Abstract
Ehrlichia chaffeensis is an obligate intracellular bacterium that causes human monocytic ehrlichiosis (HME). To determine what host components are important for bacterial replication, we performed microarray analysis on Drosophila melanogaster S2 cells by comparing host gene transcript levels between permissive and nonpermissive conditions for E. chaffeensis growth. Five-hundred twenty-seven genes had increased transcript levels unique to permissive growth conditions 24 h postinfection. We screened adult flies that were mutants for several of the “permissive” genes for the ability to support Ehrlichia replication. Three additional D. melanogaster fly lines with putative mutations in pyrimidine metabolism were also tested. Ten fly lines carrying mutations in the genes CG6479, separation anxiety, chitinase 11, CG6364 (Uck2), CG6543 (Echs1), withered (whd), CG15881 (Ccdc58), CG14806 (Apop1), CG11875 (Nup37), and dumpy (dp) had increased resistance to infection with Ehrlichia. Analysis of RNA by quantitative real-time reverse transcription-PCR (qRT-PCR) confirmed that the bacterial load was decreased in these mutant flies compared to wild-type infected control flies. Seven of these genes (san, Cht11, Uck2, Echs1, whd, Ccdc58, and Apop1) encoded proteins that had mitochondrial functions or could be associated with proteins with mitochondrial functions. Treatment of THP-1 cells with double-stranded RNA to silence the human UCK2 gene indicates that the disruption of the uridine-cytidine kinase affects E. chaffeensis replication in human macrophages. Experiments with cyclopentenyl cytosine (CPEC), a CTP synthetase inhibitor and cytosine, suggest that the nucleotide salvage pathway is essential for E. chaffeensis replication and that it may be important for the provision of CTP, uridine, and cytidine nucleotides.
INTRODUCTION
Ehrlichia chaffeensis is the causative agent of human monocytic ehrlichiosis (HME). There were 1,429 cases of HME in 2010 and 2011 (14). This represents a significant increase in the incidence of the disease since 2003 and qualifies HME as an emerging infectious disease (25). In addition to being reported in the United States, HME has also been documented in Africa, Europe, China, and Brazil (9, 16, 42, 67). E. chaffeensis is an obligate intracellular bacterium. However, little is known about the parasitized-host requirements for bacterial replication.
Drosophila melanogaster has been used to study a variety of intracellular pathogens. In particular, it has been successfully manipulated for the identification of genes involved in host-pathogen interactions. These pathogens include Listeria monocytogenes (1, 2), Chlamydia trachomatis (20), Mycobacterium marinum (1, 18, 30, 48), Francisella tularensis (51, 64), and the protozoan parasite Plasmodium gallinaceum (8, 53).
We previously demonstrated that E. chaffeensis is capable of infecting, completing its life cycle, and maintaining its pathogenicity in both Drosophila S2 cells (39) and adult flies (40). We have also identified growth conditions that were nonpermissive for the growth of E. chaffeensis infection in Drosophila S2 cells (39). Therefore, we hypothesized that a transcriptional microarray analysis of permissively infected and nonpermissively infected S2 cells would reveal host genes that contribute to the replication of Ehrlichia. We used the Affymetrix Drosophila 2.0 array to identify a subset of genes that were exclusively expressed during E. chaffeensis infection in infected S2 cells under permissive growth conditions. We screened adult flies carrying mutations in several of the candidate genes, 10 of which change the fly response to E. chaffeensis compared to wild-type flies. Gene products from 7 of the 10 genes identified are associated with mitochondrial function and/or location. We also describe follow-up experiments to investigate how Uck2 might function and its relevance to mammalian infections.
MATERIALS AND METHODS
Maintenance of cell lines and E. chaffeensis infections.
The canine macrophage cell line DH82 (ATCC CRL-10389) was cultured in Eagle's minimal essential medium supplemented with 7% fetal bovine serum (FBS) (Atlanta Biologicals, Atlanta, GA) (EMEM7). THP-1 (ATCC TIB-202) cells were cultured in RPMI 1640 medium supplemented with 10% FBS. Drosophila S2 cells, obtained from the Drosophila RNAi Screening Center (Harvard Medical School, Boston, MA), were cultivated at 25°C in Schneider's Drosophila medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum. For experiments, S2 cells were seeded at a concentration of 1 × 106 cells/well in 6-well plates.
The E. chaffeensis Arkansas isolate was continuously cultivated in DH82 cells, as described previously, at 37°C, 8% CO2 in EMEM7 (40). Purified bacteria were used to reinfect DH82, THP-1, and S2 cells.
Infections in permissive and nonpermissive Drosophila S2 cells.
To make S2 cells nonpermissive to E. chaffeensis infection, the cells were allowed to adhere for 30 min prior to adding sonicated lipopolysaccharide (LPS) from Salmonella enterica serovar Minnesota (Sigma, St. Louis, MO) at a concentration of 10 μg per ml. The S2 cells plus LPS were incubated for 5 h prior to infection with E. chaffeensis. The Ehrlichia-infected S2 cells were monitored at 24 and 96 h postinfection (p.i.). Additionally, uninfected S2 cells, inactivated and activated with LPS, were used as controls. S2 cells were centrifuged at 300 × g for 5 min prior to RNA isolation from cell pellets using 1 ml of TriReagent (Molecular Research Center, Inc., Cincinnati, OH). The TriReagent-cell mixture was transferred to 2.0-ml Heavy Phase Lock Gel tubes (5 Prime, Westbury, NY). Chloroform (200 μl) was added, and the mixture was gently mixed for 15 s. After centrifugation at 12,000 × g for 10 min at 4°C, the aqueous phase was poured into 1.5-ml tubes. The RNA was precipitated and washed with isopropanol and ethanol according to the manufacturer's instructions. The RNA pellet was suspended in 50 μl of nuclease-free water, and the RNA concentration was determined spectrophotometrically (NanoDrop Technologies, Wilmington, DE) before and after DNA digestion using a Turbo DNA-free kit (Ambion Inc., Austin, TX).
Determination of infection by RT-PCR and quantitative real-time RT-PCR (qRT-PCR).
For the microarray, infections were confirmed by reverse transcriptase (RT) PCR using the Promega Access One-Step RT-PCR kit (Madison, WI). Total RNA (750 ng) was used for each 25-μl reaction mixture, which contained the following reagents: 1× buffer, 0.2 mM deoxynucleoside triphosphates (dNTPs), 2 μM forward primer, 2 μM reverse primer, 1.5 mM MgSO4, 1 U/μl DNA polymerase, 1 U/μl reverse transcriptase, and nuclease-free water. RT-PCRs were performed in a thermocycler (Eppendorf Mastercycler Gradient, Hauppauge, NY). Primers and probes were designed with Primer Quest software (Integrated DNA Technologies, Coralville, IA) and data from NCBI reference sequences (see Table S1 in the supplemental material). RT-PCR conditions for the 16S rRNA of E. chaffeensis were 48°C for 45 min, 94°C for 4 min, and then 35 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 1 min (21). The ribosomal protein 49 gene (rp49) was used as a housekeeping gene in RT-PCR experiments with Drosophila S2 cells. The RT-PCR conditions for rp49 gene amplification were 48°C for 45 min, 94°C for 2 min, and then 35 cycles of 94°C for 45 s, 50°C for 1 min, and 72°C for 1.5 min. No-template RT-PCR controls were also included. RT-PCR product bands were identified on a ChemiImager (Protein Simple, Santa Clara, CA) after electrophoresis in 2% agarose gels and staining with ethidium bromide.
Flies were anesthetized using CO2 prior to homogenization with disposable pestles (Kimble Chase, Vineland, NJ) in 1 ml of TriReagent. The homogenates were transferred to 2.0 ml Heavy Phase Lock Gel tubes and processed as described above for RNA isolation.
Ehrlichia quantification in S2 cells and fly experiments was estimated using TaqMan-based qRT-PCR, as previously described (40, 55), using Drosophila ribosomal protein 15a as the housekeeping gene with primers described in Table S1 in the supplemental material. E. chaffeensis was detected as described previously (40, 55). The qRT-PCRs were performed in the Cepheid SmartCycler System (Sunnyvale, CA).
Analysis of gene expression based on qRT-PCR results was performed using the method described by Pfaffl (47). In short, primer efficiencies were calculated from standard dilution curves plotting threshold cycle (CT) values versus log RNA using the following equation: efficiency = 10(−1/slope of standard curve).
The change in CT values for both genes of interest and housekeeping controls was determined by calculating the ΔΔCT using the following equation: (efficiency of gene of interestGene of interest:ΔCT control − treated)/(efficiency of housekeeping geneHousekeeping gene:ΔCT control − treated).
Microarray analysis.
Microarray analysis was performed at the University of Kansas Medical Center Microarray Facility (Kansas City, KS) using Affymetrix (Santa Clara, CA) Drosophila 2.0 Gene chips according to the manufacturer's specifications. The analysis was performed on four treatment groups (each submitted in triplicate at 24 h p.i.): (i) S2 cells infected with E. chaffeensis, (ii) S2 cells incubated with LPS and then infected with E. chaffeensis, (iii) S2 cells incubated with LPS, and (iv) untreated/uninfected S2 cells. CHP files were analyzed using GeneSpring 7.3 software and normalized by “per gene: normalize to median.” The “filter on volcano plot” was applied at 1.5-fold change and one-way analysis of variance (ANOVA) at a significance level (α) of 0.05. Genes upregulated 1.5-fold or more above basal expression levels were identified under both permissive and nonpermissive conditions compared to uninfected controls at 24 h p.i. These gene sets were then compared, and those upregulated exclusively under either permissive or nonpermissive conditions at 24 h p.i. were identified. Microarray MIAMI-compliant data are available at the publicly accessible database (http://bioinformatics.kumc.edu/mdms/login.php) by accessing the experiment entitled “Differential Gene Expression in Ehrlichia chaffeensis-infected S2 cells.”
D. melanogaster.
Flies were maintained on standard dextrose-molasses-yeast medium at 18 to 29°C. For all experiments, flies with the appropriate background were used as wild-type (WT) controls. w;Hemese-Gal4 UASGFP flies (GFPHeme) (from Michael J. Williams, Umea Centre for Molecular Pathogenesis, Umea University, Umea, Sweden), yellow-white (yw) (maintained in our stock collection at Kansas State University), and/or white ocelli (wo1) (stock number 634, from the Bloomington Drosophila Stock Center at Indiana University, Bloomington, IN) were used as the WT in these experiments. withered (whd1) (FBgn0004012; stock number 441), dumpy (dpov1) (FBgn0053196; stock number 276), and tilt (tt1wo1) (FBgn0003868; stock number 623) mutants are all the result of spontaneous mutations (35, 44, 65) and were obtained from the Bloomington Drosophila Stock Center at Indiana University, Bloomington, IN. The stock numbers, genotypes, and associated genes of adult Drosophila flies screened by microinjection are listed in Table S2 in the supplemental material.
Adult Drosophila Infections.
Flies were transferred to fresh food at least 24 h prior to injection/infection. For injection/infection, adult male and female flies were anesthetized with CO2 (for no longer than 15 min at a time). Flies were injected with approximately 50 nl of sterile PBS or with >5,000 bacteria, using pulled glass capillary needles. Injections were made in the abdomen of the fly, close to the junction of the thorax and ventral to the junction between the dorsal and ventral cuticles. Following injection, the flies were maintained in clean bottles with molasses caps that were changed every other day throughout the course of the experiments. Survival of the flies was monitored daily.
CPEC treatment of S2 cells.
Cyclopentenylcytosine (CPEC) was obtained from the National Cancer Institute (Drug Synthesis and Chemistry Branch, Developmental Therapeutics Program, Division of Cancer Treatment and Diagnosis, National Cancer Institute, Bethesda, MD) through a Materials Transfer Agreement. The CPEC was prepared to a final concentration of 15.06 mM using sterile water containing 1% dimethyl sulfoxide (DMSO). Two different infection protocols were used for testing the effect of CPEC on the growth of E. chaffeensis in the S2 cells. For the first protocol, CPEC was added to S2 cells at final concentrations of 100, 10, 1, or 0.1 μM/well; E. chaffeensis was added to the treated cells 2 days later, and RNA was extracted from the cells 2 days postinfection. For the second protocol, S2 cells were infected with E. chaffeensis for 2 days, and then various concentrations of CPEC were then added and RNA extractions were performed 2 days following the CPEC treatment. S2 cells treated with diluent only and infected with Ehrlichia and uninfected S2 cells were used as controls. A qRT-PCR assay was used to analyze transcript levels of Ehrlichia 16S rRNA and Drosophila ribosomal protein 15a, as described above.
Addition of exogenous cytosine to Drosophila S2 cells.
Cytosine was obtained from Sigma-Aldrich Co. The cytosine was dissolved in 500 mM hydrochloric acid and then in sterile water to 100 mM, according to the manufacturer's recommendation. For infection experiments, S2 cells were incubated for 24 h prior to cytosine addition to a final concentration of 25 mM; the cells were further incubated for 24 h. E. chaffeensis was added to the cytosine-treated cells, to untreated S2 cells, and to S2 cells treated with the cytosine diluent. Untreated, uninfected S2 cells were used as a negative control. RNA was extracted from the cells at 24, 48, 72, and 96 h p.i. qRT-PCR was used to measure transcript levels of Ehrlichia 16S rRNA and Drosophila ribosomal protein 15a, as described above.
THP-1 cell differentiation and cell cultures.
THP-1 human macrophages were cultured as described above. For differentiation, THP-1 cells (6 × 104 THP-1 cells/well; 24-well plates) were incubated with phorbol 12-myristate 13-acetate (PMA) (Sigma-Aldrich Chemical Co., St. Louis, MO) at a final concentration of 50 μM. The cells were cultured for 24 h to allow final differentiation into macrophages in a cell culture incubator at 37°C with 8% CO2.
Measurement of transfection efficiency and UCK2 knockdown.
We selected the small interfering RNA (siRNA) transfection conditions based on the transfection efficiency in the cells of a fluorescently labeled transfection control siRNA duplex conjugated with the fluorescent marker TYE 563. Transiently transfected cells were analyzed for transfection efficiency using fluorescence microscopy.
UCK2 transcript expression was knocked down in THP-1 cells using siRNA specific to the human UCK2 transcript. We used the TriFecta RNA interference (RNAi) kit with predesigned oligonucleotide sets from IDT. The siRNA predesigned oligonucleotide query was done using the UCK2 RNA coding sequence (CDS) from NCBI reference sequence XM_846154.2 for Homo sapiens. We also used scrambled siRNA and human hypoxanthine phosphoribosyltransferase (HPRT) siRNAs as negative and positive controls, respectively, to confirm transfection and siRNA knockdown efficiency. All siRNA duplexes were dissolved in nuclease-free water (pH 7.2) to a final concentration of 2 nM. Three UCK2-specific, an HPRT-specific, and scrambled siRNA duplexes were transfected into differentiated THP-1 cells using the Lipofectamine transfection reagent (Invitrogen) following the manufacturer's protocol.
THP-1 cells were infected with E. chaffeensis 48 h posttransfection. Transfected and Ehrlichia-infected THP-1 cells were analyzed for UCK2, HPRT knockdown efficiency and the level of bacterial infection by qRT-PCR 48 h postinfection using primers and probes described in Table S1 in the supplemental material, as explained above.
TEM.
Transmission electron microscopy (TEM) analysis of uninfected and E. chaffeensis-infected DH82 cultures was performed as described previously (17).
Statistics.
Data are presented as means and standard errors of the mean (SEM). Differences in means were determined using the Mann-Whitney two-tailed rank-sum statistical test, which is independent of the underlying population distribution (StatMost statistical package; Data XIOM, Los Angeles, CA). Survival data were analyzed for significance using the log rank test of Kaplan-Meier plots and Prism (La Jolla, CA) Graphpad software. P values of < 0.05 were considered significant.
RESULTS
Microarray “permissive-exclusive” genes after infection with E. chaffeensis.
To identify host genes that are necessary for E. chaffeensis infection, we examined transcript levels of Drosophila S2 cells under four different conditions at 24 h p.i. to assess the host factors that might be needed to initiate the infection. They included (i) uninfected S2 cells, (ii) uninfected S2 cells activated with LPS, (iii) S2 cells activated with LPS and then infected with E. chaffeensis (nonpermissive conditions; this experiment, performed as in previous studies, demonstrated the inhibition of E. chaffeensis replication in LPS-activated S2 cells [43]), and (iv) S2 cells infected with E. chaffeensis (permissive conditions). E. chaffeensis replicated only under permissive conditions and not under nonpermissive conditions for as long as 96 h p.i. (Fig. 1). Uninfected cells were used to discern the basal transcript levels in the S2 cells, and comparisons across the different conditions were based on this basal level of expression. In order to understand which gene transcripts were specific to nonpermissive S2 cells, we used the S2 cells that were treated only with LPS as a comparison. Cells that received only LPS treatment revealed activation-specific gene transcripts. We compared those genes to the genes that had increased transcript levels in cells infected under nonpermissive conditions for bacterial growth to deduce transcripts that were upregulated exclusively under nonpermissive conditions. We also determined the gene transcripts that were upregulated 1.5-fold or more exclusively under permissive infection conditions for E. chaffeensis growth, and 2,128 genes were upregulated 1.5-fold more than in uninfected controls. Under nonpermissive growth conditions, 1,742 of the genes were upregulated 1.5-fold more than in uninfected S2 cells. When we compared transcripts that were upregulated under permissive growth conditions to transcripts that were upregulated under nonpermissive growth conditions, we identified 527 genes unique to permissive conditions (see Table S3 in the supplemental material). Of these, 210 had previously been ascribed some function and had some characterization. The functions of the remaining 307 genes had yet to be defined and had “CG” (computed gene) gene designations. However, a number of these genes had orthologs with some described properties. These ortholog gene descriptions were used to ascribe functions for our analyses. Viable and fertile adult fly mutant stocks were available for 118 of the 527 genes (37 stocks for defined genes and 81 stocks for undefined genes). These flies had appropriate mutations in coding exons in the genes of interest and allowed testing of how the absence of a functional gene would affect fly survival and/or bacterial replication in vivo. Our screen of mutant fly lines was loosely based on descriptions of genes that controlled functions that we guessed might be important for bacterial growth or were random choices of CG-designated genes.
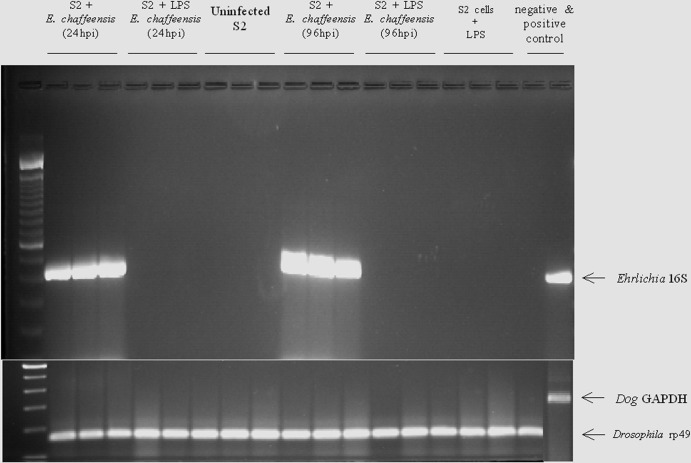
Fig 1.
Infection of microarray samples by E. chaffeensis. S2 cells with and without LPS treatment were assayed for E. chaffeensis infection after 24 or 96 h; uninfected S2 cells were also assessed. E. chaffeensis infection was confirmed by assessing the 16S ribosomal RNA as described in Materials and Methods. D. melanogaster rp49 transcript was used as a loading control.
E. chaffeensis infection in selected mutant Drosophila fly lines.
We screened 19 Drosophila lines with mutations in the genes of interest (see Table S2 in the supplemental material) for bacterial replication and fly survival. Flies (wild type and mutant) were injected with cell-free E. chaffeensis or sterile PBS and monitored for survival for 96 to 120 h p.i. (20 flies per treatment group per experiment; the experiment was repeated independently at least 3 times). We looked for the mutants that displayed increased survival of the flies after bacterial challenge compared to the wild-type flies. Our hypothesis was that the gene affected in a mutant allowed increased survival because its expression was needed for E. chaffeensis replication. We found that the flies that had mutations in Nup37, Echs1, Cht11, CG6479, Ccdc58, Apop1, san, and Uck2 displayed significantly increased survival compared to wild-type flies after infection with E. chaffeensis (Fig. 2). In contrast, several mutants did not show increased resistance to E. chaffeensis infections. They included three G3pdh mutants (stock 1124, shown in Fig. 2) and CG14434 (Fig. 2), as well as CG10672, CG4743, CG9300, tsp3A, CG10992, and Gap69C (data not shown).
Fig 2.
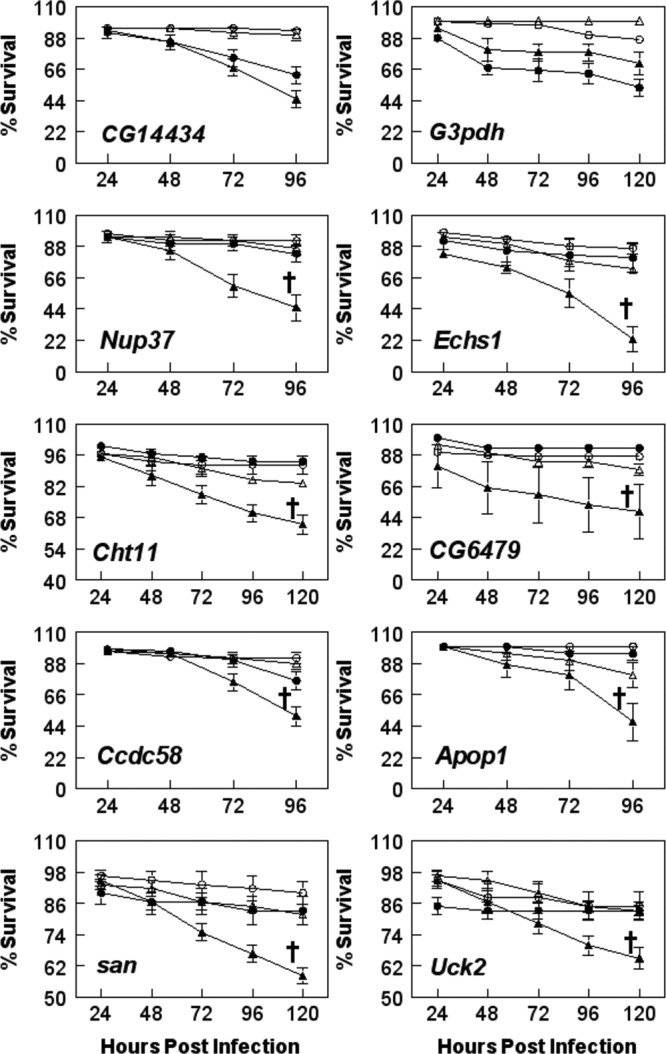
Infection of mutant flies with E. chaffeensis. D. melanogaster was mutant for the indicated genes and was observed for sensitivity to infection. Wild-type flies were injected with PBS (△) or bacteria (▲), and mutant flies were injected with PBS (○) or bacteria (●). The data represent means ± SEM of three or more independent experiments, with 20 flies per treatment group per experiment. †, survival is significantly different from that of E. chaffeensis-infected wild-type flies; P < 0.05 using the log rank test of the Kaplan-Meier plots.
To confirm that Nup37, Echs1, Cht11, CG6479, Ccdc58, Apop1, san, and Uck2 gene mutations affected E. chaffeensis infections, we also estimated bacterial replication as measured by qRT-PCR (Fig. 3). Experimental infection of flies with disruptions in all 8 genes resulted in a significant drop in the numbers of bacteria compared to those observed in wild-type flies infected with the organism (Fig. 3).
Fig 3.
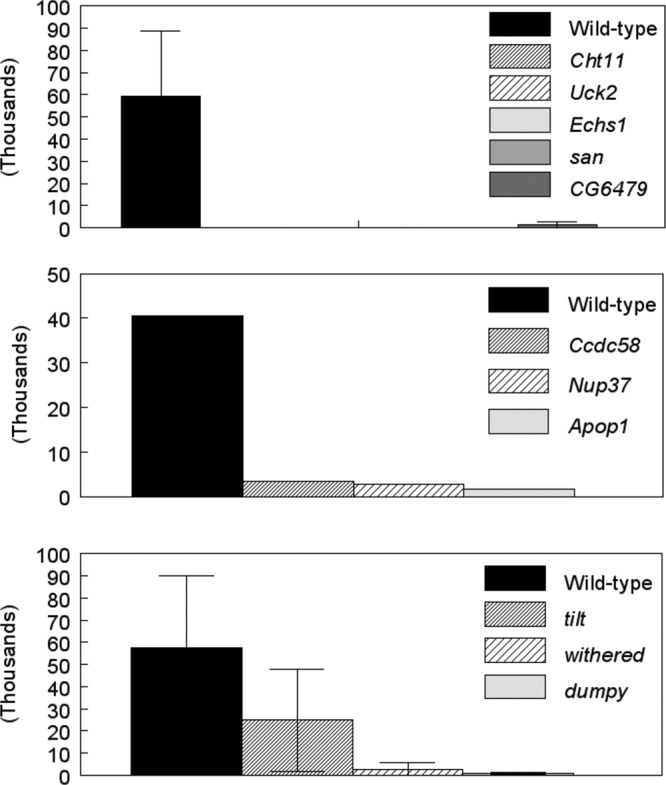
Bacterial growth in Drosophila mutants. Bacterial numbers were quantified 96 h after infection using qRT-PCR as described in Materials and Methods. The data represent means ± SEM of three assessments for Cht11, Uck2, Echs1, san, CG6479, tilt, withered, and dumpy and one assessment for Ccdc58, Nup37, and Apop1.
Uridine/cytidine kinase mutations affect fly survival and bacterial replication.
To understand the physiological relevance of disruption of gene function to E. chaffeensis growth in the host, we focused on Uck2, as an ortholog for this gene has been identified in mammalian species (43). Stroman (56) previously reported that the Drosophila dumpy, tilt, and withered stocks carried a mutation(s) that affects uridine/cytidine kinase function. To confirm the impact of Uck2 on E. chaffeensis growth, the three fly lines with putative defects in uridine/cytidine kinase function were infected with E. chaffeensis and assessed for bacterial replication. Two of these fly lines were significantly more resistant to E. chaffeensis challenge than WT flies (Fig. 4). For example, at 96 h p.i., 61% of the dumpy flies were alive compared to 42% of WT flies; similarly, 66% of withered flies survived compared to 38% of WT flies (P < 0.05). Although there was a tendency for the tilt flies (60% ± 16% survival) to be more resistant than the WT flies (46% ± 7% survival), the differences were not statistically significant (Fig. 4). The increased survival of the withered and dumpy mutants was accompanied by a significant decrease in the replication of the Ehrlichia organisms, as measured by qRT-PCR assay (Fig. 3). At 96 h p.i., WT flies contained significantly more bacteria than withered and dumpy mutants (P = 0.02) (Fig. 3, bottom). The tilt mutants averaged fewer bacteria, but there was more variation in the samples.
Fig 4.
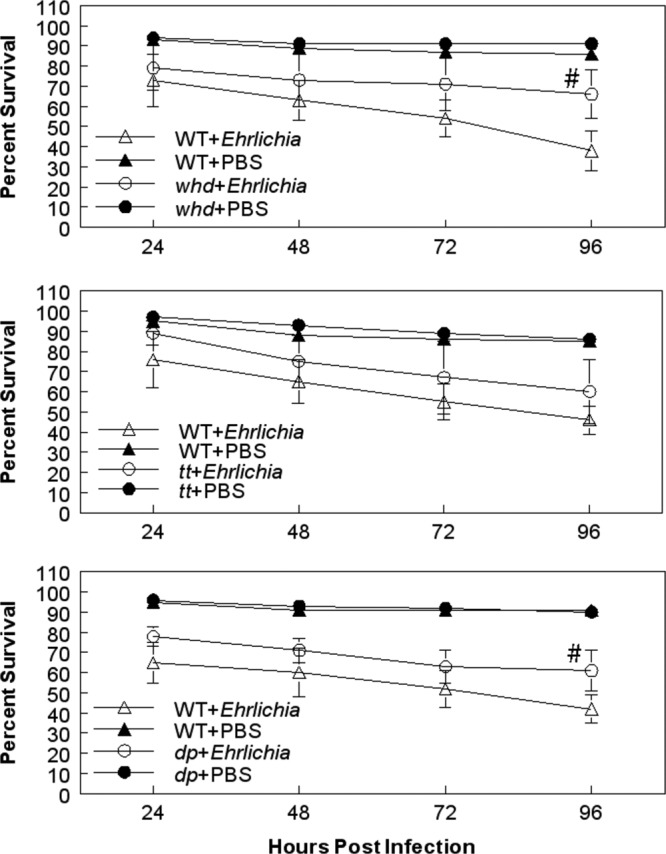
Effects of withered (top), tilt (middle), and dumpy (bottom) gene mutations on E. chaffeensis infections. withered, tilt, and dumpy flies were screened for their sensitivity to infection. The data represent means ± SEM of 4 or 5 independent experiments, with 20 flies per treatment group per experiment. #, survival is significantly different from that of E. chaffeensis-infected wild-type flies; P < 0.05 using the log rank test of the Kaplan-Meier plots.
CPEC treatment increases E. chaffeensis infection.
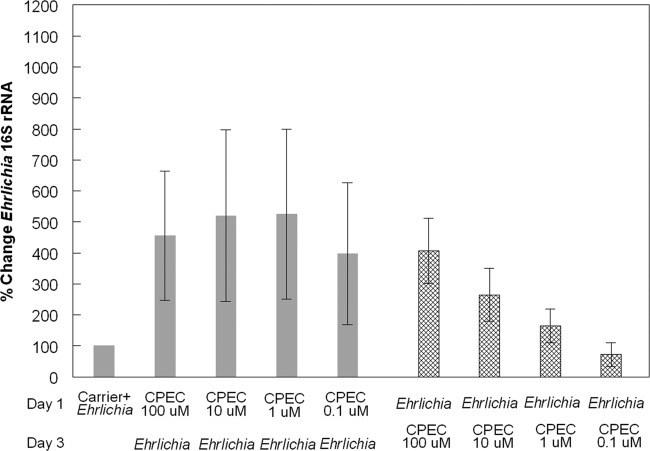
The uridine/cytidine kinase enzyme functions in pyrimidine synthesis pathways and is specifically involved in the conversion of uridine to UMP and cytidine to CMP (52, 62, 63) (Fig. 5). dCTP can also be synthesized from glutamine through the de novo synthesis pathway, as well (52, 62) (Fig. 5). To determine the importance of the de novo synthesis pathway to E. chaffeensis infections, we obtained the drug cyclopentenyl cytosine, which inhibits the conversion of [3H]UTP to [3H]CTP (28), as an inhibitor of CTP synthetase (45, 52, 62). The drug effectively inhibits de novo synthesis of pyrimidines, leaving only the salvage pathway, with cytidine as the substrate for pyrimidine synthesis. To explore the impact of CPEC on E. chaffeensis growth, S2 cells were treated with CPEC for 48 h at a final concentration of 0.1, 1, 10, or 100 μM. Subsequently, we infected those cells with E. chaffeensis for an additional 48 h. We found a significant increase in bacterial growth in all of the treated cells compared to infected cells treated with carrier only (Fig. 6, solid bars; P < 0.01). Similarly, when S2 cells were first infected with E. chaffeensis for 48 h and then treated with different concentrations of CPEC for 48 h, we observed significant increases in bacterial growth in cells treated with 10 and 100 μM CPEC compared to infected cells treated with carrier only (Fig. 6, cross-hatched bars; P < 0.01). Therefore, the de novo synthesis pathway was not needed for bacterial growth in the S2 cells and usually enhanced infection.
Fig 5.
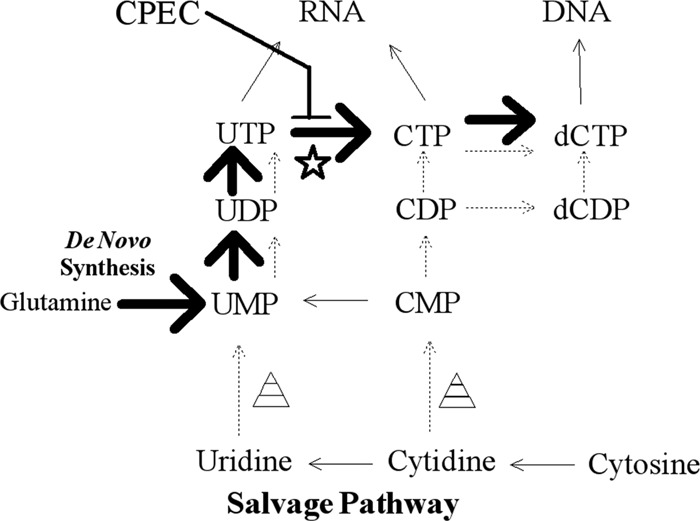
Nucleotide de novo and salvage pathways for production of dCTP. De novo synthesis through glutamine and UMP is represented by large solid arrows. The salvage pathways through uridine or cytidine are represented by thin and dashed arrows. The star represents the enzyme CTP synthetase, and the triangles represent uridine/cytidine kinase. Inhibition by CPEC occurs at the conversion of UTP to CTP.
Fig 6.
Effect of cyclopentenyl cytosine on E. chaffeensis infections. Percent change in E. chaffeensis 16S rRNA copies in S2 cells treated with CPEC compared to S2 cells treated with carrier only. There were two treatment schemes. (i) Day 1, CPEC. S2 cells were first treated with CPEC (at the indicated concentrations) for 48 h and then infected with E. chaffeensis for an additional 48 h (solid bars). (ii) Day 1, E. chaffeensis. S2 cells were first infected with E. chaffeensis for 48 h and then treated with CPEC (at the indicated concentrations) for 48 h (cross-hatched bars). RNA extraction was performed following both treatment schemes. The data presented represent the means ± SEM of 3 independent experiments.
Cytosine treatment increases E. chaffeensis infection.
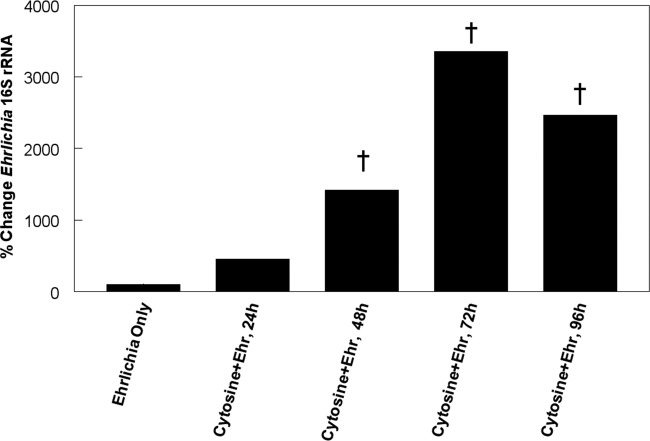
Cytosine combines with ribose to form cytidine in the nucleotide salvage pathway. Therefore, to determine if it is an important substrate for bacterial growth, we supplemented S2 cell culture medium for 24 h with 25 mM cytosine prior to infection with E. chaffeensis, and bacterial replication was assessed at up to 96 h p.i. (Fig. 7). The cytosine-treated infected cells contained averages of 456%, 1423%, 3,352%, and 2,465% more copies of Ehrlichia 16S rRNA at 24, 48, 72, and 96 h p.i., respectively, than mock-infected cells.
Fig 7.
Effect of cytosine on E. chaffeensis infections. Shown is the percent change in Ehrlichia 16S rRNA copies in S2 cells treated with cytosine and infected with E. chaffeensis organisms (Ehr) compared to S2 cells treated with carrier only and infected with bacteria. The data presented represent the means of 3 independent experiments. †, significant difference (P < 0.03). Ehrlichia only represents the baseline control, which was the same at each time, so only one control is presented.
Silencing of UCK2 in human THP-1 cells.
The uridine-cytidine kinase UCK2 is a conserved enzyme that is present in humans, as well as D. melanogaster. Therefore, to examine the contribution of the enzyme to E. chaffeensis growth in human cells, we used double-stranded RNA (dsRNA) to induce UCK2 gene-specific silencing to inhibit translation of UCK2. We used three different RNA duplex sequences to define the specificity of the UCK2 knockdown. THP-1 cells are a promyelocytic cell line that was induced to differentiate into macrophages for 24 h using phorbol ester. After being silenced for 48 h, cells were infected with E. chaffeensis. Three different siRNA duplexes targeted to the UCK2 transcript consistently inhibited E. chaffeensis growth by 48 h p.i. in the range of 47 to 69% compared to cells treated with a scrambled control siRNA (Table 1). We also detected improved E. chaffeensis growth in cells treated with siRNA for HPRT, which was used as a control for silencing-procedure effectiveness and to help show the specificity of the UCK2-specific duplexes.
Table 1.
Impact of UCK2 dsRNA silencing on E. chaffeensis growth in THP-1 cells
| Treatment | % Inhibitiona | % of E. chaffeensis compared to scrambled dsRNA controla |
|---|---|---|
| Duplex 1 | 99 ± 1b | 31 ± 21 (9) |
| Duplex 2 | 97 ± 5b | 46 ± 32 (24) |
| Duplex 3 | 96 ± 3b | 53 ± 30 (29) |
| HPRT | 100 ± 0c | 225 ± 86 (108) |
Means ± SEM (median) of 5 independent experiments. Control E. chaffeensis numbers: 26,348 ± 4,147.
Silencing efficiency compared to scrambled dsRNA.
Silencing efficiency of HPRT dsRNA. HPRT was used as a control to ensure transfection and silencing-protocol effectiveness.
Mitochondrial localization around morulae in E. chaffeensis-infected cells.
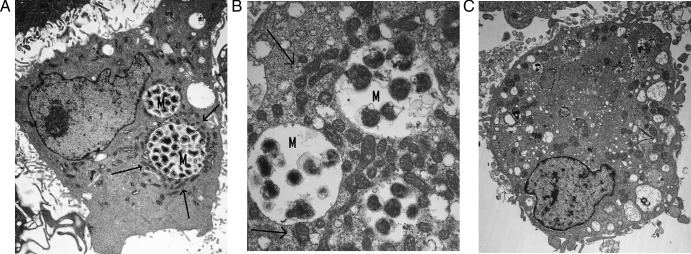
Mitochondria surround morulae in E. chaffeensis-infected cells (49). Indeed, when we examined DH82 cells after infection with E. chaffeensis by TEM, one prominent feature of the infected cells was the clustering of large numbers of mitochondria around the morulae (Fig. 8A and B). It even appears that mitochondria were closely apposed to bacteria within the parasitophorous vacuole (Fig. 8B). This contrasts with the random distribution of mitochondria in uninfected cells (Fig. 8C).
Fig 8.
Localization of mitochondria around morulae of E. chaffeensis-infected DH82 cells. (A and B) Transmission electron microscopy of DH82 cells infected with E. chaffeensis 96 (original magnification, ×5,000) (A) or 168 (original magnification, ×15,000) (B) hours after infection. (C) Uninfected DH82 cells (original magnification, ×5,000). Morulae (M) are surrounded by mitochondria, indicated by arrows.
DISCUSSION
We used D. melanogaster to investigate host factors that are important for the replication of E. chaffeensis. We combined microarray analysis and mutant fly screening to successfully identify 10 genes that contribute to the replication of E. chaffeensis in vivo. Seven genes (san, Cht11, Uck2, Echs1, whd, Ccdc58, and Apop1) encode proteins that have mitochondrial functions, serve as substrates in the mitochondria, or are associated with proteins with mitochondrial functions. For example, the Cht11 gene encodes a glycoside hydrolase with a chitinase active site (43). Cht11 is orthologous to the Chitinase 11 gene of Tribolium castaneum, which has been assigned to a separate classification group (group VII) from other identified chitinases (3) (Subbaratnam Muthukrishnan, Kansas State University, personal communication). This suggests that Cht11 has a function other than those classically described for chitinases. A two-hybrid screen associated the protein with the protein product encoded by Tiny tim 50 (22). Ttm50 (also known as Tim50) encodes a protein that is part of a transporter located on the inner mitochondrial membrane (15) (Fig. 9). Chitins and their derivatives regulate cholesterol when used as diet supplements (29, 74). Mice with high blood cholesterol had more severe Anaplasma phagocytophilum (another tick-transmitted rickettsia pathogen closely related to E. chaffeensis) infections in their blood, livers, and spleens than mice with normal cholesterol levels (72). E. chaffeensis and A. phagocytophilum cannot synthesize cholesterol, and they scavenge it from host cells during infections (34). Therefore, Cht11 may function to regulate cholesterol levels to facilitate infections. Cholesterol can also affect both the inner and outer mitochondrial membranes and affect ATPase activity (10, 19). A human homolog of Cht11 has not been identified, but homologs have been identified in ticks and body lice (43). Both of these species serve as vectors for several different rickettsia bacteria (4), and it will be interesting to explore the function of the gene for the survival of E. chaffeensis and other rickettsial agents in their respective arthropod hosts.
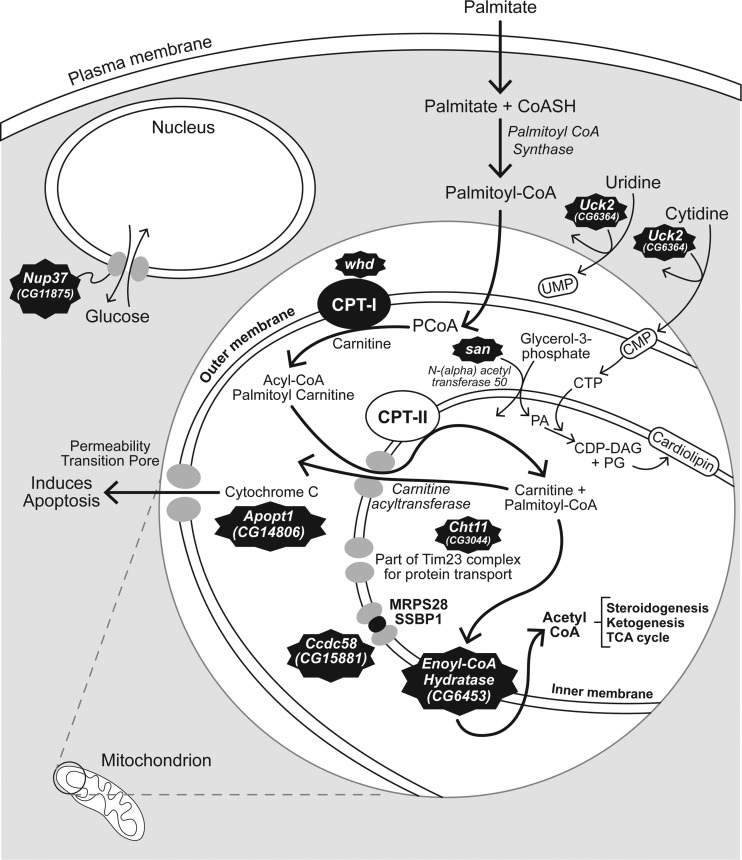
Fig 9.
Identification of mitochondrion-associated genes that impact E. chaffeensis infection. san [N-(alpha) acetyltransferase 50] and Uck2 (uridine/cytidine kinase) are needed for the synthesis of cardiolipin. Phosphatidic acid (PA), a product of san activity, and CTP, which is produced from CMP, a product of Uck2, are both necessary substrates for biosynthesis of cardiolipin, the major inner mitochondrial membrane lipid. Cht11 (chitinase 11) encodes part of a protein transporter in the inner mitochondrial membrane. whd (withered) encodes carnitine palmitoyltransferase 1, and Echs1 encodes enoyl-CoA hydratase; both enzymes are needed for the production of acetyl-CoA. Ccdc58 encodes coiled-coil domain-containing protein 58, which is found in complex with the inner mitochondrial membrane molecules MRPS28 and SSBP1. Apopt1 (Apoptogenic 1) induces the release of cytochrome c through the permeability transition pore and triggers apoptosis. Nup37 (Nucleoporin 37) is not found in the mitochondrion but is part of a nucleopore complex that transports hexose molecules, including glucose, an important molecule in energy metabolism.
Several of the genes that impact E. chaffeensis infections encode key mitochondrial enzymes needed for fatty acid metabolism and acetyl-coenzyme A (CoA) production. The whd gene was originally associated with uridine kinase (56). However, recent annotation (43) and experimentation (58) have identified whd as the palmitoyltransferase 1 (CPT 1) gene. Palmitoyltransferase 1 is the rate-limiting enzyme for palmitoyl-CoA uptake into mitochondria (24) (Fig. 9). Echs1 in Drosophila is an ortholog of the ECHS1 gene in humans and other mammals (43), which encodes an enoyl-CoA hydratase in the second step of the β-oxidation pathway in the mitochondrion (27) (Fig. 9). Therefore, when we experimentally challenged flies mutant for the Echs1 gene, the immediate downstream effect was on the supply of acetyl-CoA to the citric acid cycle, which implies that the supply of acetyl-CoA may be critical to E. chaffeensis replication. san is an ortholog of the human NAA50 gene, which encodes N(alpha)-acetyltransferase 50 (43). These acetyltransferases are involved in the conversion of glycerol-3-phosphate to phosphatidic acid (13). This biochemical step is upstream of the CDP diacylglycerol (CDP-DAG) intermediate in the synthesis of cardiolipin on the inner mitochondrial membrane. It is also possible that the acetyltransferase could be active in the β-oxidation pathway. However, this has yet to be proven experimentally.
CG15881, or Ccdc58, is an ortholog of the human gene known as coiled-coil domain-containing protein 58 (CCDC58). Although the exact function of Ccdc58 is not known, its structure likely allows it to interact with other coiled-coil molecules (41). Indeed, the STRING association tool (60) shows that CCDC58 has a direct relationship with mitochondrial ribosomal protein S28 (MRPS28) and translocase of inner mitochondrial membrane 9 homolog (TIMM9), which encode an inner mitochondrial membrane chaperone protein, SSBP1, a protein putatively thought to be involved in mitochondrial DNA replication (60). The BioGraph gene association tool (33) assigned a rank of 23 out of 18,180 gene concepts (top 0.13%) when it was assigned to “mitochondria.” Other coiled-coil domain-containing proteins have been associated with the inner mitochondrial membrane (70), and CCDC58 has also been associated with the mitochondrial single-nucleotide polymorphisms that impact the progression of AIDS (26). Therefore, it is highly likely that the impact of disruption of Ccdc58 on E. chaffeensis growth is due to its impact on mitochondrial function.
CG14806 or Apopt1 has no known function in Drosophila. It is an ortholog of the Apoptogenic 1 gene (APOPT1 or Apop1) in humans and rodents (66). It is involved in the release of cytochrome c through permeability transition pores in the outer membrane of the mitochondrion and the activation of apoptosis (59, 73) (Fig. 9). It seems plausible that inhibiting apoptosis would be to the bacterium's advantage. There is increased transcription of some apoptotic genes during Ehrlichia infections (75), and there is delayed apoptosis in neutrophils after infection with Ehrlichia ewingii (71). It is possible that Apop1 is needed to help in the release of bacteria at the end of the replicative phase. E. chaffeensis organisms disseminate by lysing host cells or by exocytosis in order to spread to uninfected cells (17). More empirical analyses will be needed to support this hypothesis.
Uck2 (CG6364) is orthologous to the mammalian UCK2 gene with the molecular function of a uridine kinase (43). Functionally, it has been associated with the phagocytosis of Candida albicans (57). Using the STRING association tool (60), we found that its predicted functional partners are all involved in nucleotide/nucleoside modification. These functions include nucleoside di- and triphosphate activity (CG5276), hydrolase activity (CG8891), uracil phosphoribosyl transferase activity (CG5537), uridine phosphorylase (CG3788, CG8349), cytidylate/uridylate kinase activity (Dak1), uridine phosphorylase (CG6330), and cytidine deaminase activity (CG8349). The BioGraph gene association tool (33) assigned a rank of 12.63% (2,297th out of 18,180 gene objects) when Uck2 was associated with mitochondria, indicating a highly probable association. Cytosine triphosphate, a downstream product of the uridine kinase (38, 62), is a substrate used for the conversion of phosphatidic acid to CDP-DAG (31, 46) (Fig. 9). CDP-DAG is synthesized in the inner mitochondrial membrane and is necessary for the synthesis of cardiolipin, a major component of the inner mitochondrial membrane (46). Therefore, we reason that the uridine kinase must be essential to providing substrates for E. chaffeensis, because it cannot be provided by the de novo synthesis pathway. When we grew E. chaffeensis in the presence of CPEC, a CTP synthetase inhibitor (52, 62) that inhibits the synthesis of CTP from glutamate (Fig. 5), bacterial replication was significantly enhanced (Fig. 6). These data suggest that the bacterium does not require the host to use glutamine as a substrate during replication. Supplementation of cytosine in cells enhanced Ehrlichia replication, which further implicates the use of the salvage pathway through cytidine. The enhanced growth of another rickettsia, Rickettsia felis, from pyrimidines in tryptose phosphate broth is also consistent with the involvement of mitochondrial enzymes in obligate intracellular bacterial growth (50). Moreover, cytidine is the least abundant nucleoside in cells (32). The depletion of cytidine pools can disrupt the balance of ribonucleotides in cells, leading to alterations in cellular homeostasis and apoptosis (52). These observations suggest that E. chaffeensis needs to regulate cytidine in order to control apoptosis until its replication is completed within a phagosome.
The enhanced growth of E. chaffeensis after treatment with CPEC instead of an inhibitory effect suggests that the bacterium requires the uridine/cytidine salvage pathway to phosphorylate cytidine and uridine (61) for its replication. More importantly, the disruption of E. chaffeensis growth in UCK2-silenced THP-1 macrophages reinforces the fact that the UCK2 target has human relevance. Therefore, we focused our attention on Uck2 because it has the potential to be a target for chemotherapy. UCK2 has been found to be more active in certain types of cancers (54) and acts to phosphorylate nucleoside analog drugs used for treatment of cancers and hepatitis C virus infections (23, 61). Uninfected host cells may be unaffected by UCK2 inhibitors, since nucleotides could be synthesized by the de novo synthesis pathway.
The clustering of mitochondria adjacent to morulae in E. chaffeensis-infected macrophages supports the hypothesis that it has physiological relevance and is consistent with previous observations (37, 49). Mitochondrial membrane permeability and membrane potential were not disrupted by either E. chaffeensis (37) or E. ewengii (71) infection. However, Liu et al. found that mitochondrial biochemical activity was reduced, as evidenced from the measurement of mitochondrial DNA synthesis or the transcription of several mitochondrial genes (37). There is also evidence that the E. chaffeensis type IV secretion system is used to insert bacterial effector proteins into host mitochondria (36). These observations suggest that the organism inhibits mitochondrial activity, perhaps to inhibit the generation of mitochondrially produced oxidative products that would be harmful to the bacteria (5, 68). In contrast, we have disrupted several genes that are associated with mitochondrial function that negatively affect E. chaffeensis infections. The gene products of san and Uck2 affect cardiolipin synthesis on the inner mitochondrial membrane (Fig. 9). Cht11 encodes protein for part of a transport molecule on the same inner mitochondrial membrane. The gene products of whd and Echs1 affect steroidogenesis, ketogenesis, and the Krebs cycle (Fig. 9), the last of which has clear mitochondrial dependence. Although Ccdc58 and Apopt1 are associated with mitochondria (Fig. 9), we have yet to fully understand their significance. It is also interesting that genes like Nup37 and dumpy, which do not have direct mitochondrial connections, also affect E. chaffeensis infections. The NUP37 gene product is part of nucleopores. One of the molecules it transports, glucose, is also important for mitochondrial functions. The dumpy gene has been characterized genetically and appears to impact wing and other body part development (11, 12). It has no known mammalian orthologs (43), and dumpy encodes a large extracellular molecule needed for structural integrity in wing epithelial tissue (12, 69). Interestingly, one of the properties listed for dumpy is that it binds iron and sulfur (43), properties of mitochondrial proteins involved in oxidation-reduction reactions (6). Clearly, more work is necessary to understand these molecules and their host-bacterium interactions. Nevertheless, our findings provide clues about the mitochondrial processes that are needed by the bacteria. Interestingly, at least one alphaproteobacterium closely related to Ehrlichia, designated IricES1, infects the mitochondria of ovarian cells of Ixodes ricinus (7). Perhaps E. chaffeensis is one step away from a similar symbiotic relationship.
Supplementary Material
ACKNOWLEDGMENTS
We thank Rishi Drolia, Taylor Kinney, and Rachel Nichols for their help in the laboratory with the flies, screening, and bacteria. We thank Lloyd Willard for his help with the electron microscopy. We thank Nanyan Lu and the K-INBRE bioinformatics center for help in analyzing the microarray data, Mal Rooks Hoover for her design of Fig. 9, and David Rintoul for helpful comments concerning the manuscript. We thank the Kansas University Medical Center-Microarray Facility (KUMC-MF) for generating array data sets.
The Microarray Facility is supported by the Kansas University School of Medicine, the KUMC Biotechnology Support Facility, the Smith Intellectual and Developmental Disabilities Research Center (HD02528), and the Kansas IDeA Network of Biomedical Research Excellence (RR016475). This project was supported by NIH grants AI088070, AI55052, AI052206, AI070908, RR16475, and RR17686; Kansas Agriculture Experiment Station Animal Health Project grant 481848; the Kansas Space Grant Consortium; NASA grants NAG2-1274 and NNX08BA91G; American Heart Association grant 0950036G; the Terry C. Johnson Center for Basic Cancer Research; and the Kansas Agriculture Experiment Station.
Footnotes
Published ahead of print 30 July 2012
This is Kansas Agriculture Experiment Station publication 12-447-J.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1. Agaisse H, et al. 2005. Genome-wide RNAi screen for host factors required for intracellular bacterial infection. Science 309:1248–1251 [DOI] [PubMed] [Google Scholar]
- 2. Alvarez-Dominguez C, Barbieri AM, Beron W, Wandinger-Ness A, Stahl PD. 1996. Phagocytosed live Listeria monocytogenes influences Rab5-regulated in vitro phagosome-endosome fusion. J. Biol. Chem. 271:13834–13843 [DOI] [PubMed] [Google Scholar]
- 3. Arakane Y, Muthukrishnan S. 2010. Insect chitinase and chitinase-like proteins. Cell. Mol. Life Sci. 67:201–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Azad A, Beard C. 1998. Rickettsial pathogens and their arthropod vectors. Emerg. Infect. Dis. 4:179–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Basu Ball W, et al. 2011. Uncoupling protein 2 negatively regulates mitochondrial reactive oxygen species generation and induces phosphatase-mediated anti-inflammatory response in experimental visceral leishmaniasis. J. Immunol. 187:1322–1332 [DOI] [PubMed] [Google Scholar]
- 6. Beinert H, Holm RH, Münck E. 1997. Iron-sulfur clusters: nature's modular, multipurpose structures. Science 277:653–659 [DOI] [PubMed] [Google Scholar]
- 7. Beninati T, et al. 2004. A novel alpha-proteobacterium resides in the mitochondria of ovarian cells of the tick Ixodes ricinus. Appl. Environ. Microbiol. 70:2596–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brandt SM, Jaramillo-Gutierrez G, Kumar S, Barillas-Mury C, Schneider DS. 2008. Use of a Drosophila model to identify genes regulating Plasmodium growth in the mosquito. Genetics 180:1671–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brouqui P, Lecam C, Olson J, Raoult D. 1994. Serologic diagnosis of human monocytic ehrlichiosis by immunoblot analysis. Clin. Diagn. Lab. Immunol. 1:645–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Calanni Rindina F, Baracca A, Solaini G, Rabbi A, Parenti Castelli G. 1986. Effects of cholesterol on the kinetics of mitochondrial ATPase. FEBS Lett. 198:353–356 [DOI] [PubMed] [Google Scholar]
- 11. Carmon A, Guertin MJ, Grushko O, Marshall B, MacIntyre R. 2010. A molecular analysis of mutations at the complex dumpy locus in Drosophila melanogaster. PLoS One 5:e12319 doi:10.1371/journal.pone.0012319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carmon A, Topbas F, Baron M, MacIntyre RJ. 2010. Dumpy interacts with a large number of genes in the developing wing of Drosophila melanogaster. Fly 4:117–127 [DOI] [PubMed] [Google Scholar]
- 13. Castorena K, Stapleford K, Miller D. 2010. Complementary transcriptomic, lipidomic, and targeted functional genetic analyses in cultured Drosophila cells highlight the role of glycerophospholipid metabolism in Flock House virus RNA replication. BMC Genomics 11:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Centers for Disease Control and Prevention 2012. Notifiable diseases and mortality tables. MMWR Morb. Mortal. Wkly. Rep. 60:1729–177622217621 [Google Scholar]
- 15. Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. 2009. Importing mitochondrial proteins: machineries and mechanisms. Cell 138:628–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cinco M, et al. 2004. Seroprevalence of tick-borne infections in forestry rangers from northeastern Italy. Clin. Microbiol. Infect. 10:1056–1061 [DOI] [PubMed] [Google Scholar]
- 17. Dedonder SE, Cheng C, Willard LH, Boyle DL, Ganta RR. 2012. Transmission electron microscopy reveals distinct macrophage- and tick cell-specific morphological stages of Ehrlichia chaffeensis. PLoS One 7:e36749 doi:10.1371/journal.pone.0036749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dionne MS, Ghori N, Schneider DS. 2003. Drosophila melanogaster is a genetically tractable model host for Mycobacterium marinum. Infect. Immun. 71:3540–3550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Echegoyen S, et al. 1993. Cholesterol increase in mitochondria: its effect on inner-membrane functions, submitochondrial localization and ultrastructural morphology. Biochem. J. 289:703–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Elwell C, Engel JN. 2005. Drosophila melanogaster S2 cells: a model system to study Chlamydia interaction with host cells. Cell. Microbiol. 7:725–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ganta RR, Cheng C, Wilkerson MJ, Chapes SK. 2004. Delayed clearance of Ehrlichia chaffeensis infection in CD4+ T-cell knockout mice. Infect. Immun. 72:159–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Giot L, et al. 2003. A protein interaction map of Drosophila melanogaster. Science 302:1727–1736 [DOI] [PubMed] [Google Scholar]
- 23. Golitsina NL, Danehy FT, Jr, Fellows R, Cretton-Scott E, Standring DN. 2010. Evaluation of the role of three candidate human kinases in the conversion of the hepatitis C virus inhibitor 2′-C-methyl-cytidine to its 5′-monophosphate metabolite. Antiviral Res. 85:470–481 [DOI] [PubMed] [Google Scholar]
- 24. Gomez LA, Heath SH, Hagen TM. 2012. Acetyl-L-carnitine supplementation reverses the age-related decline in carnitine palmitoyltransferase 1 (CPT1) activity in interfibrillar mitochondria without changing the L-carnitine content in the rat heart. Mech. Ageing Dev. 133:99–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hall-Baker PA, et al. 2007. Summary of notifiable diseases—United States, 2007. MMWR Morb. Mortal. Wkly. Rep. 56:1–94 [PubMed] [Google Scholar]
- 26. Hendrickson SL, et al. 2010. Genetic variants in nuclear-encoded mitochondrial genes influence AIDS progression. PLoS One 5:e12862 doi:10.1371/journal.pone.0012862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Janssen U, Davis EM, Le Beau MM, Stoffel W. 1997. Human mitochondrial enoyl-CoA hydratase gene (ECHS1): structural organization and assignment to chromosome 10q26.2-q26.3. Genomics 40:470–475 [DOI] [PubMed] [Google Scholar]
- 28. Kang GJ, et al. 1989. Cyclopentenylcytosine triphosphate. Formation and inhibition of CTP synthetase. J. Biol. Chem. 264:713–718 [PubMed] [Google Scholar]
- 29. Koide S. 1998. Chitin-chitosan: properties, benefits and risks. Nutr. Res. 18:1091–1101 [Google Scholar]
- 30. Koo IC, et al. 2008. Role for lysosomal enzyme beta-hexosaminidase in the control of mycobacteria infection. Proc. Natl. Acad. Sci. U. S. A. 105:710–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kopka J, Ludewig M, Muller-Rober B. 1997. Complementary DNAs encoding eukaryotic-type cytidine-5′-diphosphate-diacylglycerol synthases of two plant species. Plant Physiol. 113:997–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Korte D, Haverkort W, van Gennip A, Roos D. 1985. Nucleotide profiles of normal human blood cells determined by high-performance liquid chromatography. Anal. Biochem. 147:197–209 [DOI] [PubMed] [Google Scholar]
- 33. Liekens A, et al. 2011. BioGraph: unsupervised biomedical knowledge discovery via automated hypothesis generation. Genome Biol. 12:R57 doi:10.1186/gb-2011-12-6-r57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lin M, Rikihisa Y. 2003. Ehrlichia chaffeensis and Anaplasma phagocytophilum lack genes for lipid A biosynthesis and incorporate cholesterol for their survival. Infect. Immun. 71:5324–5331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lindsley D, Zimm G. 1992. The genome of Drosophila melanogaster. Academic Press, Inc., San Diego, CA [Google Scholar]
- 36. Liu H, Bao W, Lin M, Niu H, Rikihisa Y. 2012. Ehrlichia type IV secretion effector ECH0825 is translocated to mitochondria and curbs ROS and apoptosis by upregulating host MnSOD. Cell. Microbiol. 14:1037–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu Y, et al. 2011. Obligate intracellular bacterium Ehrlichia inhibiting mitochondrial activity. Microbes Infect. 13:232–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Loffler M, Fairbanks LD, Zameitat E, Marinaki AM, Simmonds HA. 2005. Pyrimidine pathways in health and disease. Trends Mol. Med. 11:430–437 [DOI] [PubMed] [Google Scholar]
- 39. Luce-Fedrow A, Von Ohlen T, Boyle D, Ganta RR, Chapes SK. 2008. Drosophila S2 cells as a model for studying Ehrlichia chaffeensis infections. Appl. Environ. Microbiol. 74:1886–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Luce-Fedrow A, Von Ohlen T, Chapes SK. 2009. Ehrlichia chaffeensis infections in Drosophila melanogaster. Infect. Immun. 77:4815–4826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lupas AN, Gruber M. 2005. The structure of α-helical coiled coils. Adv. Protein Chem. 70:37–38 [DOI] [PubMed] [Google Scholar]
- 42. Machado RZ, Duartes JM, Dagnone AS, Szabo MP. 2006. Detection of Ehrlichia chaffeensis in Brazilain marsh deer (Blastocerus dichotomus). Vet. Parasitol. 139:262–266 [DOI] [PubMed] [Google Scholar]
- 43. McQuilton P, St Pierre SE, Thurmond J, FlyBase Consortium 2012. FlyBase 101—the basics of navigating FlyBase. Nucleic Acids Res. 40:D706–D714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Morgan T. 1929. Contributions to the genetics of Drosophila simulans and Drosophila melanogaster. Pub. Carnegie Inst. 399:169–199 [Google Scholar]
- 45. Moyer J, Malinowski N, Treanor S, Marquez V. 1986. Antitumor activity and biochemical effects of cyclopentenyl cytosine in mice. Cancer Res. 46:3325–3329 [PubMed] [Google Scholar]
- 46. Osman C, Haag M, Wieland FT, Brugger B, Langer T. 2010. A mitochondrial phosphatase required for cardiolipin biosynthesis: the PGP phosphatase Gep4. EMBO J. 29:1976–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45 doi:10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Philips JA, Rubin EJ, Perrimon N. 2005. Drosophila RNAi screen reveals CD36 family member required for mycobacterial infection. Science 309:1251–1253 [DOI] [PubMed] [Google Scholar]
- 49. Popov VL, Chen S-M, Feng H-M, Walker OH. 1995. Ultrastructural variation of cultured Ehrlichia chaffeensis. J. Med. Microbiol. 43:411–421 [DOI] [PubMed] [Google Scholar]
- 50. Saisongkorh W, et al. 2012. Tryptose phosphate broth improves Rickettsia felis replication in mammalian cells. FEMS Immunol. Med. Microbiol. 64:111–114 [DOI] [PubMed] [Google Scholar]
- 51. Santic M, et al. 2009. Intracellular fate of Francisella tularensis within arthropod-derived cells. Environ. Microbiol. 11:1473–1481 [DOI] [PubMed] [Google Scholar]
- 52. Schimmel KJ, Gelderblom H, Guchelaar HJ. 2007. Cyclopentenyl cytosine (CPEC): an overview of its in vitro and in vivo activity. Curr. Cancer Drug Targets 7:504–509 [DOI] [PubMed] [Google Scholar]
- 53. Schneider D, Shahabuddin M. 2000. Malaria parasite development in a Drosophila model. Science 288:2376–2379 [DOI] [PubMed] [Google Scholar]
- 54. Shimamoto Y, et al. 2002. Sensitivity of human cancer cells to the new anticancer ribo-nucleoside TAS-106 is correlated with expression of uridine-cytidine kinase 2. Jpn. J. Cancer Res. 93:825–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sirigireddy KR, Ganta RR. 2005. Multiplex detection of Ehrlichia and Anaplasma species pathogens in peripheral blood by real-time reverse transcriptase-polymerase chain reaction. J. Mol. Diagn. 7:308–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Stroman P. 1974. Pyrimidine-sensitive drosophila wing mutants: withered (whd), tilt (tt) and dumpy (dp). Hereditas 78:157–168 [DOI] [PubMed] [Google Scholar]
- 57. Stroschein-Stevenson SL, Foley E, O'Farrell PH, Johnson AD. 2006. Identification of Drosophila gene products required for phagocytosis of Candida albicans. PLoS Biol. 4:e4 doi:10.1371/journal.pbio.0040004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Strub BR, et al. 2008. Mutations of the withered (whd) gene in Drosophila melanogaster confer hypersensitivity to oxidative stress and are lesions of the carnitine palmitoyltransferase I (CPT I) gene. Genome 51:409–420 [DOI] [PubMed] [Google Scholar]
- 59. Sun X, et al. 2008. Akt activation prevents Apop-1-induced death of cells. Biochem. Biophys. Res. Commun. 377:1097–1101 [DOI] [PubMed] [Google Scholar]
- 60. Szklarczyk D, et al. 2011. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 39:D561–D568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Van Rompay AR, Norda A, Linden K, Johansson M, Karlsson A. 2001. Phosphorylation of uridine and cytidine nucleoside analogs by two human uridine-cytidine kinases. Mol. Pharmacol. 59:1181–1186 [DOI] [PubMed] [Google Scholar]
- 62. Verschurr AC. 2007. Cytidine triphosphate synthetase (CTP synthetase) as a druggable target in cancer. Drugs Future 32:1071–1080 [Google Scholar]
- 63. Verschuur AC, et al. 2000. In vitro inhibition of cytidine triphosphate synthetase activity by cyclopentenyl cytosine in paediatric acute lymphocytic leukaemia. Br. J. Haematol. 110:161–169 [DOI] [PubMed] [Google Scholar]
- 64. Vonkavaara M, Telepnev MV, Ryden P, Sjostedt A, Stoven S. 2008. Drosophila melanogaster as a model for elucidating the pathogenicity of Francisella tularensis. Cell Microbiol. 10:1327–1338 [DOI] [PubMed] [Google Scholar]
- 65. Waddington C. 1940. The genetic control of wing development in Drosophila. J. Genet. 41:75–139 [Google Scholar]
- 66. Waterhouse RM, Zdobnov EM, Tegenfeldt F, Li J, Kriventseva EV. 2011. OrthoDB: the hierarchical catalog of eukaryotic orthologs in 2011. Nucleic Acids Res. 39:D283–D288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wen B, Cao W, Pan H. 2003. Ehrlichiae and ehrlichial diseases in China. Ann. N. Y. Acad. Sci. 990:45–53 [DOI] [PubMed] [Google Scholar]
- 68. West AP, et al. 2011. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature 472:476–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wilkin MB, et al. 2000. Drosophila dumpy is a gigantic extracellular protein required to maintain tension at epidermal-cuticle attachment sites. Curr. Biol. 10:559–567 [DOI] [PubMed] [Google Scholar]
- 70. Xie J, Marusich MF, Souda P, Whitelegge J, Capaldi RA. 2007. The mitochondrial inner membrane protein Mitofilin exists as a complex with SAM50, metaxins 1 and 2, coiled-coil-helix coiled-coil-helix domain-containing protein 3 and 6 and DnaJC11. FEBS Lett. 581:3545–3549 [DOI] [PubMed] [Google Scholar]
- 71. Xiong Q, Bao W, Ge Y, Rikihisa Y. 2008. Ehrlichia ewingii infection delays spontaneous neutrophil apoptosis through stabilization of mitochondria. J. Infect. Dis. 197:1110–1118 [DOI] [PubMed] [Google Scholar]
- 72. Xiong Q, Wang X, Rikihisa Y. 2007. High-cholesterol diet facilitates Anaplasma phagocytophilum infection and up-regulates macrophage inflammatory protein-2 and CXCR2 expression in apolipoprotein E-deficient mice. J. Infect. Dis. 195:1497–1503 [DOI] [PubMed] [Google Scholar]
- 73. Yasuda O, et al. 2006. Apop-1, a novel protein inducing cyclophilin D-dependent but Bax/Bak-related channel-independent apoptosis. J. Biol. Chem. 281:23899–23907 [DOI] [PubMed] [Google Scholar]
- 74. Zacour A, Silva M, Cecon P, Bambirra E, Vieira E. 1992. Effect of dietary chitin on cholesterol absorption and metabolism in rats. J. Nutr. Sci. Vitaminol. 32:609–613 [DOI] [PubMed] [Google Scholar]
- 75. Zhang JZ, Sinha M, Luxon BA, Yu XJ. 2004. Survival strategy of obligately intracellular Ehrlichia chaffeensis: novel modulation of immune response and host cell cycles. Infect. Immun. 72:498–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.