170 [DOI] [PubMed] [Google Scholar]
- 9. Frazer DM, et al. 2004. Delayed hepcidin response explains the lag period in iron absorption following a stimulus to increase erythropoiesis. Gut 53:1509–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ganne-Carrié N, et al. 2000. Liver iron is predictive of death in alcoholic cirrhosis: a multivariate study of 229 consecutive patients with alcoholic and/or hepatitis C virus cirrhosis: a prospective follow up study. Gut 46:277–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gerjevic LN, Liu N, Lu S, Harrison-Findik DD. 2012. Alcohol activates TGF-beta but inhibits BMP receptor-mediated Smad signaling and Smad4 binding to hepcidin promoter in the liver. Int. J. Hepatol. 2012:459278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gordeuk VR, et al. 2011. Chuvash polycythemia VHLR200W mutation is associated with down-regulation of hepcidin expression. Blood 118:5278–5282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Haase VH. 2005. The VHL tumor suppressor in development and disease: functional studies in mice by conditional gene targeting. Semin. Cell Dev. Biol. 16:564–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harrison-Findik DD. 2009. Is the iron regulatory hormone hepcidin a risk factor for alcoholic liver disease? World J. Gastroenterol. 15:1186–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Harrison-Findik DD, et al. 2007. Iron-mediated regulation of liver hepcidin expression in rats and mice is abolished by alcohol. Hepatology 46:1979–1985 [DOI] [PubMed] [Google Scholar]
- 16. Harrison-Findik DD, Klein E, Evans J, Gollan J. 2009. Regulation of liver hepcidin expression by alcohol in vivo does not involve Kupffer cell activation or TNF-alpha signaling. Am. J. Physiol. Gastrointest. Liver Physiol. 296:G112–G118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Harrison-Findik DD, et al. 2006. Alcohol metabolism-mediated oxidative stress down-regulates hepcidin transcription and leads to increased duodenal iron transporter expression. J. Biol. Chem. 281:22974–22982 [DOI] [PubMed] [Google Scholar]
- 18. He C, et al. 2012. Downregulating hypoxia-inducible factor-2alpha improves the efficacy of doxorubicin in the treatment of hepatocellular carcinoma. Cancer Sci. 103:528–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Heritage ML, et al. 2009. Hepcidin regulation in wild-type and Hfe knockout mice in response to alcohol consumption: evidence for an alcohol-induced hypoxic response. Alcohol. Clin. Exp. Res. 33:1391–1400 [DOI] [PubMed] [Google Scholar]
- 20. Iqbal T, et al. 2009. Is iron overload in alcohol-related cirrhosis mediated by hepcidin? World J. Gastroenterol. 15:5864–5866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kapitsinou PP, Haase VH. 2008. The VHL tumor suppressor and HIF: insights from genetic studies in mice. Cell Death Differ. 15:650–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kautz L, et al. 2008. Iron regulates phosphorylation of Smad1/5/8 and gene expression of Bmp6, Smad7, Id1, and Atoh8 in the mouse liver. Blood 112:1503–1509 [DOI] [PubMed] [Google Scholar]
- 23. Kohgo Y, et al. 2005. Iron accumulation in alcoholic liver diseases. Alcohol. Clin. Exp. Res. 29:189S–193S [DOI] [PubMed] [Google Scholar]
- 24. Krause A, et al. 2000. LEAP-1, a novel highly disulfide-bonded human peptide, exhibits antimicrobial activity. FEBS Lett. 480:147–150 [DOI] [PubMed] [Google Scholar]
- 25. Liu Q, et al. 2009. Role of iron deficiency and overload in the pathogenesis of diabetes and diabetic complications. Curr. Med. Chem. 16:113–129 [DOI] [PubMed] [Google Scholar]
- 26. Mastrogiannaki M, et al. 2012. Hepatic HIF-2 down-regulates hepcidin expression in mice through epo-mediated increase in erythropoiesis. Haematologica 97:827–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Matak P, et al. 2009. Activated macrophages induce hepcidin expression in HuH7 hepatoma cells. Haematologica 94:773–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nemeth E, et al. 2004. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science 306:2090–2093 [DOI] [PubMed] [Google Scholar]
- 29. Nicolas G, et al. 2001. Lack of hepcidin gene expression and severe tissue iron overload in upstream stimulatory factor 2 (USF2) knockout mice. Proc. Natl. Acad. Sci. U. S. A. 98:8780–8785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nicolas G, et al. 2002. The gene encoding the iron regulatory peptide hepcidin is regulated by anemia, hypoxia, and inflammation. J. Clin. Invest. 110:1037–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ohtake T, et al. 2007. Hepcidin is down-regulated in alcohol loading. Alcohol. Clin. Exp. Res. 31:S2–S8 [DOI] [PubMed] [Google Scholar]
- 32. Pabst T, et al. 2001. Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-alpha (C/EBPalpha), in acute myeloid leukemia. Nat. Genet. 27:263–270 [DOI] [PubMed] [Google Scholar]
- 33. Park CH, Valore EV, Waring AJ, Ganz T. 2001. Hepcidin, a urinary antimicrobial peptide synthesized in the liver. J. Biol. Chem. 276:7806–7810 [DOI] [PubMed] [Google Scholar]
- 34. Paz-Priel I, Friedman A. 2011. C/EBPalpha dysregulation in AML and ALL. Crit. Rev. Oncog. 16:93–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Peyssonnaux C, et al. 2007. Regulation of iron homeostasis by the hypoxia-inducible transcription factors (HIFs). J. Clin. Invest. 117:1926–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pinto JP, et al. 2008. Erythropoietin mediates hepcidin expression in hepatocytes through EPOR signaling and regulation of C/EBPalpha. Blood 111:5727–5733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Piperno A, et al. 2011. Modulation of hepcidin production during hypoxia-induced erythropoiesis in humans in vivo: data from the HIGHCARE project. Blood 117:2953–2959 [DOI] [PubMed] [Google Scholar]
- 38. Qu A, et al. 2011. Hypoxia-inducible transcription factor 2alpha promotes steatohepatitis through augmenting lipid accumulation, inflammation, and fibrosis. Hepatology 54:472–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rapisarda C, et al. 2010. Transferrin receptor 2 is crucial for iron sensing in human hepatocytes. Am. J. Physiol. Gastrointest. Liver Physiol. 299:G778–G783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Safran M, et al. 2006. Mouse model for noninvasive imaging of HIF prolyl hydroxylase activity: assessment of an oral agent that stimulates erythropoietin production. Proc. Natl. Acad. Sci. U. S. A. 103:105–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schuler M, Dierich A, Chambon P, Metzger D. 2004. Efficient temporally controlled targeted somatic mutagenesis in hepatocytes of the mouse. Genesis 39:167–172 [DOI] [PubMed] [Google Scholar]
- 42. Semenza GL. 2010. Oxygen homeostasis. Wiley Interdiscip. Rev. Syst. Biol. Med. 2:336–361 [DOI] [PubMed] [Google Scholar]
- 43. Semenza GL, Wang GL. 1992. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol. Cell. Biol. 12:5447–5454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shayakhmetov DM, Li ZY, Ni S, Lieber A. 2004. Analysis of adenovirus sequestration in the liver, transduction of hepatic cells, and innate toxicity after injection of fiber-modified vectors. J. Virol. 78:5368–5381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Silvestri L, et al. 2008. The serine protease matriptase-2 (TMPRSS6) inhibits hepcidin activation by cleaving membrane hemojuvelin. Cell Metab. 8:502–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Taylor M, et al. 2011. Hypoxia-inducible factor-2alpha mediates the adaptive increase of intestinal ferroportin during iron deficiency in mice. Gastroenterology 140:2044–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tomita S, Sinal CJ, Yim SH, Gonzalez FJ. 2000. Conditional disruption of the aryl hydrocarbon receptor nuclear translocator (Arnt) gene leads to loss of target gene induction by the aryl hydrocarbon receptor and hypoxia-inducible factor 1alpha. Mol. Endocrinol. 14:1674–1681 [DOI] [PubMed] [Google Scholar]
- 48. Vokurka M, Krijt J, Sulc K, Necas E. 2006. Hepcidin mRNA levels in mouse liver respond to inhibition of erythropoiesis. Physiol. Res. 55:667–674 [DOI] [PubMed] [Google Scholar]
- 49. Volke M, et al. 2009. Evidence for a lack of a direct transcriptional suppression of the iron regulatory peptide hepcidin by hypoxia-inducible factors. PLoS One 4:e7875 doi:10.1371/journal.pone.0007875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang GL, et al. 2010. Elimination of C/EBPalpha through the ubiquitin-proteasome system promotes the development of liver cancer in mice. J. Clin. Invest. 120:2549–2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang XL, et al. 2009. Ablation of ARNT/HIF1beta in liver alters gluconeogenesis, lipogenic gene expression, and serum ketones. Cell Metab. 9:428–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang Y, Liu Y, Malek SN, Zheng P. 2011. Targeting HIF1alpha eliminates cancer stem cells in hematological malignancies. Cell Stem Cell 8:399–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wu D, Cederbaum AI. 2009. Oxidative stress and alcoholic liver disease. Semin. Liver Dis. 29:141–154 [DOI] [PubMed] [Google Scholar]
- 54. Xue X, et al. 2012. HIF-2alpha activation promotes colorectal cancer progression by dysregulating iron homeostasis. Cancer Res. 72:2285–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.