Abstract
Hydration is known to affect molecular recognition processes, such as between protein and ligand. Now, theoretical simulations have provided thermodynamic insight into cavity–ligand binding, and revealed how it is predominantly driven by the behaviour of the few surrounding water molecules.
Water is a crucial, yet challenging participant in virtually all ligand-binding reactions in biology. The association of a ligand with its binding partner requires at least a partial desolvation of the ligand, the removal of water molecules from the binding site, and the rearrangement of water in the vicinity (Fig. 1a and b). These changes in the hydration of the two binding partners occur on a molecular scale, with only a countable number of water molecules directly affected. Nevertheless, the contributions of water to the binding energetics can be large. Seemingly subtle changes in the water hydrogen-bonding network are often associated with large changes in the interaction energy, with gains of about 10 kBT per hydrogen bond formed (where T is the absolute temperature and kB is Boltzmann's constant, with kBT≈0.6 kcal mol−1). As a result, the contributions of water in ligand binding tend to be large and not easily quantifiable, unlike the contributions of simpler apolar solvents. Consequently, water continues to challenge the development of quantitative descriptions of ligand-binding energetics.
Figure 1. Schematic representation of a ligand-binding reaction in water.
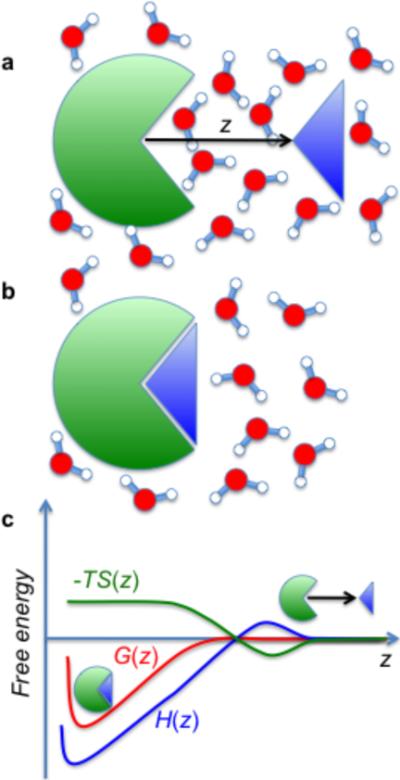
a, b, As the distance z of the ligand from the binding site shrinks, water is displaced and reorganized. c, Thermodynamics of the binding process. For an apolar ligand binding to an apolar pocket, it is the hydration contribution that dominates the binding. In particular, a surprising finding is that the free energy G(z) that drives such hydrophobic bindings can be dominated by gains in the enthalpy H(z) that outweigh losses in the entropy S(z)1,2. These effects are associated with, respectively, an overall increase in water–water hydrogen bonds and the suppression of solvent fluctuations in the ligand-binding interface.
To quantify the role of water in ligand binding, and to dissect the free energy of this association process into contributions from enthalpy and entropy, Baron, Setny and McCammon have studied1,2 a simple model system using molecular dynamics simulations. The thermodynamic signatures of ligand binding emerging from their studies, described in the Journal of the American Chemical Society and the Journal of Chemical Theory and Computation, are remarkable. For a model receptor–ligand system, they show that the free energy of the binding process is dominated not by the direct interaction between the ligand and its binding pocket, but by the contributions of water. Moreover, contrary to the common belief that it is the entropy that dominates molecular-scale hydrophobic interactions, in their model system the association between an apolar ligand and an apolar binding pocket is driven by enthalpy, and opposed by entropy.
In the simulations, a spherical methane-size ligand binds from bulk water into a hemispherical pocket within an apolar surface.1,2 For such a simple model system the competing contributions from solvent entropy and enthalpy can be disentangled, and the polarity of the ligand and binding site can be varied in a controlled way. Specifically, both the charge of the ligand and the charge at the centre of the binding pocket were changed from −e, to 0, to +e. The binding process was then characterized in each case by calculating the Gibbs free energy surface G(z) of the ligand as a function of its distance (z) from the pocket. This `potential of mean force' was then further decomposed into enthalpic and entropic contributions, G(z)=H(z)-TS(z), where S is the entropy and H the enthalpy.
Although this may be counter-intuitive, the results revealed that ion pairs that formed in the binding pocket (between solutes of charge ±e and pockets of opposite charge) were bound in a less stable manner than apolar ligands in a hydrophobic pocket (where both participants were uncharged)1. Moreover, distinct asymmetries were observed between positive–negative and negative–positive combinations of pocket–ligand charges. The most unexpected finding, however, may be the fact that the hydrophobic binding between uncharged ligands and pockets is strongly driven by enthalpy, and opposed to by entropy2 (Fig. 1c). This gain in enthalpy upon binding was explained by the release of water from the pocket, and the loss in entropy was interpreted in terms of elimination of solvent fluctuations3–5 inside the pocket.
The studies by Baron, Setny and McCammon1,2 reflect both the growing awareness of the importance of water in ligand binding, and the increasing efforts to develop quantitative descriptions of hydration effects.4–7 The traditional approach of calculating binding free energies by combining continuum electrostatics for polar interactions with surface area models for hydrophobic interactions is often more successful than one might expect — but it is almost bound to fail in cases where rearrangements of individual water molecules matter. Friesner and collaborators6 have recently developed an approach to deal with the contributions of the solvent to the free energy of ligand binding process, and to capture the effects of the displacement of water from the binding site. In this approach, the energy and entropy of water removal from the binding site were efficiently calculated by feeding data obtained from molecular dynamics simulations into an approximate statistical-mechanical formalism. In particular, the important entropic contributions are estimated from low-order structural correlations of the water molecules. This method relies on the fact that the binding of ligands with apolar groups tends to be particularly strong if water is either absent from the binding site at equilibrium, or if it can be easily removed.4,6,8,9 Indeed, Baron et al.1 observe the strongest binding between an apolar ligand and an apolar pocket, in which partial dewetting takes place even at equilibrium,2 indicating that water can be easily displaced by the approaching ligand.
Understanding and quantifying the role of water in ligand binding is not only of academic interest but also of great practical importance. Computer-assisted drug design relies on accurate scoring functions to provide reliable estimates of the binding free energies of potential drug molecules to their target sites. There is a growing realization that gaining insight into the behaviour of water at a molecular level is a key requirement for more reliable scoring, in particular if the binding site has extended hydrophobic regions. The recent studies based on molecular simulations1,2 point the way toward a more accurate modeling of hydration contributions to ligand binding.
References
- 1.Baron R, Setny P, McCammon JA. J. Am. Chem. Soc. 2010;132:12091–12097. doi: 10.1021/ja1050082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Setny P, Baron R, McCammon JA. J. Chem. Theory Comput. 2010;6:2866–2871. doi: 10.1021/ct1003077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chandler D. Nature. 2005;437:640–647. doi: 10.1038/nature04162. [DOI] [PubMed] [Google Scholar]
- 4.Rasaiah JC, Garde S, Hummer G. Annu. Rev. Phys. Chem. 2008;59:713–740. doi: 10.1146/annurev.physchem.59.032607.093815. [DOI] [PubMed] [Google Scholar]
- 5.Siebert X, Hummer G. Biochemistry. 2002;41:2956–2961. doi: 10.1021/bi0158526. [DOI] [PubMed] [Google Scholar]
- 6.Abel R, Young T, Farid R, Berne BJ, Friesner RA. J. Am. Chem. Soc. 2008;130:2817–2831. doi: 10.1021/ja0771033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michel J, Tirado-Rives J, Jorgensen WL. J. Phys. Chem. B. 2009;113:13337–13346. doi: 10.1021/jp9047456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mobley DL, et al. J. Mol. Biol. 2007;371:1118–1134. doi: 10.1016/j.jmb.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qvist J, Davidovic M, Hamelberg D, Halle B. Proc. Natl. Acad. Sci. USA. 2008;105:6296–6301. doi: 10.1073/pnas.0709844105. [DOI] [PMC free article] [PubMed] [Google Scholar]


