Abstract
Yersinia pestis, the causative agent of plague, evolved from the gastrointestinal pathogen Yersinia pseudotuberculosis. Both species have numerous type Va autotransporters, most of which appear to be highly conserved. In Y. pestis CO92, the autotransporter genes yapK and yapJ share a high level of sequence identity. By comparing yapK and yapJ to three homologous genes in Y. pseudotuberculosis IP32953 (YPTB0365, YPTB3285, and YPTB3286), we show that yapK is conserved in Y. pseudotuberculosis, while yapJ is unique to Y. pestis. All of these autotransporters exhibit >96% identity in the C terminus of the protein and identities ranging from 58 to 72% in their N termini. By extending this analysis to include homologous sequences from numerous Y. pestis and Y. pseudotuberculosis strains, we determined that these autotransporters cluster into a YapK (YPTB3285) class and a YapJ (YPTB3286) class. The YPTB3286-like gene of most Y. pestis strains appears to be inactivated, perhaps in favor of maintaining yapJ. Since autotransporters are important for virulence in many bacterial pathogens, including Y. pestis, any change in autotransporter content should be considered for its impact on virulence. Using established mouse models of Y. pestis infection, we demonstrated that despite the high level of sequence identity, yapK is distinct from yapJ in its contribution to disseminated Y. pestis infection. In addition, a mutant lacking both of these genes exhibits an additive attenuation, suggesting nonredundant roles for yapJ and yapK in systemic Y. pestis infection. However, the deletion of the homologous genes in Y. pseudotuberculosis does not seem to impact the virulence of this organism in orogastric or systemic infection models.
INTRODUCTION
Yersinia pestis and Yersinia pseudotuberculosis are close enough genetically for Y. pestis to be considered a subspecies of Y. pseudotuberculosis (1, 36), but a clear separation remains in the classification of these two species because of their vastly different life cycles and virulence potentials. Y. pestis is maintained predominantly in enzootic reservoirs by a continuous vector-borne transmission cycle between rodents and fleas (23). The continuation of this cycle is predicated on the development of a high-level septicemic infection allowing for the inoculation of feeding fleas and the potential for lung colonization, resulting in direct host-to-host aerosol transmission (47). In contrast, Y. pseudotuberculosis can be found in soil and water as well as in association with domestic and wild animals, where it typically causes self-limiting gastroenteritis that deposits organisms back into the environment, where they can infect new hosts via the fecal-oral route (21).
Despite these striking differences in life-styles, it was calculated that Y. pestis evolved from Y. pseudotuberculosis biotype O:1b only within the last 1,500 to 20,000 years (1). The major changes that distinguish Y. pestis include the acquisition of plasmids necessary for virulence and vector-borne transmission, the loss of specific genes needed for enteric colonization (52), and a dramatic increase in the number of insertion sequence (IS) elements (13, 46). Despite these changes, Y. pestis has remained relatively monomorphic since its emergence (36), with most of the genetic changes being the result of IS element rearrangement and locally adaptive microevolution (1, 25, 44).
While several comparative studies have been undertaken to find areas of genetic difference between Y. pestis and Y. pseudotuberculosis (13, 18, 29, 46), limited evidence exists regarding how the genes in these regions contribute to the virulence of either organism (18, 46). An understanding of the differences in genetic content between these species is one important step toward explaining their observed differences in virulence. Among the loci that have been noted to undergo significant changes between these two species are several containing autotransporters, a large family of surface-exposed proteins implicated in the virulence of numerous Gram-negative pathogens (recently reviewed in reference 11). These changes include the loss or inactivation of the yapA, yapB, and YPO0765 genes as well as the acquisition of a region that contains the autotransporter yapJ (YPO1672) (18, 29).
In our previous work characterizing the conventional autotransporters found in Y. pestis CO92 (yap's), we observed that the members of this group shared little similarity in their passenger domains, with the exception of yapJ and yapK (YPO0309), which share 98% nucleotide identity across 55% of their passenger domains (38). This similarity was also observed previously by Derbise et al., who noted the high level of identity between yapJ, yapK, and YPO0765 (18). The similarity between the sequences of these autotransporters implies the potential for an evolutionary linkage between yapJ and yapK. In addition, the absence of yapJ in Y. pseudotuberculosis IP32953, and the variation in the numbers and locations of other homologous genes, implies that these loci are among those that have changed during the emergence of Y. pestis. Together, these observations warrant further investigation to determine how yapJ arose and what its potential precursors could have been in Y. pseudotuberculosis. The relatedness of these genes also raises the question of whether they have evolved to serve similar or different functions in Y. pestis virulence. The deletion of the locus encompassing YPO1668 to YPO1672 (containing yapJ) was determined not to impact the survival of 5-week-old OF1 mice following subcutaneous or aerosol infection with Y. pestis CO92, but no tissue burdens were examined (18). However, it was shown that the expression of yapJ and yapK is induced during mammalian infection in both bubonic and pneumonic models of plague in C57BL/6 mice (38). In addition, antibodies against YapJ were detected following experimental infection in a sublethal rabbit model of bubonic plague when the sera of surviving rabbits were screened with an Escherichia coli library expressing Y. pestis proteins (7). While expression in a host implies a role in virulence, it remained unknown what role these genes might be playing during the infectious process.
In this study, we outline a previously unappreciated evolutionary relationship between yapJ and yapK as well as heretofore-unidentified homologs in other Y. pestis strains and homologous genes in Y. pseudotuberculosis strains. To begin to understand if these genes contribute to the pathogenesis of either Y. pestis or Y. pseudotuberculosis, we investigated the contribution of these autotransporters using mouse models of bubonic, pneumonic, and septicemic Y. pestis infections as well as gastrointestinal and septicemic Y. pseudotuberculosis infections. We show that yapK and yapJ each play measureable roles in systemic (but not bubonic) Y. pestis infection and that a Y. pestis mutant lacking both yapK and yapJ is more attenuated than each of these individual mutants. This indicates that the contributions of yapK and yapJ are unique and that they are likely performing nonredundant functions during infection. Furthermore, we show that the contribution of yapK in Y. pestis is not shared by the homologous genes during disseminated Y. pseudotuberculosis infection, nor do these genes exhibit unique contributions to gastrointestinal infection.
MATERIALS AND METHODS
Bioinformatics and sequence comparisons.
YapJ and YapK homolog sequences were identified by pairwise sequence similarity searches using the NCBI (http://blast.ncbi.nlm.nih.gov/) and ExPASy (http://web.expasy.org/blast/) BLAST services (6). Alignments of partial or entire genes and proteins were performed by using MUSCLE (22), which also provided the percent identity scores. Graphical comparisons of percent identity were based on the above-described alignments and were generated with Geneious Pro 5.3.6 (Biomatters Ltd.). For the generation of trees, sequences were aligned by using MUSCLE, with indels being manually edited for clarity. A phylogenetic tree was calculated by ClustalX v. 2.0 using the bootstrapped neighbor-joining method with default parameters (33). The major nodes defining the phylogenetic relationships between the YapJ, YapK, YPTB3285, and YPTB3286 protein families were found 100% of the time in the bootstrapped trees. The gene content across Y. pestis and Y. pseudotuberculosis genomes was assessed by using annotated whole-genome sequence data available from the NCBI. Gene content comparisons were done by aligning sequenced genomes with Progressive MAUVE (17), using MAUVE Multiple Genome Alignment software (http://asap.ahabs.wisc.edu/mauve/).
Bacterial strains, growth conditions, and animal infections.
All bacterial strains and plasmids used in this study are listed in Table 1. Y. pestis CO92, a pCD1+ pgm+ sequenced clinical isolate, was used as the wild-type strain for all Y. pestis infections and as the parent strain for all gene deletions. Y. pseudotuberculosis IP32953, a sequenced clinical isolate with an intact phoPQ locus, was used as the wild-type strain for all Y. pseudotuberculosis infections and as the parent strain for all gene deletions. All animal experiments were approved by the University of North Carolina Institutional Animal Care and Use Committee (protocols 11-127 and 11-128). Four- to six-week-old female C57BL/6J mice (The Jackson Laboratory, Bar Harbor, ME) were used for all Y. pestis infections, and 4- to 6-week-old female BALB/cJ mice (The Jackson Laboratory) were used for all Y. pseudotuberculosis infections. All animals were allowed free access to sterilized food and water throughout the course of the experiment. For subcutaneous (s.c.) and intraperitoneal (i.p.) infections, Y. pestis CO92 cells were cultivated on brain heart infusion (BHI) agar (BD Biosciences, Bedford, MA) at 26°C for 48 h followed by culturing in BHI broth (BD Biosciences) overnight with aeration at 26°C. s.c. and i.p. inoculations with Y. pestis were performed as described previously (12), with doses of ∼102 CFU/mouse and ∼103 CFU/mouse, respectively. For intranasal (i.n.) inoculations, Y. pestis cells were cultured in BHI broth (BD Biosciences) with aeration at 26°C for 8 to 10 h prior to subculturing into BHI broth (BD Biosciences) containing 2.5 mM CaCl2 for growth overnight at 37°C. Cultures were then diluted in sterile phosphate-buffered saline (PBS) to achieve an inoculum of ∼104 CFU/mouse, and inoculations were performed as described previously (34). For all Y. pestis inoculations, groups of 4 to 6 mice were euthanized by i.p. injection with sodium pentobarbital (150 mg/kg of body weight) at the time points indicated in the figure legends. At each time point, tissues were harvested, weighed, and homogenized manually in sterile PBS, and serial dilutions were plated onto BHI agar to determine the bacterial load.
Table 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Descriptiona | Reference |
|---|---|---|
| Strains | ||
| Y. pestis | ||
| CO92 | Wild type; PolyBr | 19 |
| YP302 | CO92 ΔyapK | This work |
| YP424 | CO92 ΔyapJ | This work |
| YP405 | CO92 ΔyapK yapK complemented | This work |
| YP432 | CO92 ΔyapJ yapJ complemented | This work |
| YP307 | CO92 ΔyapK ΔyapJ | This work |
| Y. pseudotuberculosis | ||
| IP32953 | Wild type; Irgr | 13 |
| YPTB029 | IP32953 ΔYPTB0365 ΔYPTB3285 ΔYPTB3286 | This work |
| E. coli S17-1 λpir | Tpr Strr recA thi pro hsdR hsdM+ RP4::2-Tc::Mu::Km Tn7 λpir lysogen | 43 |
| Plasmids | ||
| pSR47S | Kanr MobRP4 oriR6K sacB suicide vector | 42 |
| pSR47S::ΔyapJ | Kanr; yapJ in-frame deletion in pSR47S | This work |
| pSR47S::ΔyapK/YPTB0365 | Kanr; yapK (YPTB0365) in-frame deletion in pSR47S | This work |
| pSR47S:: ΔYPTB3285 | Kanr; YPTB3285 in-frame deletion in pSR47S | This work |
| pSR47S:: ΔYPTB3286 | Kanr; YPTB3286 in-frame deletion in pSR47S | This work |
| pSR47S::yapJ-comp | Kanr; yapJ with upstream and downstream sequences in pSR47S | This work |
| pSR47S::yapK-comp | Kanr; yapK with upstream and downstream sequences in pSR47S | This work |
PolyBr, polymyxin B resistant; Irgr, Irgasan resistant; Kanr, kanamycin resistant.
For oral and i.p. inoculations with Y. pseudotuberculosis, the strains were cultivated on Luria-Bertani (LB) agar (BD Biosciences) at 26°C for 48 h, followed by culturing overnight in LB broth at 26°C with aeration. For oral inoculations, cultures grown overnight were normalized in sterile PBS to an optical density at 600 nm (OD600) reading of 3.0 to achieve an inoculum of 2 × 108 to 5 × 108 CFU/mouse, which was administered intragastrically via a 22-gauge ball-tipped feeding needle. For i.p. inoculations, cultures grown overnight were normalized in sterile PBS to an OD600 reading of 3.0 and diluted in sterile PBS to achieve an inoculum of ∼103 CFU/mouse. For all Y. pseudotuberculosis inoculations, groups of 4 to 6 mice were euthanized by CO2 asphyxiation at the time points detailed below. Tissues were harvested, weighed, and manually homogenized in sterile PBS. Serial dilutions of spleens and mesenteric lymph nodes were plated onto LB agar (BD Biosciences) containing 2 μg ml−1 Irgasan, and Peyer's patches were plated onto Yersinia selective agar (YSA) (BD Biosciences) to determine the bacterial load. In all experiments, animals were regularly monitored for signs of distress, and moribund animals were euthanized in accordance with approved protocols.
Plasmid and strain construction.
Gene deletions and complementations in Y. pestis CO92 and Y. pseudotuberculosis IP32953 were made by using pSR47S (42) and the primers listed in Table 2, essentially as described previously (53). Briefly, ∼500 bp of DNA 5′ to the coding sequence of yapK (and including the start codon) was amplified from Y. pestis CO92 genomic DNA by using primers JDL65 and JDL66, and this fragment was digested with SalI and BamHI. Primers JDL67 and JDL68 were used to generate a similarly sized fragment containing the sequence 3′ of yapK (including the stop codon), and this fragment was digested with BamHI and NotI. Each of these fragments was cloned into pSR47S that was digested with SalI and NotI, resulting in plasmid pSR47S::ΔyapK, which was subsequently transformed into E. coli S17λpir by electroporation. Transformants were selected on LB agar (BD Biosciences) containing 50 μg ml−1 kanamycin and screened for insertion by restriction digestion. An identical methodology was used to generate pSR47S::ΔyapJ using primers JDL69/JDL70 and JDL71/JDL72. Complementation constructs were generated by using a similar methodology, except that the internally directed flanking primers, which result in the amplification of both flanking regions and the intervening full-length gene, were used to generate inserts into pSR47S (JDL65 and JDL68 for yapK and JDL69 and JDL72 for yapJ). All deletion and complementation constructs were confirmed by primer-extension sequencing to ensure that no errors were generated during amplification.
Table 2.
Primers used in this work
| Primer | Description | Sequencea (5′–3′) |
|---|---|---|
| JDL65 | yapK 5′ forward | ACGCGTCGACTGTTAGAAACTTCCATTACCCCCCCC |
| JDL66 | yapK 5′ reverse | CGGATCCCATAAATGTACTCTATTAATTGCCTGAATTTATCGC |
| JDL67 | yapK 3′ forward | GAAGATCTTAAATCGTTTTATATTAGCGGGTAAATCGTTTTATAGC |
| JDL68 | yapK 3′ reverse | ATAAGAATGCGGCCGCATACGGAGTACCGCTATTTTGTTGC |
| JDL69 | yapJ 5′ forward | ATAAGAATGCGGCCGCGTCTCCGCAAGTCTGGGGATTAGTCCGGT |
| JDL70 | yapJ 5′ reverse | CGGATCCCATAGAATTACCCTATTAGCTGCCCC |
| JDL71 | yapJ 3′ forward | CGGATCCTAATACAGGACGGTTTTGTTATATCGGG |
| JDL72 | yapJ 3′ reverse | ACGCGTCGACGCGTTTATTAAGTAAAAAATTCTCTATGAGATAACCCTA |
| JDL168 | YPTB3286 5′ forward | ACGCGTCGACGATAGATCGTGAACCCGATTTGAAC |
| JDL169 | YPTB3286 5′ reverse | CGGATCCCATAAATGTACTCTATTAATTGCCTGAATTTATCG |
| JDL170 | YPTB3286 3′ forward | CGGATCCTAAGTCGTTTTTATGTGTAAAGGGTATCAG |
| JDL171 | YPTB3286 3′ reverse | ATAAGAATGCGGCCGCGGTATAAGGCAGAGTACAGAAGGA |
| JDL172 | YPTB3285 5′ forward | ACGCGTCGACCAGGGGAGAATTTCAACATCCTG |
| JDL173 | YPTB3285 5′ reverse | CGGATCCCATAGGGTACCTTATTCATTGTAACTGTC |
| JDL174 | YPTB3285 3′ forward | CGGATCCTAAATCGTTTTTATGTGTAAAGGGTATCAGT |
| JDL175 | YPTB3285 3′ reverse | ATAAGAATGCGGCCGCCTGTCAGCAAACATATCTGCTCC |
Restriction sites are underlined.
Deletion strains of Y. pestis and Y. pseudotuberculosis were generated by allelic exchange, and conjugation was used to introduce the suicide plasmid constructs carrying the mutation into the strain of interest for integration onto the chromosome, essentially as described previously for Y. enterocolitica (53). Briefly, equal volumes of cultures of the recipient (Y. pestis CO92 or Y. pseudotuberculosis IP32953) and the donor (E. coli S17λpir with a pSR47S-based plasmid) grown overnight were mixed, plated, and incubated overnight at 26°C. For Y. pestis, bacteria were collected in 1 ml PBS with a cell scraper, and serial dilutions were plated onto BHI agar (BD Biosciences) containing 25 μg ml−1 polymyxin (to select against E. coli) and 50 μg ml−1 kanamycin (to select for plasmid insertion), followed by incubation for 2 days at 26°C. Individual kanamycin-resistant colonies were then struck for isolation on BHI agar (BD Biosciences) containing 25 μg ml−1 polymyxin and 5% sucrose, followed by incubation for 2 to 3 days at 26°C to force plasmid excision. Colonies from sucrose plates were then patched in parallel onto BHI agar with 25 μg ml−1 polymyxin and BHI agar with both 25 μg ml−1 polymyxin and 50 μg ml−1 kanamycin, to test for colonies that had excised the deletion construct. Kanamycin-sensitive colonies were grown overnight at 26°C with aeration, and genomic DNA was prepared in order to screen for gene deletions by PCR. Potential deletion mutants were struck for isolation on BHI agar (BD Biosciences), and multiple independent colonies were retested by PCR for deletions as well as for the retention of virulence genes in the high-pathogenicity island and on each of the plasmids (yopH, yopT, ymt, pst, and hms). For Y. pseudotuberculosis, transconjugants were selected on YSA with 50 μg ml−1 kanamycin and counterselected on YSA with 5% sucrose, and the resulting colonies were patched in parallel onto both LB agar with 2 μg ml−1 Irgasan and LB agar with 50 μg ml−1 kanamycin, to test for colonies that excised the deletion construct. Strains in which multiple genes were deleted were subjected to sequential rounds of the same methodology as that described above and, when complete, were checked by PCR for the deletion of all targeted genes as well as virulence factor retention. Complementation strains of Y. pestis CO92 were generated by using essentially the same allelic exchange methodology as that described above for deletion strains. Conjugation was used to introduce suicide plasmid pSR47S::yapJ-comp or pSR47S::yapK-comp (carrying native copies of the gene to be complemented and flanking sequences) into the Y. pestis CO92 ΔyapJ or Y. pestis CO92 ΔyapK mutant, respectively, for integration onto the chromosome at the site of the deletion. The insertion of the full-length gene at its native site, in the correct direction, was confirmed by PCR. All deletion and complementation strains were tested for a normal growth rate in liquid cultures at both 26°C and 37°C prior to use in animal experiments; all mutants and complemented strains showed wild-type growth rates in vitro in liquid cultures (data not shown).
RESULTS
yapJ and yapK from Y. pestis evolved from three autotransporter genes in Y. pseudotuberculosis.
Within the genome of Y. pestis CO92, there are nine functional, conventional autotransporters, and nearly all of these genes can be matched with a putative functional autotransporter in an equivalent genomic location in Y. pseudotuberculosis IP32953 (38). yapJ (YPO1672) is the notable exception, appearing in a new location with a cluster of genes not found in the equivalent location in Y. pseudotuberculosis (18). Because yapJ and yapK share a high level of similarity, we wanted to determine how divergent the sequence of yapJ was from yapK and how closely each of these genes resembled homologs in Y. pseudotuberculosis IP32953 (YPTB0365, YPTB3285, and YPTB3286). Thus, each of these gene sequences was compared to each other in a series of alignments. The comparison of yapK with yapJ reiterated previous findings (38) that these two genes share a great deal of identity (83.3% at the nucleotide level and 79.9% at the amino acid level), with the majority of the divergence being located at the 5′ end of the gene (Fig. 1A). This same pattern of similarity and divergence was seen when yapK and yapJ were compared to the homologous Y. pseudotuberculosis genes. While it was not surprising that yapK from CO92 was virtually identical to YPTB0365 (its positional ortholog) from IP32953, alignments revealed that yapK was more similar to YPTB3285 than it was to the neighboring gene, YPTB3286 (Fig. 1B). The yapJ gene shares similar percent identities with all of the Y. pseudotuberculosis homologs, with a slightly higher percent identity to YPTB3286, making it difficult to distinguish whether this gene arose by a duplication or a rearrangement event with resulting drift from one of these existing genes (Fig. 1C). In examining these comparisons, we note that the greatest divergence is seen in the 5′ halves of the genes, while the 3′ halves share >96.1% identity over a region that extends well beyond the conserved β-domains (the overall β-domain identity is >96.7% [data not shown]).
Fig 1.
Comparison of yapK and yapJ sequences from Y. pestis CO92 with homologs in Y. pseudotuberculosis IP32953. (A) Schematic diagram showing the predicted domains common to yapK and yapJ (top) and histogram showing nucleotide similarity over the length of their sequences (bottom). The percent nucleotide identities followed by translated amino acid identities (in parentheses) are given for regions indicated by lines and hatch marks. (B and C) The alignment of yapK and yapJ was used to delineate regions of the greatest difference/similarity, which were applied to the comparison of yapK (B) and yapJ (C) to YPTB0365, YPTB3285, and YPTB3286.
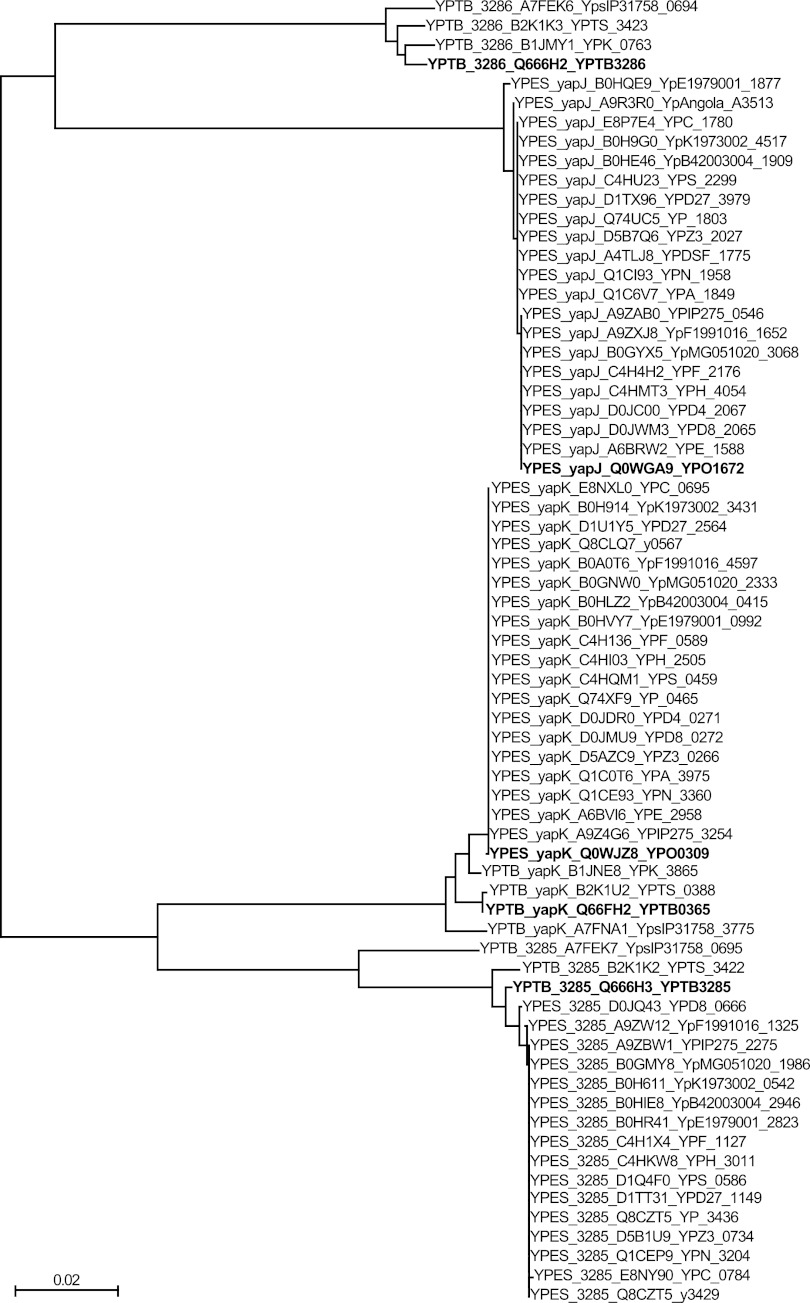
While it appears that yapK has not diverged much since the emergence of Y. pestis, the origin of yapJ is not discernible from the comparison of only representative strains of Y. pestis and Y. pseudotuberculosis. To determine which Y. pseudotuberculosis gene is the most likely precursor of yapJ, all predicted protein sequences similar to YapJ or YapK were obtained from the complete genome sequences and unassembled whole-genome shotgun sequences of all Y. pestis and Y. pseudotuberculosis sequences available from the NCBI. These sequences were aligned by using MUSCLE (22), and a phylogenetic tree was generated by using the bootstrapped neighbor-joining method to compare these sequences (Fig. 2). By aligning amino acid sequences rather than nucleotides, differences were limited to nonsynonymous changes, which should better reflect functional differences rather than random drift. This analysis revealed that these proteins fell into one of two classes, designated YapJ-like and YapK-like, with the majority being the latter. This is in part because YPTB3285 is closer in sequence to YapK, and the genes encoding homologs of each of these proteins are found more often across both species than those in the YapJ-like class. YapJ clusters with other Y. pestis YapJ proteins and with YPTB3286-like sequences, revealing a closer relationship of YPTB3286 to YapJ than of YPTB3285 to YapJ.
Fig 2.
Neighbor-joining phylogenetic tree of YapJ- and YapK-like protein sequences. Sixty-eight genes predicted to produce protein products similar to YapJ, YapK, YPTB0365, YPTB3285, or YPTB3286 were identified in sequenced genomes of Y. pestis and Y. pseudotuberculosis available through the NCBI. Translated sequences were aligned by using MUSCLE and used to generate a bootstrapped phylogenetic tree (ClustalX v. 2.0). Major nodes defining the phylogenetic relationships between the YapJ, YapK, YPTB3285, and YPTB3286 protein families were found 100% of the time in the bootstrapped trees. Proteins are listed first by their species of origin, then by the reference protein to which they are closest (YapJ, YapK, YPTB3285, or YPTB3286), then by their UniProt identification, and finally by their NCBI accession number. YPTB0365 falls within the same group as YapK due to their extremely high level of identity. The proteins investigated in this study (YapJ and YapK from Y. pestis CO92 and YPTB0365, YPTP3285, and YPTB3286 from Y. pseudotuberculosis IP32953) are identified in boldface type for ease of reference. YPES, Y. pestis sequences; YPTB, Y. pseudotuberculosis sequences. The scale bar indicates the number of amino acid substitutions per site for each branch.
YapK is conserved in Y. pestis and Y. pseudotuberculosis, while YapJ is unique to Y. pestis, and the YPO0765 locus varies from strain to strain.
In obtaining sequences of YapJ- and YapK-like proteins for evolutionary comparisons, more complete protein sequences of YPTB3285-like proteins were found than sequences of YPTB3286-like proteins. This indicated possible differences in genome contents or alternative arrangements of these genes from their positions in Y. pestis CO92 and Y. pseudotuberculosis IP32953. Since these differences could reveal evolutionary intermediates, the location of each of the genes encoding the proteins that were used in the evolutionary analysis (Fig. 2) was determined, and the genetic context was compared to that of CO92 or IP32953 by first using the available annotations. Differences between strains were then confirmed by aligning the available assembled genomes with MAUVE (17) and performing alignments of genes or gene fragments with MUSCLE (22) when genomes were available but not yet fully assembled. This analysis confirmed that yapK is present in the same location in the chromosomes of all sequenced Y. pestis and Y. pseudotuberculosis strains (adjacent to ubiA and ubiC) (Fig. 3A) and that yapJ is conserved in its location in Y. pestis but is absent from that location in Y. pseudotuberculosis (Fig. 3B). This finding agrees with the previously reported observation that yapJ (YPO1672) is found in a region containing five open reading frames (ORFs) unique to Y. pestis (YPO1668 to YPO1672) that were inserted as a block between YPTB2403 and YPTB2404 (18).
Fig 3.
Location and distribution of related autotransporter sequences among Y. pestis and Y. pseudotuberculosis strains. (A) Genomic context of yapK (red) and surrounding genes (gray) as they are found in sequenced Y. pseudotuberculosis and Y. pestis strains. (B) Genomic context of yapJ (blue) and surrounding genes (gray) as they are found in sequenced Y. pestis strains. (C) Comparison of the locus containing YPTB3285 (light red) and YPTB3286 (light blue) in Y. pseudotuberculosis IP32953 to the same locus in other strains. YPTB3285 and genes that grouped with it in the phylogenetic analysis (Fig. 2) are depicted in light red to indicate their phylogenetic clustering with yapK (red). YPTB3286 and all similar sequences (including genes, gene fragments, and pseudogenes with a high level of sequence identity) are depicted in light blue to indicate their close relationship to yapJ (blue). Loci from Y. pseudotuberculosis are separated from Y. pestis sequences by a dotted line. Full locus tags are provided where space permits, and numerical portions of locus tags are provided in all other cases.
Of the loci where these related genes are found, the area with the most variability between Y. pestis CO92 and Y. pseudotuberculosis IP32953 is the region containing the tandem genes YPTB3285 and YPTB3286 (Fig. 3C). The YPTB3285 sequence is absent from CO92, while the pseudogene (YPO0765) near that location appears to be a remnant of YPTB3286 (sharing 97% nucleotide identity). Among the analyzed Y. pestis strains, Antiqua and CA88-4125 are similar to CO92, lacking a YPTB3285-like gene. In most of the other analyzed strains, however, a gene related to YPTB3285 is present and apparently intact. In addition, as in CO92, many of these strains have an inactivated version of YPTB3286 (adjacent to YPTB3285, as in IP32953). This distribution of genes does not directly correlate to the classical designations of the three Y. pestis biovars.
The Y. pestis homologs of YPTB3286 appear to be nonfunctional in all strains examined. Of the sequenced Y. pestis strains analyzed for this work, the annotation of the YPTB3286 equivalent in Y. pestis varies from strain to strain, and we wanted to determine if this difference reflects different inactivating mutations. By aligning each of the YPTB3286-like sequences from Y. pestis to YPTB3286 from Y. pseudotuberculosis, we found that each of these sequences (regardless of annotation) contains the same single-base deletion at position 1583, resulting in a frameshift mutation. All of these YPTB3286-like genes (whether annotated as a pseudogene or as two ORFs) each contain a conserved single-point mutation (74 bp downstream), which, once the reading frame is shifted, changes a CAG (glutamine) to TAG (stop). The only difference between whether a locus was annotated as a pseudogene or as two smaller ORFs is whether or not a nearby GAG (in the original reading frame prior to mutation) is called as an alternative start codon. Interestingly, the C-to-T mutation does not appear to be a “second hit” that prematurely stops any of these genes after the frameshift; this base change is found in Y. pseudotuberculosis strains PB1/+ and YPIII (but not IP32953 or IP31758), neither of which contains the frameshift common to the Y. pestis strains. Even without the C-to-T change, several other stop codons exist in the newly established frame. These findings suggest that the splitting of the YPTB3286-like gene in Y. pestis strains does not represent a rearrangement or recombination event but rather a common mutation that likely results in inactivation. Thus, these data indicate that many Y. pestis strains carry not only yapK and yapJ but also a third related autotransporter gene similar to YPTB3285 from Y. pseudotuberculosis. However, this YPTB3285-like gene does not seem to be essential to Y. pestis, as it is absent from some strains, notably, the two strains isolated from a North American source. It also appears that YPTB3286 is dispensable for (or detrimental to) the life cycle of Y. pestis, since all analyzed strains have inactivated this gene. The equivalent region containing YPTB3285 and YPTB3286 was not found in the available Y. pestis Angola, Antiqua UG05-0454, or Pestoides F genome sequences at the time when these data were compiled, although yapK and yapJ were present in these strains.
YapJ and YapK are involved in systemic dissemination of Y. pestis CO92 but not lymph node colonization in C57BL/6 mice.
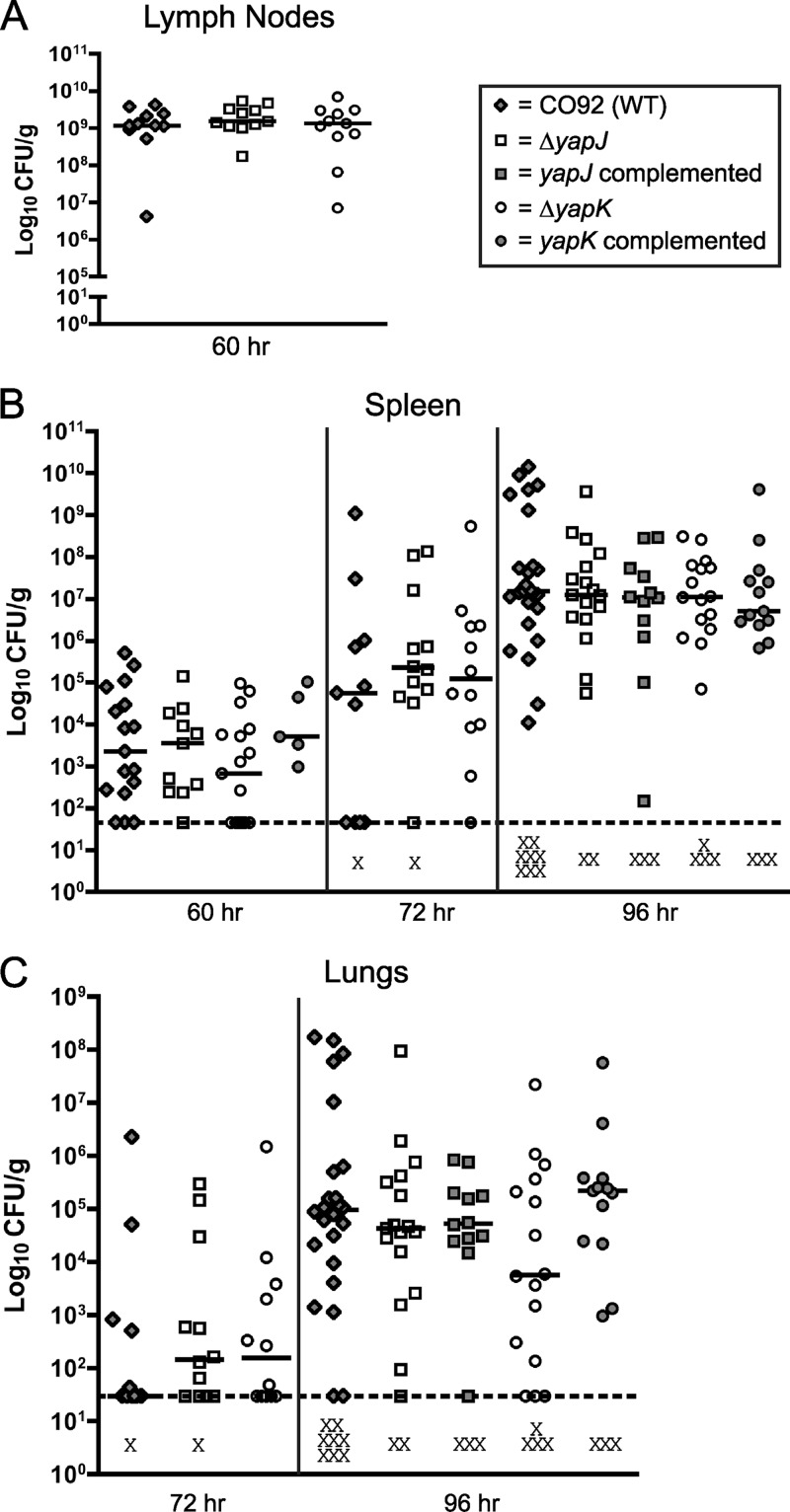
Proteins in the autotransporter family have often been linked to virulence-associated functions, but only a subset of these genes have been shown to make contributions to bacterial virulence in animal models of infection (2–5, 20, 24, 45, 48). Of the conventional autotransporters identified in Y. pestis CO92 (38), only the adhesin YapE has so far been shown to contribute significantly to infection, with the loss of yapE resulting in the less efficient colonization of cervical lymph nodes and delayed dissemination to spleen and lungs following subcutaneous (s.c.) infection (37). Previous work to determine the expression of yap's during infection after s.c. and intranasal (i.n.) inoculation, however, indicated that yapJ and yapK are induced during mouse infection (38). To explore the possibility that yapJ and yapK contribute to the disease-causing potential of Y. pestis CO92, we tested strains with in-frame deletions of each of these genes with established animal models of bubonic and pneumonic plague (12, 34). Following the s.c. inoculation of C57BL/6 mice with ∼102 bacteria of either wild-type strain CO92 or the ΔyapJ or ΔyapK strain, no differences were observed in cervical lymph node colonization at 24, 36, 48, or 60 h postinfection (hpi) (Fig. 4A and data not shown). At sites of disseminated infection, there is a small decrease (relative to that of the wild type) in the median CFU of the ΔyapK strain recovered from spleens at 60 hpi and lungs at 96 hpi (Fig. 4B and C). These differences are not statistically significant, most likely due to a large variance in the bacterial load in these tissues. However, both of these differences appear to be complemented by restoring an intact copy of yapK to its original location. Infection by the i.n. route showed no evidence of differences in lung colonization between the wild type and the ΔyapJ or ΔyapK mutant (data not shown). A double mutant (ΔyapK ΔyapJ) also showed no statistically significant difference from the wild type by the subcutaneous or intranasal route of infection (data not shown). These results indicate that the ΔyapJ, ΔyapK, and ΔyapK ΔyapJ mutants colonize comparably to wild-type Y. pestis at the initial site of colonization but suggested a possible role for YapK in the systemic phase of infection, which requires further investigation. To accomplish this, the spread of Y. pestis to deeper tissues and growth at these sites were evaluated in a systemic infection model by intraperitoneal (i.p.) inoculation (12).
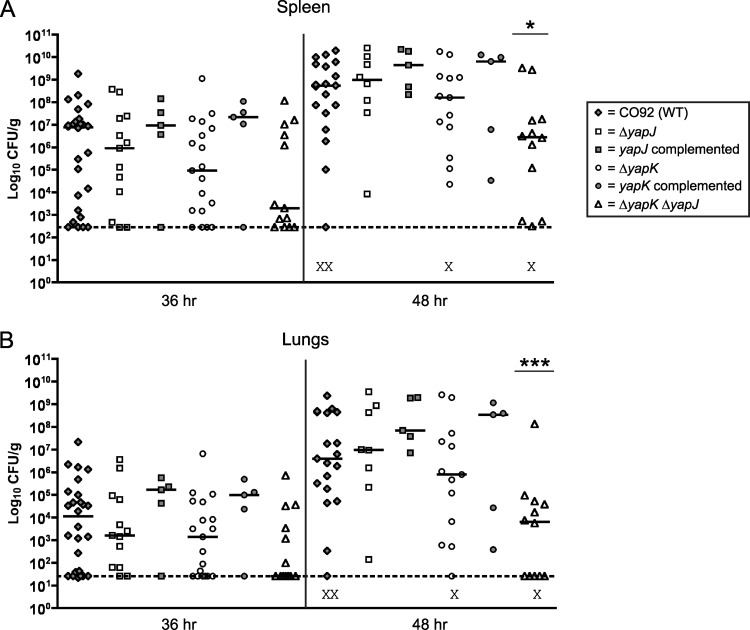
Fig 4.
Subcutaneous inoculation of C57BL/6 mice with wild-type (WT) Y. pestis CO92, the ΔyapJ mutant, the complemented yapJ mutant, the ΔyapK mutant, or the complemented yapK mutant. Mice were injected with ∼102 CFU of the above-described strains in the scruff of the neck and were sacrificed at 60, 72, and 96 hpi to determine the colonization of cervical lymph nodes (A), spleen (B), and lungs (C). Each symbol represents a single mouse. Lymph node data are the composites of data from two independent experiments, and spleen and lung data are the composites of data from five independent experiments. Black bars indicate the median CFU recovered per gram of tissue for each group, dashed lines indicate the limit of detection for each tissue, and the X symbols indicate dead mice. No significant differences between groups were present when subjected to a Mann-Whitney t test with a two-tailed nonparametric analysis.
The i.p. route of inoculation can be used to initiate an earlier form of systemic infection, with detectable bacterial counts being recoverable from the spleen and lungs a full 24 h earlier than by the s.c. route. i.p. inoculation has been used to study other virulence factors of Y. pestis CO92 (12) and has been used specifically to bypass the colonization of the lymphatics in Y. enterocolitica infections (49, 54). The i.p. route bypasses the need for the bacteria to transit from the infection site and replicate in the draining lymph node, steps which do not seem to be impacted by the presence of yapJ and yapK. Following the i.p. inoculation of C57BL/6 mice with ∼103 CFU of either wild-type CO92 or the ΔyapJ, ΔyapK, or ΔyapK ΔyapJ (double deletion) strain, spleens and lungs were harvested at 36 and 48 hpi. At 36 hpi, the median level of colonization of the spleen and lungs by the ΔyapK mutant was noticeably lower than that by the wild-type strain, although this did not quite achieve statistical significance (Fig. 5). Wild-type colonization levels were restored by the introduction of an intact copy of this gene into its native site on the chromosome. Likewise, the ΔyapJ mutant also appeared to be recovered more often at lower levels than the wild-type strain at 36 hpi (and restored to wild-type levels by complementation), although this difference was not as large as that seen for the ΔyapK mutant. At 48 hpi, neither of these deletion strains was significantly different from the wild type. The double mutant strain, however, displayed lower median levels of colonization of both spleen and lungs at 36 hpi than the wild-type or single mutant strain, including a higher proportion of uncolonized spleens and lungs at this time point. While this decrease in colonization by the double mutant relative to colonization by the wild type did not reach statistical significance in either the spleen or lungs at 36 hpi, a statistically significant difference in colonization was observed at 48 hpi for the double mutant in both spleen (P < 0.05) and lungs (P < 0.001) (Fig. 5). These data suggest that while yapK and yapJ are likely acting in the systemic stages of Y. pestis infection, their virulence contributions are unique. Despite their similarities, each gene appears to have a nonredundant function, as indicated by the individual phenotypes of the single mutant strains and the additive phenotype of a double mutant.
Fig 5.
Intraperitoneal inoculation of C57BL/6 mice with wild-type Y. pestis CO92, the ΔyapJ mutant, the complemented yapJ mutant, the ΔyapK mutant, the complemented yapK mutant, or the ΔyapK ΔyapJ double mutant. Mice were injected with ∼103 CFU of the above-mentioned strains intraperitoneally and were sacrificed at 36 and 48 hpi to determine the colonization of spleen (A) and lungs (B). Each symbol represents a single mouse, and data are the composites of data from five independent experiments. Each mutant was tested at least two independent times, and wild-type-infected groups were run for comparison in all experiments. Black bars indicate the median CFU recovered per gram of tissue for each group, dashed lines indicate the limit of detection for each tissue, and the X symbols indicate dead mice. Asterisks and bars over a column denote a statistically significant difference in the median CFU/g recovered from the indicated group relative to the wild type, as determined by a Mann-Whitney t test with a two-tailed nonparametric analysis (*, P < 0.05; ***, P < 0.001).
The YapJ and YapK orthologs of Y. pseudotuberculosis do not contribute significantly to oral or intraperitoneal infection models.
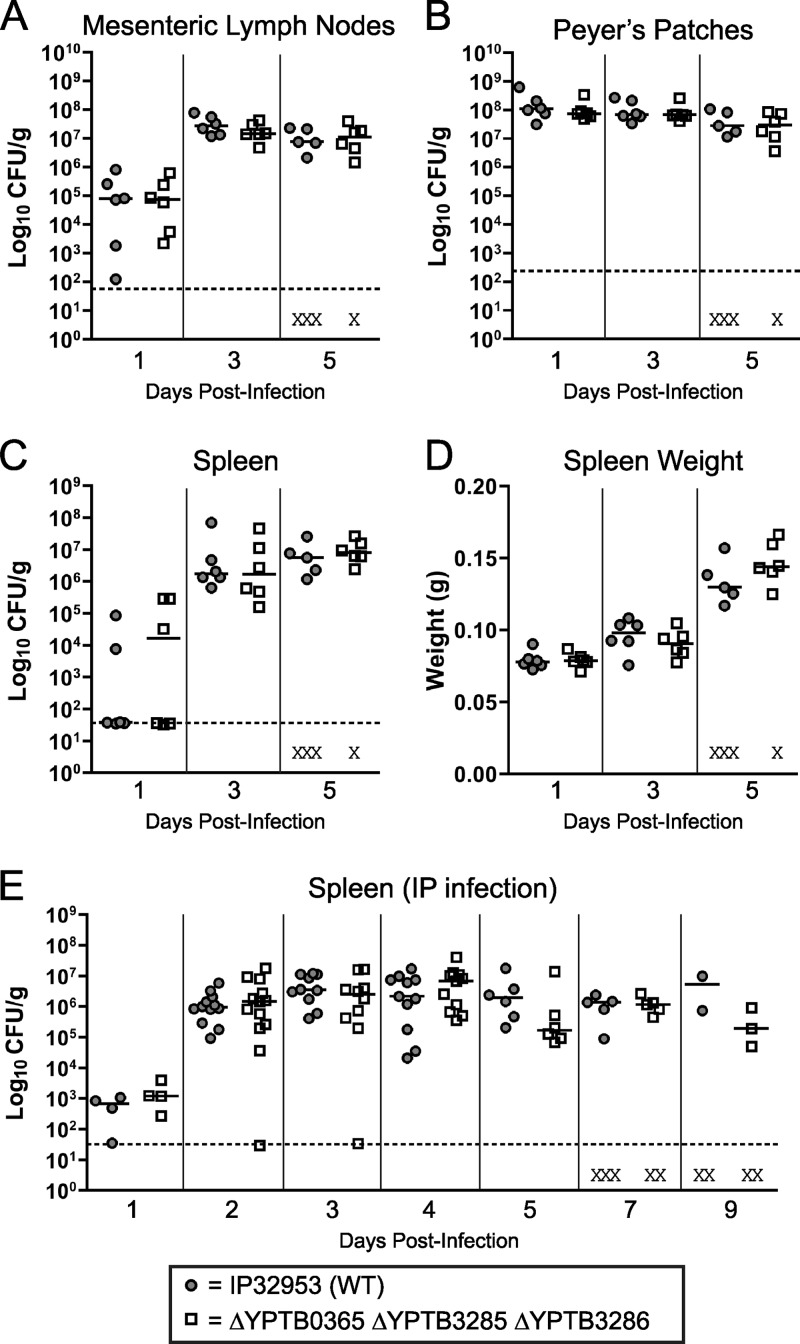
After evaluating the contribution of yapJ and yapK to Y. pestis infection, we wanted to determine if the related orthologs of these genes had similar roles as virulence factors in mouse models of Y. pseudotuberculosis infection. Despite genetic similarities, the diseases typically caused by Y. pestis and Y. pseudotuberculosis differ greatly, which is reflected in the animal models most commonly used to evaluate virulence factors possessed by these two species. For Y. pestis, the subcutaneous infection model has been well characterized (12, 16), but the failure of Y. pseudotuberculosis to sustain a successful infection by this route makes this model of little use for evaluating factors involved in systemic dissemination (27). Because Y. pseudotuberculosis is a gastrointestinal pathogen, the most commonly used route of experimental infection is the oral route (8, 41, 50). A mutant lacking all three autotransporters (YPTB0365 [yapK], YPTB3285, and YPTB3286) was shown to colonize Peyer's patches, mesenteric lymph nodes, and spleens similarly to wild-type strain IP32953 following oral infections of BALB/c mice (Fig. 6A to C). The wild-type and mutant strains also caused similar changes in spleen weight (a marker of inflammation) during infection (Fig. 6D). Similar results were observed when a double mutant of YPTB3285 and YPTB3286 was tested in BALB/c mice and when a single mutant of YPTB0365 (yapK) was tested in both the BALB/c and C57BL/6 mouse backgrounds (data not shown). Also, no differences were observed in the colonization of the small intestine or cecum or in fecal shedding following single infections or coinfections of any of these mutant strains with the wild type (data not shown).
Fig 6.
Oral and intraperitoneal inoculation of BALB/c mice with wild-type Y. pseudotuberculosis strain IP32953 or the ΔYPTB0365 (yapK) ΔYPTB3285 ΔYPTB3286 mutant. (A to D) For oral inoculation, groups of six mice were administered 2 × 108 to 5 × 108 CFU intragastrically of either the wild-type or mutant strain and were sacrificed at day 1, 3, or 5 postinfection to determine the colonization of the mesenteric lymph nodes (A), Peyer's patches (B), and spleen (C) as well as the spleen weight (D). Data from one representative experiment are shown. (E) For intraperitoneal infections, mice were injected with ∼103 CFU of the above-mentioned strains and were sacrificed at days 1, 2, 3, 4, 5, 7, and 9 postinfection to determine spleen colonization. Intraperitoneal infection data are the composites of data from three independent experiments. For both inoculation routes, each symbol represents a single mouse. Black bars indicate the median CFU recovered per gram of tissue for each group, dashed lines indicate the limit of detection for each tissue, and the X symbols indicate dead mice. No significant differences between any groups by either inoculation route were present when subjected to a Mann-Whitney t test with a two-tailed nonparametric analysis.
As with Y. pestis, i.p. inoculation has been used to evaluate the potential for the colonization of systemic sites by Y. pseudotuberculosis, and both species can be used at similar infectious doses (8, 12). Since our Y. pestis yapJ and yapK mutants differ the most from the wild-type strain in colonization when mice are challenged by the i.p. route of inoculation, we tested deletions in the autotransporter genes of Y. pseudotuberculosis IP32953 by the same route of inoculation. Since examples of Y. pseudotuberculosis i.p. inoculations from previously reported work used either strain IP2666 or YPIII (8, 10), we first performed a pilot infection with IP32953 and determined that a dose of ∼103 CFU produced a reliable infection comparable to that reported previously for IP2666 (data not shown). Using the i.p. route, we then inoculated separate groups of BALB/c mice with wild-type Y. pseudotuberculosis strain IP32953 or the triple mutant lacking the three autotransporters (YPTB3285, YPTB3286, and YPTB0365 [yapK]) and compared the CFU recovered from spleens and mesenteric lymph nodes at 1 to 9 days postinfection. There was no significant difference in spleen colonization or spleen weight between the wild-type and mutant strains (Fig. 6E and data not shown).
DISCUSSION
The genome of Y. pestis CO92 contains genes encoding two closely related autotransporters that are evolutionarily related to three genes in the gastrointestinal pathogen Y. pseudotuberculosis IP32953. We have shown that these five genes exist as part of an extended family, with one gene being conserved in all sequenced Y. pestis and Y. pseudotuberculosis strains (yapK), two that vary from strain to strain, and one new gene unique to Y. pestis (yapJ). By comparing these genes both on an individual basis and across sequenced strains of Y. pestis and Y. pseudotuberculosis, we determined that these genes fall into two major evolutionary branches, the yapK (YPTB3285) branch and the yapJ (YPTB3286) branch. The first major branch includes the highly conserved yapK (found in both species) and YPTB3285 (found in most isolates of both species) genes. The second major branch seems more divergent, with sequenced Y. pseudotuberculosis strains retaining YPTB3286 and most Y. pestis strains inactivating YPTB3286, perhaps in favor of the Y. pestis-specific yapJ gene.
In a comparison of gene content that considers only the representative Y. pseudotuberculosis strain IP32953 and the representative Y. pestis strain CO92, it would be reasonable to conclude that YPTB3286 became the pseudogene YPO0765 in CO92 (they share >98% nucleotide identity) and that YPTB3285 was lost, perhaps as a result of the gene reduction that occurred during the emergence of Y. pestis (46). The origin of yapJ, however, remains unclear, as it seems nearly equivalent in nucleotide identity to both YPTB3285 and YPTB3286 in IP32953. However, by taking into consideration the gene content from 26 Y. pseudotuberculosis and Y. pestis strains and by performing a phylogenetic analysis of the protein sequences from the total set of 68 yapJ- and yapK-like genes, we can conclude that yapJ is more likely to have arisen from YPTB3286. This is an observation that must be considered in the context of the Y. pestis-specific distribution of yapJ and the apparent inactivation of YPTB3286 in Y. pestis strains.
Gene loss can be considered of nearly equal importance to gene gain in the emergence of highly virulent Y. pestis from Y. pseudotuberculosis. Genes critically important for intestinal colonization, such as yadA and inv, are inactivated in all Y. pestis isolates, and as more strains are sequenced, the amount of evidence that gene reduction continues to be under positive selective pressure increases (14). The existence of intact copies of YPTB3285, YPTB3286, and YPTB0365 (yapK) in several Y. pseudotuberculosis strains implies that these genes are each under some selective pressure to be maintained, even if their shared sequence identity reflects a high likelihood of having similar functions. It is therefore likely that YPTB3286 has not been maintained after the emergence of Y. pestis because its function is not advantageous in the new life cycle of Y. pestis.
Our comparison of sequences and evolutionary analysis implicate a YPTB3286-like gene as the most likely of the three Y. pseudotuberculosis orthologs to be the precursor of yapJ. This analysis, however, does not explain how this event may have occurred. One of the major features of the Y. pestis genome that distinguishes it from Y. pseudotuberculosis is the significant expansion of insertion elements, which subsequently resulted in isolate-specific intrachromosomal rearrangements (25). The way in which Y. pestis handles its genome informs how we think about the origin of yapJ in Y. pestis. The larger genomic region containing yapJ is conserved back to Y. enterocolitica, where several uninterrupted motility/chemotaxis systems exist (including flhDC, motAB, and cheAWDRBYZ). In Y. pseudotuberculosis, two genes with homology to YPO1673 and YPO1674 were inserted between cheW and cheD. In Y. pestis, the additional insertion of genes of unknown origin (YPO1668 to YPO1671) along with yapJ (YPO1672) occurs at the same location, downstream of cheW and upstream of YPO1673. There are no clear signs of the presence (or remnants) of a transposable element in that location in Y. enterocolitica, Y. pseudotuberculosis, or Y. pestis, and there is no evidence of the acquisition of yapJ through horizontal gene transfer from an unrelated bacterium, as no homologs are found in any other sequenced bacterial species outside Y. pestis and Y. pseudotuberculosis. This leaves the mechanism of yapJ insertion an open question but should tell us something about its potential function, namely, that if it represents a duplication of YPTB3286, the strict maintenance of yapJ in Y. pestis (while YPTB3286 is lost) implies a Y. pestis-specific function for yapJ.
One approach to the question of how yapJ and yapK contribute to the virulence of Y. pestis (and how the related genes contribute to the virulence of Y. pseudotuberculosis) is to use established animal models of disease for each of these organisms to detect if and where these genes contribute during infection. This strategy has been applied to the autotransporter YapE, which is required for full virulence via the bubonic route (but not the pneumonic route) of Y. pestis infection and is currently being explored for its role in Y. pseudotuberculosis and Y. enterocolitica (37; M. B. Lawrenz and V. L. Miller, unpublished data). Since yapK is conserved in both Y. pseudotuberculosis and Y. pestis, a reasonable hypothesis would be that it is retained because it serves a function useful to both species. In this regard, it is not surprising that yapK contributes less to bubonic infection (a capability exclusive to Y. pestis) and more to septicemic infection (which results frequently from Y. pestis infection and rarely from Y. pseudotuberculosis infection). The inability to detect a similar phenotype for systemic Y. pseudotuberculosis infection serves as a reminder that the diseases caused by these two species vary greatly and that these organisms have different ecological needs that guide their pathogenesis. This contrast is evident following i.p. infection, where the same dose of these two bacteria results in vastly different levels of spleen colonization and infection outcomes. Wild-type Y. pestis reaches ∼109 CFU/gram in the spleen by 2 to 2.5 days postinfection, and death is observed as early as 2 days after infection. The same inoculum of Y. pseudotuberculosis results in spleen colonization that peaks at ∼106 CFU/gram and holds steady between days 2 and 4 before mice begin to die on day 6.
While some of the regions lost in Y. pestis turn out to be essential in Y. pseudotuberculosis (46), this does not appear to be the case with the YPTB3285-YPTB3286 locus, since the deletion of this locus does not measurably impact virulence after oral or i.p. inoculation. Because YPTB3285 is retained by many Y. pestis strains, it seemed less likely to play a role specific to the gastrointestinal infection caused by Y. pseudotuberculosis. However, the inactivation of YPTB3286 in Y. pestis strains made it a better candidate for the contribution to the fecal-oral life cycle that distinguishes Y. pseudotuberculosis from Y. pestis. The functional redundancy among the paralogs does not appear to be masking any phenotypes, since single, double, and triple mutants lacking these genes are no different from the wild type (or each other) in either of the Y. pseudotuberculosis infection models (data not shown). The idea that these genes have nonredundant functions is supported by the observation that in Y. pestis infection, deletions of yapK and yapJ display some individual attenuation. This may indicate that these two genes have at least partially nonredundant functions which both contribute to the systemic phase of Y. pestis infection. Competition (coinfection) experiments with wild-type Y. pseudotuberculosis and the triple mutant in oral and i.p. models also showed no obvious attenuation of the mutant (data not shown). This finding indicates that perhaps the current mouse models (or mouse strains) being used are not the ideal models for discovering how these particular genes benefit Y. pseudotuberculosis (and particularly IP32953).
Whether or not these autotransporter-containing loci have evolved from precursors in Y. pseudotuberculosis to fit new requirements in Y. pestis infection or are serving similar functions that manifest differently in the different diseases caused by these organisms, it should also be considered that the regulation of their expression may have changed following speciation. While outside the scope of this work, evidence is accumulating that regulatory differences exist between Y. pestis and Y. pseudotuberculosis, resulting in the altered expression of virulence factors (15); some of these differences may be related to the small RNA-stabilizing regulator Hfq and alterations in the compositions of sRNAs in Y. pestis versus Y. pseudotuberculosis (9, 32). Just as the coding sequences of yapK in Y. pestis and Y. pseudotuberculosis share a high level of sequence identity, the promoter regions of these genes are also nearly identical. Although the promoter regions of yapK in Y. pestis and Y. pseudotuberculosis are nearly identical, (in the 831 bases between the stop codon of the upstream gene ubiC and the start codon of yapK, there are only 3 nucleotide changes between CO92 and IP32953), it remains possible that yapK is expressed differently in these two species. Likewise, potential differences in the regulation of yapJ and yapK might also account for the additive attenuation of a double mutant. Since it is not yet clear what the functions of YapJ and YapK are, it is possible that the small attenuation of the single mutants and the additive attenuation of the double mutant are the result of a stepwise loss of the expression levels of redundant virulence factors. The region upstream of yapJ differs significantly from yapK and from both YPTB3285 and YPTB3286, again reinforcing the idea that yapJ has a regulatory pattern or function distinct from those of yapK and the Y. pseudotuberculosis homologs.
When characterizing virulence factors in Y. pestis CO92, the route of infection can be a critical determinant of the ability to detect attenuation. This has been observed for the global regulator rovA, where a mutant was severely attenuated in a bubonic model of infection but was less attenuated in pneumonic and systemic models (12). This is also the case for a mutant lacking the autotransporter yapE, which was attenuated in bubonic infection but not in pneumonic infection (37). In nature, variation undoubtedly exists in the ways by which plague is transmitted from vector to host, and an organism that is dependent on this cycle must be adapted to accommodate this variation. While it is generally accepted that bubonic plague is spread by infected fleas in which the proventriculus has been blocked and repeated attempts to feed inoculate the host with bacteria (30), there is also evidence that for a portion of intradermal inoculations, a primary septicemic infection is initiated without prior lymph node involvement (51). In that same work, it was found that the Y. pestis-specific virulence factor Pla, which is critical for the progression of both bubonic and pneumonic plague (35, 51), is not required for intravenously initiated septicemic infections. It may be that yapK and yapJ have the opposite specificities, being dispensable for bubonic infection but serving some role in septicemic spread.
Numerous in vitro assays were used to probe the activities of YapJ and YapK, which do not appear to have the clear functions of many previously characterized autotransporters. It is unlikely that they conjugate the residues commonly found in serine protease autotransporters of the Enterobacteriaceae (SPATES) or lipase/esterase autotransporters such as EstA (28, 55). To confirm this, they have no detectable proteolytic activity on immunoglobulins, hemoglobin, or casein, nor do they have any lipase/esterase activity on emulsified tributyrin (J. D. Lenz and V. L. Miller, unpublished data). They do not significantly increase the association of Y. pestis with common lines of cultured epithelial cells or macrophages from various species and do not promote autoaggregation, and the deletion of these genes has no impact on the survival of Y. pestis in mouse or rabbit serum (Lenz and Miller, unpublished). Predictions of functional properties of these proteins have also been largely unhelpful, with no conserved functional domains being identified by CDD (40), Pfam (26), SMART (39), or InterPro (56) searches, aside from the conserved autotransporter domain and some homology to pertactin within the C-terminal regions shared by most conventional autotransporters. Attempts to perform homology modeling of YapJ onto known autotransporter crystal structures work best when fit to E. coli hemoglobin protease (M. Redinbo and M. Frazier, personal communication), but the majority of the similarity exists in the C-terminal end of the passenger domain, where parallel-beta-helix repeats likely make up a common pertactin-like structural core needed for proper folding (31). The N terminus of the molecule does not map well to known structures, which may simply reflect the few passenger domain crystal structures available to model against. It is this N terminus of the protein that is of most interest, as it is the portion that varies the most among all of the genes of the yapJ-yapK family and is the portion most likely to be engaged in interactions with the host. Based on our previous work, we know that yapJ and yapK are induced during bubonic and pneumonic infections of mice (38), and reactivity against either YapJ or YapK (and two other Yaps) has also been detected in convalescent-phase sera from experimentally infected rabbits (7), indicating that at least one of these genes is induced following subcutaneous infections of rabbits. As surface-exposed proteins expressed during infection and recognized by the adaptive immune response, these proteins of unknown function represent potential targets for new vaccines.
As more genomes are sequenced from the numerous isolates of Y. pestis and Y. pseudotuberculosis that have been collected, more of these homologous genes will be discovered, and a picture of their relatedness, their origin, and, possibly, their function will grow only clearer. The evolutionary analysis presented in this work outlines a previously unrecognized relationship among members of a family of autotransporters that span the gap between Y. pestis and Y. pseudotuberculosis, adding some definition to the blurred line between these genetically similar but phenotypically different species. We also provide the first detailed comparison of these genes from both species, using several animal models, in order to begin to explore their unique functions and determine the selective pressures that have driven their gain, loss, or retention in the evolutionary history of Y. pestis.
ACKNOWLEDGMENTS
We thank Nikki Wagner, Eric Weening, M. Chelsea Lane, and Rodrigo Gonzalez for assistance with animal infections and tissue processing; Monica Frazier and Matthew Redinbo for homology modeling of YapJ and related structural discussions; and Matthew Lawrenz (University of Louisville) and Kimberly Walker for critical reading and evaluation of the manuscript.
This study was supported by funds from National Institutes of Health grants R56AI078930, R21AI64313, and U54AI057157 (Southeast Regional Center for Biodefense and Emerging Infectious Diseases, project 006) to V.L.M. and a Morse/Berg fellowship from the Department of Molecular Microbiology, Washington University, to J.D.L.
Footnotes
Published ahead of print 16 July 2012
REFERENCES
- 1. Achtman M, et al. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. U. S. A. 96:14043–14048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alamuri P, et al. 2010. Adhesion, invasion, and agglutination mediated by two trimeric autotransporters in the human uropathogen Proteus mirabilis. Infect. Immun. 78:4882–4894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alamuri P, Mobley HL. 2008. A novel autotransporter of uropathogenic Proteus mirabilis is both a cytotoxin and an agglutinin. Mol. Microbiol. 68:997–1017 [DOI] [PubMed] [Google Scholar]
- 4. Allsopp LP, et al. 2012. Molecular characterization of UpaB and UpaC, two new autotransporter proteins of uropathogenic Escherichia coli CFT073. Infect. Immun. 80:321–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Allsopp LP, et al. 2010. UpaH is a newly identified autotransporter protein that contributes to biofilm formation and bladder colonization by uropathogenic Escherichia coli CFT073. Infect. Immun. 78:1659–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Andrews GP, et al. 2010. Identification of in vivo-induced conserved sequences from Yersinia pestis during experimental plague infection in the rabbit. Vector Borne Zoonotic Dis. 10:749–756 [DOI] [PubMed] [Google Scholar]
- 8. Auerbuch V, Isberg RR. 2007. Growth of Yersinia pseudotuberculosis in mice occurs independently of Toll-like receptor 2 expression and induction of interleukin-10. Infect. Immun. 75:3561–3570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bai G, Golubov A, Smith EA, McDonough KA. 2010. The importance of the small RNA chaperone Hfq for growth of epidemic Yersinia pestis, but not Yersinia pseudotuberculosis, with implications for plague biology. J. Bacteriol. 192:4239–4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Balada-Llasat JM, Panilaitis B, Kaplan D, Mecsas J. 2007. Oral inoculation with type III secretion mutants of Yersinia pseudotuberculosis provides protection from oral, intraperitoneal, or intranasal challenge with virulent Yersinia. Vaccine 25:1526–1533 [DOI] [PubMed] [Google Scholar]
- 11. Benz I, Schmidt MA. 2011. Structures and functions of autotransporter proteins in microbial pathogens. Int. J. Med. Microbiol. 301:461–468 [DOI] [PubMed] [Google Scholar]
- 12. Cathelyn JS, Crosby SD, Lathem WW, Goldman WE, Miller VL. 2006. RovA, a global regulator of Yersinia pestis, specifically required for bubonic plague. Proc. Natl. Acad. Sci. U. S. A. 103:13514–13519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chain PS, et al. 2004. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. U. S. A. 101:13826–13831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chain PS, et al. 2006. Complete genome sequence of Yersinia pestis strains Antiqua and Nepal516: evidence of gene reduction in an emerging pathogen. J. Bacteriol. 188:4453–4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chauvaux S, et al. 2011. In silico comparison of Yersinia pestis and Yersinia pseudotuberculosis transcriptomes reveals a higher expression level of crucial virulence determinants in the plague bacillus. Int. J. Med. Microbiol. 301:105–116 [DOI] [PubMed] [Google Scholar]
- 16. Comer JE, et al. 2010. Transcriptomic and innate immune responses to Yersinia pestis in the lymph node during bubonic plague. Infect. Immun. 78:5086–5098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Darling AE, Mau B, Perna NT. 2010. ProgressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147 doi:10.1371/journal.pone.0011147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Derbise A, et al. 2010. Delineation and analysis of chromosomal regions specifying Yersinia pestis. Infect. Immun. 78:3930–3941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Doll JM, et al. 1994. Cat-transmitted fatal pneumonic plague in a person who traveled from Colorado to Arizona. Am. J. Trop. Med. Hyg. 51:109–114 [DOI] [PubMed] [Google Scholar]
- 20. Dorsey CW, Laarakker MC, Humphries AD, Weening EH, Baumler AJ. 2005. Salmonella enterica serotype Typhimurium MisL is an intestinal colonization factor that binds fibronectin. Mol. Microbiol. 57:196–211 [DOI] [PubMed] [Google Scholar]
- 21. Dube P. 2009. Interaction of Yersinia with the gut: mechanisms of pathogenesis and immune evasion. Curr. Top. Microbiol. Immunol. 337:61–91 [DOI] [PubMed] [Google Scholar]
- 22. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Eisen RJ, Gage KL. 2009. Adaptive strategies of Yersinia pestis to persist during inter-epizootic and epizootic periods. Vet. Res. 40:01. doi:10.1051/vetres:2008039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Elder KD, Harvill ET. 2004. Strain-dependent role of BrkA during Bordetella pertussis infection of the murine respiratory tract. Infect. Immun. 72:5919–5924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Eppinger M, et al. 2010. Genome sequence of the deep-rooted Yersinia pestis strain Angola reveals new insights into the evolution and pangenome of the plague bacterium. J. Bacteriol. 192:1685–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Finn RD, et al. 2010. The Pfam protein families database. Nucleic Acids Res. 38:D211–D222 doi:10.1093/nar/gkp985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guinet F, Ave P, Jones L, Huerre M, Carniel E. 2008. Defective innate cell response and lymph node infiltration specify Yersinia pestis infection. PLoS One 3:e1688 doi:10.1371/journal.pone.0001688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Henderson IR, Nataro JP. 2001. Virulence functions of autotransporter proteins. Infect. Immun. 69:1231–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hinchliffe SJ, et al. 2003. Application of DNA microarrays to study the evolutionary genomics of Yersinia pestis and Yersinia pseudotuberculosis. Genome Res. 13:2018–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jarrett CO, et al. 2004. Transmission of Yersinia pestis from an infectious biofilm in the flea vector. J. Infect. Dis. 190:783–792 [DOI] [PubMed] [Google Scholar]
- 31. Junker M, et al. 2006. Pertactin beta-helix folding mechanism suggests common themes for the secretion and folding of autotransporter proteins. Proc. Natl. Acad. Sci. U. S. A. 103:4918–4923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Koo JT, Alleyne TM, Schiano CA, Jafari N, Lathem WW. 2011. Global discovery of small RNAs in Yersinia pseudotuberculosis identifies Yersinia-specific small, noncoding RNAs required for virulence. Proc. Natl. Acad. Sci. U. S. A. 108:E709–E717 doi:10.1073/pnas.1101655108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Larkin MA, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 34. Lathem WW, Crosby SD, Miller VL, Goldman WE. 2005. Progression of primary pneumonic plague: a mouse model of infection, pathology, and bacterial transcriptional activity. Proc. Natl. Acad. Sci. U. S. A. 102:17786–17791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lathem WW, Price PA, Miller VL, Goldman WE. 2007. A plasminogen-activating protease specifically controls the development of primary pneumonic plague. Science 315:509–513 [DOI] [PubMed] [Google Scholar]
- 36. Laukkanen-Ninios R, et al. 2011. Population structure of the Yersinia pseudotuberculosis complex according to multilocus sequence typing. Environ. Microbiol. 13:3114–3127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lawrenz MB, Lenz JD, Miller VL. 2009. A novel autotransporter adhesin is required for efficient colonization during bubonic plague. Infect. Immun. 77:317–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lenz JD, et al. 2011. Expression during host infection and localization of Yersinia pestis autotransporter proteins. J. Bacteriol. 193:5936–5949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Letunic I, Doerks T, Bork P. 2009. SMART 6: recent updates and new developments. Nucleic Acids Res. 37:D229–D232 doi:10.1093/nar/gkn808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Marchler-Bauer A, et al. 2011. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 39:D225–D229 doi:10.1093/nar/gkq1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mecsas J, Bilis I, Falkow S. 2001. Identification of attenuated Yersinia pseudotuberculosis strains and characterization of an orogastric infection in BALB/c mice on day 5 postinfection by signature-tagged mutagenesis. Infect. Immun. 69:2779–2787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Merriam JJ, Mathur R, Maxfield-Boumil R, Isberg RR. 1997. Analysis of the Legionella pneumophila fliI gene: intracellular growth of a defined mutant defective for flagellum biosynthesis. Infect. Immun. 65:2497–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Miller VL, Mekalanos JJ. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulaton of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Morelli G, et al. 2010. Yersinia pestis genome sequencing identifies patterns of global phylogenetic diversity. Nat. Genet. 42:1140–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Noofeli M, et al. 2011. BapC autotransporter protein is a virulence determinant of Bordetella pertussis. Microb. Pathog. 51:169–177 [DOI] [PubMed] [Google Scholar]
- 46. Pouillot F, Fayolle C, Carniel E. 2008. Characterization of chromosomal regions conserved in Yersinia pseudotuberculosis and lost by Yersinia pestis. Infect. Immun. 76:4592–4599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Prentice MB, Rahalison L. 2007. Plague. Lancet 369:1196–1207 [DOI] [PubMed] [Google Scholar]
- 48. Roy K, et al. 2011. Adhesin degradation accelerates delivery of heat-labile toxin by enterotoxigenic Escherichia coli. J. Biol. Chem. 286:29771–29779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ruiz-Bravo A, Moreno E, Sampedro A, Jimenez-Valera M. 1999. Experimental infection of mice with Yersinia enterocolitica serotype O9 by oral and parenteral routes: spreading and enterotropism of virulent yersiniae. Curr. Microbiol. 38:257–263 [DOI] [PubMed] [Google Scholar]
- 50. Schiano CA, Bellows LE, Lathem WW. 2010. The small RNA chaperone Hfq is required for the virulence of Yersinia pseudotuberculosis. Infect. Immun. 78:2034–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sebbane F, Jarrett CO, Gardner D, Long D, Hinnebusch BJ. 2006. Role of the Yersinia pestis plasminogen activator in the incidence of distinct septicemic and bubonic forms of flea-borne plague. Proc. Natl. Acad. Sci. U. S. A. 103:5526–5530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Simonet M, Riot B, Fortineau N, Berche P. 1996. Invasin production by Yersinia pestis is abolished by insertion of an IS200-like element within the inv gene. Infect. Immun. 64:375–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Walker KA, Miller VL. 2004. Regulation of the Ysa type III secretion system of Yersinia enterocolitica by YsaE/SycB and YsrS/YsrR. J. Bacteriol. 186:4056–4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Westphal S, et al. 2008. Resistance of chemokine receptor 6-deficient mice to Yersinia enterocolitica infection: evidence of defective M-cell formation in vivo. Am. J. Pathol. 172:671–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wilhelm S, Tommassen J, Jaeger KE. 1999. A novel lipolytic enzyme located in the outer membrane of Pseudomonas aeruginosa. J. Bacteriol. 181:6977–6986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zdobnov EM, Apweiler R. 2001. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17:847–848 [DOI] [PubMed] [Google Scholar]