Abstract
Cancer initiation, progression, and the emergence of therapeutic resistance are evolutionary phenomena of clonal somatic cell populations. Studies in microbial experimental evolution and the theoretical work inspired by such studies are yielding deep insights into the evolutionary dynamics of clonal populations, yet there has been little explicit consideration of the relevance of this rapidly growing field to cancer biology. Here, we examine how the understanding of mutation, selection, and spatial structure in clonal populations that is emerging from experimental evolution may be applicable to cancer. Along the way, we discuss some significant ways in which cancer differs from the model systems used in experimental evolution. Despite these differences, we argue that enhanced prediction and control of cancer may be possible using ideas developed in the context of experimental evolution, and we point out some prospects for future research at the interface between these traditionally separate areas.
Introduction
In the 36 years since Peter Nowell’s seminal paper on the clonal evolution of tumor cell populations [1], it has become apparent that a comprehensive theory of cancer must include a substantial amount of the theory of evolution (reviews [2-5]). Roughly the same time interval has seen the remarkable rise of direct experimental approaches to evolution in which microbial populations are propagated for many generations and their evolution in real time is observed and analysed (reviewed in [6]). The burgeoning field of microbial experimental evolution has generated a rich empirical and theoretical literature that is largely focused on the evolution of clonal populations: i.e., populations lacking intergenomic recombination. Because recombination between somatic cells is rare to nonexistent, theory and experimental work in asexually evolving microbial populations can, in principle, be applied to the understanding of cancer. Although connections between cancer and the phenomena observable in microbial evolution experiments have occasionally been drawn [7-17], to date there has been no attempt to assess the overall relevance of experimental evolution to cancer biology. We believe such an assessment is timely: on the one hand, increased awareness of experimental evolution and its related theory on the part of cancer researchers and clinicians may hold benefits for the understanding and treatment of cancers; on the other hand, there is value in anticipating the limits of experimental evolution approaches as applied to cancer.
Early studies of evolution assumed, perhaps following Darwin [18], that its course is too slow to observe directly and must therefore be inferred indirectly. Indeed, the very fact of evolution and the major outlines of its history were established largely by indirect inference from fossils, biogeographical patterns, and anatomical comparisons among extant taxa (reviewed in [19]). Into the second half of the 20th century, most empirical studies of evolution focused on comparison of patterns of variation and divergence within and between populations and species rather than on real-time analysis of evolutionary change. There was, however, a growing awareness that evolution could also be observed and studied directly, both in the field with sufficient effort [20] and in the laboratory [21-26]. The past two decades have seen a rapid expansion of experimental studies of evolution of many kinds, but in particular of those using microbial populations in the laboratory (reviews in [6,27,28]). Microbial evolution experiments have, in turn, stimulated advances in theory related to the evolution of clonal populations (e.g.,[10,13,29-41]). Our goal here is to relate the increasingly sophisticated literature on microbial experimental evolution to cancer biology.
Cancer certainly has organism-level evolutionary consequences for humans and other taxa. Selection at the organismal level, for example, has arguably favored the maintenance of low somatic mutation rates (except in the immune system) and the evolution of tumour suppression mechanisms [42-46]. However, in this review we focus on processes at a lower level of organisation: namely, the evolutionary dynamics that occur within the somatic cell populations of an individual organism and give rise to cancer. At this level, cancers and experimental microbial populations are potentially quite similar in that each evolves under mutation, selection, genetic drift, migration, and varying amounts of spatial structure in the absence of intergenomic recombination. (See Table 1 for a glossary of some common terms in evolution and cancer.)
Table 1.
Common terms in evolution and cancer.
| Cell lineage: A group of cells related by a common ancestral cell. A particular lineage may accumulate multiple mutations during cancer progression. |
| Clone: A group of cells, descended from a single common ancestral cell, that can share the same genetic lesions. |
| Beneficial mutation: A mutation that increases the fitness of the cell/organism. Note that in the case of cancer, mutations that are beneficial to cancer cells are detrimental to the host. |
| Deleterious mutation: A mutation that decreases the fitness of the cell/organism. Lethal mutations are one type of deleterious mutation. |
| Driver: A beneficial mutation in a neoplasm that increases the fitness of the cell, causing the cell’s lineage to spread. |
| Epistasis: Interaction between genes. In a practical sense, this means that the effect of a particular mutation is dependent upon the genetic background on which it occurs. For a thorough review of the many definitions and implications of epistasis in evolution, see [169]. |
| Fitness (relative): Fitness is a function of an organism’s ability to survive and reproduce. Usually, relative fitness is measured in microbial experimental evolution: for example, the growth rate of one genotype compared to that of a second, competing genotype. |
| Fixation: When a mutation in a population of cells/organisms spreads such that all individuals have the mutation. |
| Genetic drift: In genetic drift, clones (or genotypes) carrying a mutation change in frequency due to stochastic processes rather than selection. Drift is of particular importance in small populations. |
| Hitchhiking: The process by which a mutation that is genetically linked to a beneficial mutation can rise toward fixation with the beneficial mutation as it expands through the population. Hitchhikers are usually neutral or of weak selective effect. |
| Mutator: A defective allele at a locus controlling genomic fidelity (e.g., replication or repair). Mutator alleles raise the mutation rate. |
| Neoplasm: A collection of abnormal somatic cells. In genetic terms, a neoplasm contains mutations compared to the germ line that render the cells precancerous or cancerous. |
| Neutral mutations: Mutations that have such small effects on fitness that they are not under selection. This occurs when, approximately, Nes < 1, where Ne represents the size of the genetically evolving population (effective population size) and s represents the strength of selection for/against the mutation. |
| Passenger: A neutral or slightly deleterious mutation that is genetically linked to a driver mutation. As the driver mutation sweeps to fixation in the neoplasm, the passenger mutation goes along for the ride. In this way, genetic lesions that are not selectively advantageous may fix in a population of neoplastic cells. See also ‘hitchhiking’. |
| Selective sweep: The process by which a beneficial mutation increases in frequency until it reaches fixation in a population. |
In a typical microbial evolution experiment, replicate populations of an experimental organism (e.g., bacteria or yeast) are founded from a single ancestral cell and propagated either by transferring them periodically to fresh medium or by supplying them continuously with fresh medium (Figure 1A,B). The ancestor and intermediate stages in the history of the populations are archived as frozen stocks and revived for later analyses of fitness and phenotype evolution. The investigator can control the environment, the population size, the initial genetic state of the populations, and other variables of interest, in contrast to the situation in most natural populations. Because of the ease with which experimental microbial populations can be propagated and maintained, investigations of this kind can be carried out for many thousands of generations: the iconic example in the field is a set of 12 replicate Escherichia coli populations that have been propagated since the late 1980s for a current total of more than 50,000 generations [47].
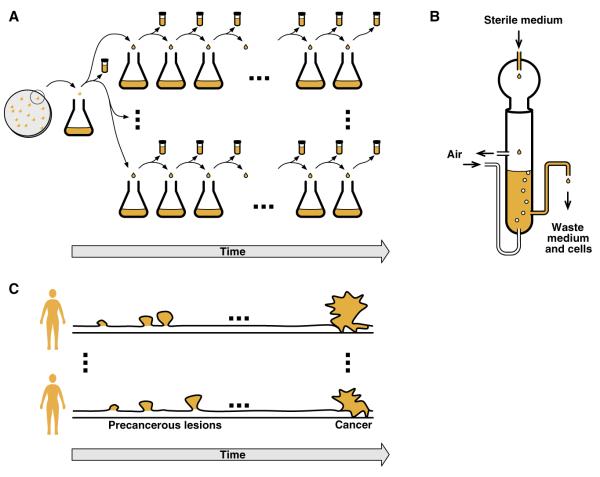
Figure 1.
Experimental evolution and cancer.
(A) In serial transfer evolution experiments, several replicate microbial populations are seeded from a culture originating from a single ancestral colony (derived from a single cell). Thereafter, a small subsample of each replicate population is regularly transferred into fresh medium. Aliquots of the evolving populations are preserved at regular time intervals for future analysis. (B) In evolution experiments conducted in a chemostat, the number of cells remains essentially constant. As fresh medium is fed into the chemostat, waste medium is removed. Cells can be sampled repeatedly from the waste medium, allowing for analysis of the population over time (adapted from [170]). (C) Somatic cells within individuals may evolve over time to become cancerous. The ancestral genotype for each neoplasm is the germ line genotype of the individual with the neoplasm.
There are some strong parallels between experimental populations of microbes and cancer cell populations (Figure 1). Like cancers, experimental populations initiate from clonal ancestors: an initial tumourigenic mutation in the case of a cancer; a single ancestral cell in the case of an experimental population. Cancers evolve independently in different individuals over hundreds or thousands of somatic cell divisions [48]; as noted above, the evolution of replicate experimental populations can be studied over similar time spans. In the absence of intergenomic recombination, all genetic variation in both situations must arise as a consequence of mutation, and evolution can be broadly characterised as a process of sorting among lineages representing different genomic sequences (Figure 2). Deeper and even more interesting phenomenological parallels also seem to apply. For example, there is evidence for the evolution of high genomic mutation rates in experimental microbial populations and in some cancers [8].
Figure 2.
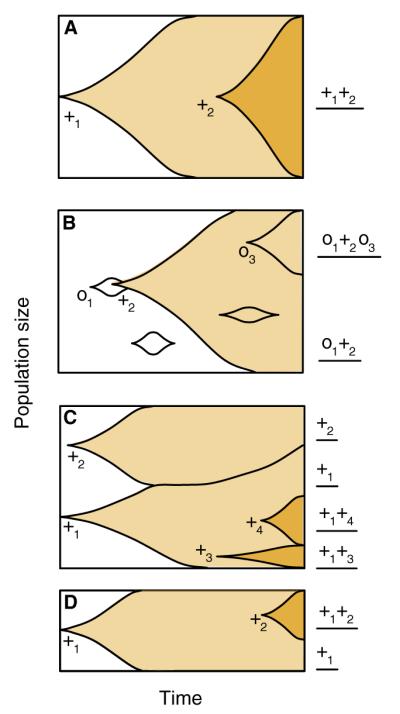
Dynamics of clonal evolution.
Several variations on a visualisation of clonal evolution originated by Muller in 1932 [53] and reinterpreted by Crow and Kimura [171]. We consider the fate of new mutations in a finite population of constant size. Time is depicted on the x-axis beginning at an arbitrary instant at which we assume that the population has no competing clones; the population sizes of clones harbouring mutations that have arisen since that instant are depicted on the y-axis. The total number of evolving cells in this neoplasm is constant (as might occur in the early stages of neoplastic progression), but the fraction of cells that have a given genotype varies as mutations arise and then either expand or are lost. The genotypes of clones are depicted to the right; darker colours indicate clones harbouring increasing numbers of beneficial mutations. (A) When new, beneficial mutations (‘+’) are rare, they are likely to sweep to fixation in the population before the next beneficial mutation arises. In this case, all the cells in the final population will have the +1 and +2 mutations. (B) Beneficial mutations are thought to be rarer than neutral mutations (‘o’); neutral mutations may hitchhike to fixation with a beneficial mutation. In this case, all of the cells in the population will have the neutral o1 and the beneficial +2 mutations, as indicated on the lower right margin of the panel. Additional neutral mutations may arise and expand in the population (e.g., o3), leading to intrapopulation heterogeneity. It is also possible for neutral mutations to arise and go extinct: two such mutations are illustrated here. (C) When the beneficial mutation supply rate is high, several beneficial mutations may arise in separate clones and compete, as depicted here by the +1 and +2 beneficial mutations (and also the +3 and +4 beneficial mutations). The competition between the clones may delay the fixation of any one of the beneficial mutations and thus prolong intrapopulation heterogeneity. Here, there are four distinct genotypes in the neoplasm. (D) The time between new beneficial mutations will tend to be longer in small populations than in large populations, given the same beneficial mutation rate.
Below, we discuss parallels between experimental evolution and cancer in more detail, placing empirical results from both fields into the general framework provided by evolutionary theory, especially as developed for clonal populations in the past two decades. We begin with a broad discussion of cancer and evolution in clonal populations. We then focus, in turn, on methods of detecting selection in neoplasms, mutation rates and their consequences for cancer, spatial structure and cell–cell interactions, and the evolution of drug resistance. We conclude by briefly describing a potential new approach to predicting cancer progression based on general evolutionary theory.
Overview: Cancer and the Nature of Evolution in Clonal Populations
The accumulation of genetic variants that originated initially as mutations — whether single nucleotide changes or largerscale genomic alterations such as deletions, insertions, and chromosomal rearrangements — underlies genetic evolution in all populations, including the populations of somatic cells that can give rise to cancer. The most important mutations for the study of cancer are those that release somatic cells from constraints on proliferation and migration and thereby allow their lineage to outcompete others in a multicellular organism; these are often called ‘driver mutations’ in the cancer literature (e.g., [49-51]). From a general evolutionary perspective, such mutations increase the capacity of an individual cell to survive and reproduce (i.e., its relative fitness) within a somatic cell population, and thus they may accurately be regarded as beneficial mutations for the individual cell despite their potential detrimental effects on the whole organism.
The central fact of evolution in clonal populations is that genetic variants from different genomic backgrounds cannot be combined into a single genotype as in a sexual population[52,53]; a clonal population can only give rise to such genotypes via the occurrence of multiple mutations on a common genomic background (Figure 2). The way in which beneficial mutations in particular accumulate within clonal lineages thus depends strongly on the rate at which they arise in a population. If the supply rate of beneficial mutations is low, then new mutations will tend to be temporally isolated from one another in a clonal population (Figure 2A). Under these circumstances, a lineage bearing a single new beneficial mutation may spread through a somatic cell population, replacing its members in what is termed a periodic selection event or selective sweep in the evolution literature (reviewed in [33]). Some time later, another such sweep may occur, completely separate in time from the first sweep. On the other hand, if the supply rate of beneficial mutations is high, then multiple beneficial mutations will be present in the population at any given time and the dynamics of clonal evolution can be considerably more complicated (Figure 2C). In either case, the progressive accumulation of mutations beneficial to individual cells within a somatic cell lineage can cause that lineage to proliferate, resulting in a neoplasm and potentially in cancer.
In populations of microbes or of somatic cells, the supply rate of beneficial mutations is given by the product of the population size and the rate at which beneficial mutations arise. Both of these parameters are controllable in microbial evolution experiments, and this has facilitated sophisticated experimental investigations of clonal evolution in such populations that greatly exceed the current phenomenological power of in vivo cancer studies. Neither population size nor mutation rate is under experimenter control in cancer investigations. Indeed, the size of the evolving population in a neoplasm is an unresolved question and may be one of the more important differences between experimental microbial populations and cancer. In an experimental microbial population, all cells have an equal opportunity to contribute to the next generation. In a neoplasm, however, the evolving population consists only of those cells that can continue dividing, either because they have the capacity to self-renew as ‘cancer stem cells’ ([54-58], but see [59-61]), or because they have not terminally differentiated (e.g.,[5,62-64]). Despite these caveats, it appears that the absolute size of the evolving cell population in many neoplasms can be quite large [65], and this suggests that, in at least some cases, the evolutionary dynamics of cancer involves multiple simultaneous driver mutations. We discuss some further implications of this possibility below.
Of course, not all mutations are beneficial: the great majority are likely to be either deleterious (decrease fitness) or neutral (have no effect on fitness). In both neoplasms and experimental populations, cell lineages bearing strongly deleterious mutations are very unlikely to persist and spread. However, cell lineages bearing neutral mutations or weakly deleterious mutations may persist and even spread in such populations, either individually through genetic drift or as a byproduct of fortuitous association with beneficial mutations. The latter effect is particularly important in clonal populations because all new mutations that occur in a genome are genetically linked. Thus, a spreading beneficial mutation (driver) can carry mutations present or arising on its genetic background to high frequency in a process of ‘genetic hitchhiking’ (Figure 2C). In a further extension of the transportation metaphor, hitchhiking mutations are often referred to as ‘passenger mutations’ in the cancer literature (e.g., [50,51]).
It is important to note here that whether and to what degree a given mutation is neutral, deleterious, or beneficial can depend on the genetic background, on the environment, and even on interactions between the genetic background and the environment. In this way, for example, mutations that accumulate neutrally in a population may later be beneficial if the environment changes [66]; or a mutation that would not spread in a neoplasm on one genetic background may do so in a different background. We discuss some possible examples of such epistasis in cancer in the next section. Finally, it is also possible that the beneficial effect of a mutation could depend negatively on the number of cells carrying that mutation. Such density dependence is suggested by the view of cancer cells as social cheaters in a restraint-based economy [14].
Selection in Neoplasms
Given the central importance of selection on new mutations to evolutionary theories of cancer, we turn next to a brief discussion of methods for detecting selection and the nature of the evidence that they provide. Perhaps the most direct method for detecting selection — and certainly the most directly relevant to cancer — is to look for parallel, functionally convergent mutational changes in populations that have been evolving independently in identical conditions. Experimental populations and cancers are well suited to this method; because each experimental population and each individual’s cancer evolves uniquely, we can look across different populations or cancers to identify common genetic lesions. This approach can distinguish mutations directly under selection (drivers in cancer) from hitchhiker mutations (passengers). The fact that p53 mutations occur in many different types of cancers across different individuals [67], for example, indicates that mutations at this locus are not merely hitchhikers but actually confer a selective advantage to cancer cells. Studies in experimental evolution have taken the lead in using genome sequencing to identify parallel mutational changes in replicate populations that have been subjected to a common selective regime (e.g., [68-70]). As human genome sequencing becomes more and more affordable, the prevalence of this approach is increasing in cancer research. Several recent studies, for example, have used various deep sequencing and single-cell sequencing approaches to investigate the clonal origin and genetic heterogeneity of tumours and to identify candidate driver mutations [71-76]. It is worth noting some caveats, however. First, in contrast to replicate experimental populations, individuals represent different genotypes. Such genotypic differences could well result in different individuals taking quite different genetic pathways to cancer. Second, and also in contrast to the situation in experimental evolution, individuals may represent very different environments for tumour evolution (smokers vs. nonsmokers, for example), again potentially affecting the pathway to cancer. Finally, when common mutations (whether point mutations or chromosomal rearrangements) are found across replicate populations or cancers, these could in principle be due to increased susceptibility to mutation in certain regions rather than to selection. For example, it is well known that fragile sites [77] tend to be mutated in certain cancers [77-79], but it is unclear as to whether this is simply because of their high mutation rates or also because such mutations are selectively favored in neoplasms.
Various statistical tests for detecting selection on point mutations in nucleotide sequence data sampled from natural populations have been in use by evolutionary biologists since around 1990 [80] (reviewed in [81,82]). In principle, ratios of nonsynonymous to synonymous nucleotide substitutions (dN/dS) and/or polymorphisms observed in homologous sequences within and between populations can be used to infer ‘purifying selection’ against deleterious mutations (dN/dS < 1), neutral evolution (dN/dS = 1), or “positive selection” in favor of beneficial mutations (dN/dS *gt; 1). Such tests, however, are quite sensitive to certain demographic assumptions, are often lacking in statistical power [83,84], and are generally not suited to pinpointing the exact functional changes under selection — something which is obviously of great interest in cancer research. Furthermore, most of these methods are based on detecting deviations from equilibrium expectations under the forces of mutation, selection, and genetic drift and thus may not apply well to growing clonal populations such as cancers. An example of the difficulty of applying these tests to cancer comes from the work of Pleasance et al. [85], who observed an elevated dN/dS ratio in known colon cancer cell lineages yet found this to be nonsignificant as evidence for selection and noted that the approach is insensitive to small numbers of selected mutations. Some more recent methods for detecting selection may be more promising: Fraser [82] has recently described approaches employing gene expression data to detect selection in cancer, and Illingworth et al. [86] have proposed novel theoretical methods for distinguishing driver and passenger mutations using time series sequence data from an evolving clonal population that may also be applicable to cancer.
In evolution experiments with microbes, the strength of selection, i.e. the magnitude of the fitness difference between genotypes, can be estimated directly by measuring the relative growth of two genotypes in direct competition under replicated, controlled conditions (e.g., [87-89]). The power and precision afforded by these approaches have directly demonstrated that even mutations of very small selective effect can spread through large populations (e.g., [90]) as predicted by evolutionary theory. Moreover, longitudinal fitness data of this kind from experimental populations have provided strong evidence for competition among multiple beneficial mutations in clonal populations (reviewed in [33]), a subject with implications for cancer which we discuss below. The magnitude of selection on cancer-causing mutations remains largely unexplored and clearly requires further work (but see [51]). It may be possible to estimate the selective advantage of cancer cells in vitro using competition experiments similar to those developed in experimental evolution. An obvious concern with such an approach, however, is that in vitro cell culture conditions need not correspond to the in vivo conditions that would favor certain mutations in a neoplasm. This is in contrast to the situation in microbial experimental evolution, where in vitro conditions can be in vivo conditions.
A further complication in assessing the relative effects of mutations that cause cancer is that the magnitude and even the sign of selection affecting a given mutation can vary depending upon other mutations that have occurred in the same genetic background. Such epistatic effects are likely to be especially important in clonal populations, where mutations are effectively trapped in the genetic background on which they have arisen. There is rapidly growing evidence from studies in experimental evolution for epistasis among beneficial mutations (e.g., [66,91-95]). In cancer, there is emerging evidence for epistasis from studies of the temporal sequence of mutations in clones. One example comes from Barrett’s esophagus, a chronic condition in which the normal squamous lining of the esophageal epithelium is replaced by columnar metaplasia which can lead to esophageal adenocarcinoma. In Barrett’s patients who progress to cancer, a CDKN2A (p16) mutation tends to be an initiating lesion and TP53 (p53) mutations are usually found closer to the development of cancer [96,97]. This implies that a p53 mutation is more beneficial to cells in the context of a p16 mutation (or perhaps some other, undefined ‘early’ mutation) than it is alone. Further specific evidence for epistasis in cancer is provided by a very recent study of clonal ordering and prognosis in acute myeloid leukemia (AML) [98]. In a large cohort of AML patients for whom outcomes were known, Patel et al. sequenced all of the mutations known to be relevant to prognosis: they showed that NPM1 mutations, which have been associated with improved clinical outcomes, improve prognosis only in the presence of wild-type FLT3, and then only if a particular mutation is present in IDH2 [98]. More broadly, the potential importance of epistasis in cancer is illustrated by the phenomenon of ‘oncogene addiction’ [99], in which the presence of a prior oncogenic mutation determines whether a later mutation will act as a driver (be selectively favoured) or, in contrast, be highly deleterious to the tumour. Interestingly, a somewhat parallel phenomenon has been observed in bacterial populations, in which certain mutations which compensate for the cost of antibiotic resistance are deleterious in the absence of the resistance mutation [100]. In general, epistasis may well play an important role in cancer evolution, but because the total number of mutations involved in cancers and their epistatic effects have yet to be fully characterised, that role largely remains to be determined.
Ideally, analyses of selection in cancer would lead to an understanding of both the exact mutations involved in cancer development and the likely order in which those mutations accumulate as a cancer develops. Although there have been some influential attempts to identify predictable mutational pathways to carcinogenesis [101], simulations of cancer evolution indicate that the temporal order of even a known set of mutational substitutions during cancer evolution can be quite variable and need not be replicated across independent cancers in the absence of epistasis [102]. Moreover, some empirical studies suggest that there are multiple redundant pathways to cancer [103] with only minimal overlap in the mutations that occur between different tumors, such that identifying common pathways to cancer may be more difficult than originally supposed.
Mutagenesis and Carcinogenesis: Evolutionary Considerations
The relationship between mechanisms of mutation and cancer has obviously been the subject of an enormous amount of research for many decades. It is well known that cancer cells often harbour extensive changes in genomic architecture (such as major chromosomal rearrangements and aneuploidies) in addition to point mutations [85,104-106], and recent genomic sequence data suggest the possibility that different cancer types have different mutational spectra [106,107]. Experimental evolution studies in yeast have provided evidence for the role that rearrangements, duplications, and other major chromosomal alterations can play in the adaptation of eukaryotic genomes to novel environments (e.g., [15-17]), suggesting a useful parallel between these studies and cancer evolution. Nonetheless, there is a limit to what microbial experimental evolution can reveal about the mechanistic importance of the specific mutations that are found in cancer cells because the nature and functional significance of mutational changes in experimental populations of single-celled microbes can never completely parallel those in a complex multicellular organism. Microbial evolution approaches have, however, contributed significantly to our broad understanding of how genomic mutation rates evolve in clonal populations, with some important implications for understanding cancer. In this section, we focus at this general level on mutation rates, their evolution, and their consequences for evolution in clonal populations, including cancers.
Lynch [108] has provided a comprehensive review of mutation rates in normal human cells. In keeping with the deleterious nature of most mutations that affect the phenotype, estimated per nucleotide mutation rates per cell division in the human germ line are quite low (0.06 × 10−9) — lower than those, for example, in E. coli (0.26. × 10−9) and Saccharomyces cerevisiae (0.33 × 10−9). However, when the number of germ line generations is taken into account the human per nucleotide mutation rate per organismal generation (12.8 × 10−9) is far higher than that of microbes. Remarkably, the per nucleotide per cell division mutation rate (0.77 × 10−9) in human somatic cells appears to be much higher than in germ cells; as yet, the molecular basis for this difference between somatic and germ line mutation rates is unknown.
Despite the widely accepted association between genome instability and cancer, it is unclear whether mutation rates in cancer cells are always elevated above those in normal somatic cells [109]. Difficulties arise here in that it is more challenging to measure the actual somatic mutation rate than to measure the rate at which mutations are substituted in a population: while the origination of cancer-causing mutations is a haphazard process, their substitution depends on the dynamics of selection within a tumour. Good evidence supporting a role for elevated somatic mutation rates in cancer [1,110] comes, for example, from the observed hereditary predisposition toward cancer in individualsbearing one or another defect in DNA repair (reviewed in[111]). On the other hand, a recent sequence-based study[112] estimated a point mutation rate in colorectal cancer that is comparable to that measured in normal cells by similar means (4.6 × 10−10 vs. 10 × 10−10), supporting the notion that an elevated mutation rate is not needed for cancer evolution [109,113].
Experimental evolution studies in E. coli have shown that high genomic mutation rates can evolve in clonal populations by a process in which spontaneously originated mutator alleles hitchhike to fixation with beneficial mutations[7,114-116]; the results of these studies are broadly supported by theoretical work modeling the mutator hitchhiking process [31,41,117-119]. Taken together, this work suggests the possibility that mutation rates may indeed become elevated by a similar process in cancers. However, whether the rate of adaptation (or cancer progression) is significantly increased at a higher mutation rate is a more subtle issue [33]. If beneficial mutations are rare within a tumour (Figure 2A), then an increase in the mutation rate can shorten the waiting time between them and greatly increase their rate of incorporation into a clonal population. However, if beneficial mutations are quite common within a tumour (Figure 2C), then the rate of adaptation (or cancer progression) can be limited by the sorting process that takes place amongst competing clones rather than by the mutation supply rate [120]. Studies in experimental bacterial populations have shown that as the supply rate of beneficial mutations is increased to very high values, further increase in the rate of adaptation is stymied by this process of ‘clonal interference’ (reviewed in [33], but see [121]). Thus, an increase in the genomic mutation rate need not necessarily translate into faster adaptation or faster cancer progression.
There is a growing body of evidence that tumours are genetically heterogeneous and that this reflects the presence of multiple cancer-related lesions in some cases. Such heterogeneity can arise as a result of a high beneficial mutation supply rate but may also be contributed to by spatial structure within a tumour (see below). Cancer progression in such circumstances may well be slowed by the kind of clonal interference dynamics described above, and this perhaps offers avenues for treatment of early stage cancers: it is conceivable that the application of multiple benign external selection pressures to an early-stage cancer via drug treatment could delay further progression as a consequence of clonal interference. A test of this idea in experimental yeast populations has provided some encouraging results [122].
Increasing the genomic mutation rate increases the supply of deleterious mutations as well as of those that are neutral and beneficial. A considerable body of theoretical work in evolutionary genetics is devoted to understanding the effects of deleterious mutation accumulation on the fitness and persistence of clonal (asexual) populations [123-129], and there has been particular interest recently in the possibility that mutagenesis could be used to cure or inhibit viral infections by decreasing viral fitness within individual hosts. For example, the antiviral ribavirin is widely used to treat hepatitis C and is thought to decrease viral titer by increasing the mutation rate [130]. Similarly, in tumour cell populations that already have high mutation rates it is conceivable that an appropriate therapy could drive the tumour extinct or greatly suppress its growth by raising the mutation rate even higher [131-133]. Interestingly, several widely used anticancer therapies (examples include 5-fluorouracil and temozolomide) are mutagenic. Perhaps some of their effectiveness is due to the indirect effect they have in increasing the supply of deleterious mutations in the tumour population. On the other hand, it has been suggested that a tumour could garner additional driver mutations and spread even more rapidly with an artificially elevated mutation rate, so caution is clearly warranted in considering such an approach [133].
Evolution of Drug Resistance
A fundamental tenet of evolutionary theory is that populations do not respond to a selection pressure by producing new, ‘directed’ beneficial mutations specifically suited to that selection pressure; instead, evolutionary adaptation occurs via sorting among genetic variants that have arisen without regard to adaptive utility. This is, in fact, one clear area where experimental evolution should inform cancer biology. Whether or not mutations are directed has been extensively debated in the context of experimental evolution (reviewed in [134,135]), and the prevailing consensus continues to be that mutations arise indifferently with respect to selective need (although the possibility of directed mutation has been raised again in recent experiments [11]). In this respect, it is important to remember that there is also no evidence that therapeutic resistance in cancer arises in response to the selective pressure of therapy; instead, mutations conferring resistance are likely to exist within a population of cancer cells and are subsequently selected for when cancer therapy is applied. This principle has been demonstrated in the case of several therapies (e.g. [71,136-140]). It is possible, of course, that some mutations may arise during the course of therapy if the therapy is not completely effective, and population genetics theory has been developed that can distinguish between the two possibilities[141,142]. A few studies have suggested that resistance may develop in some naïve populations of bacteria during antibiotic application [143] while work in HIV has elegantly demonstrated that, most of the time, resistance mutations are present before therapy is applied [144].
Experimental evolution also provides insights into how therapy regimens should be designed to reduce the incidence and development of resistance. The development of resistance depends critically on parameters such as population size, mutation rate, and selective effect of the adaptive mutations [142]. For instance, lower-dose, pulsed therapy can drive the development of resistance, as has been demonstrated for evolution of antibiotic resistance [145], particularly with suboptimal dosing (e.g., [146]), and with resistance to malaria [147]. Theoretical evolutionary models suggest that this is also likely to be a problem for cancer therapies and that we should reevaluate the common dosing schedules of chemotherapeutic agents [148,149].
The Microenvironment: Spatial Structure and Cell-Cell Interactions
To date, most experimental evolution and the theoretical work it has stimulated have assumed a set of environmental conditions that might be termed the ‘planktonic ideal’: well-mixed (zero-dimensional) liquid cultures (Figure 1A,B). The planktonic ideal has been invaluable for working out fundamental evolutionary dynamics. But cancers do not necessarily conform to the well-mixed, cell-autonomous ideal; neoplasms are three-dimensional objects with a relatively stable spatial structure (but see [150,151]) and cells can interact with each other and their microenvironment[152,153]. Recent theoretical studies show that spatial structure can have important consequences for the speed of adaptation or cancer progression. Neoplasms in which cells remain in one place are predicted to take longer to progress to cancer than those in which cells are highly motile [9]. This is because the limiting effect of clonal interference on the rate of adaption is augmented in the presence of spatial structure [9]; long-range migration can mitigate this effect and increase the rate of adaptation or neoplastic progression [10].
As mentioned in a previous section, high levels of genetic heterogeneity within tumours and pre-neoplasias are increasingly reported [71,106,154-158]. Evolution experiments with microbes have provided insights into how spatial structure and interactions between cells may explain intrapopulation heterogeneity. For example, Kerr et al. [159] demonstrated that three bacterial clones could coexist in a stable nontransitive, competitive relationship (rock-paperscissors) in a spatially structured environment whereas this relationship was unstable in a well mixed environment. In Pseudomonas fluorescens, populations grown in spatially structured environments showed sustained levels of heterogeneity, while those grown in homogeneous environments did not [160]. Even in the well mixed environment of serial culture or the chemostat (Figure 1A,B), homogeneous ancestor populations of bacteria have evolved into distinct, stable subclones in which each subclone depends on the metabolic byproducts of another in a process called cross-feeding[161-164]. Bacteria can also live in matrix-encased, cooperative, multicellular communities called biofilms (described in [165]). In one study, several different heritable phenotypes of clones were observed in Pseudomonas aeruginosa biofilms, including one cancer-like clone that developed faster and was more resistant to treatment than the other clones measured in the biofilm [166]. Intriguingly, biofilm communities often exhibit increased resistance to treatment by anti-microbial agents [167]. Cell-cell interactions and spatial structure are clearly important aspects of neoplasms, and so it is worth considering whether general interclonal relationships of the kind we have discussed here apply within neoplasms as well.
Concluding Perspective
So far, the evolutionary approach to understanding cancer has focused mainly on the analysis of patterns of standing genetic variation within tumours. In this review, we have tried to draw insights on cancer biology from the large and rapidly growing field of microbial experimental evolution, which studies evolution in real time. In making this broad connection, our rationale has been that cancer may be usefully understood as an evolutionary phenomenon of clonal populations with important dynamical similarities to microbial experimental evolution and that the phenomenological power of experimental evolution can provide insights into these dynamics that are not as readily available from in vivo studies of cancer.
Fundamentally, of course, microbial experimental evolution can only serve as a model system or analogy for cancer development. In any analogy, the differences between entities being compared can be as important as the similarities. We have not specifically discussed the evolution of metastasis in this review because there is very little in the experimental evolution literature to date that is directly applicable to this problem. Another obvious topic that we have ignored in this review is the role of epigenetic changes in cancer development. There has not been a significant body of work in experimental evolution in this area so far. If epigenetic changes are completely stable in cancers, then the dynamics of ordinary mutations as described above provide a useful analogy; however, if epigenetic changes are unstable, then their theoretical and experimental study will require new approaches differing from those we have discussed — this may well be an important area in which experimental evolution and theory can contribute hand-in-hand in the near future.
We end our review on a more speculative note. A vast amount of research has focused on the molecular genetic bases of cancer for many decades now and enormous advances have clearly been made, yet the progression of any individual cancer remains difficult to predict [2,3,168]. The problem of predicting cancer based on explicit knowledge of its genetic bases can be regarded more generally as the problem of predicting evolution given the large number of mutations that may occur in a population and their potential interactions in determining fitness. Here, we briefly describe new theoretical work by two of us [12] that explores a framework for forecasting the near-future evolution of adapting populations that does not rely on detailed knowledge of the underlying genetics of adaptation.
A commonplace metaphor in the evolution literature is the ‘adaptive landscape’, which is conceptually a topographic mapping of fitness (vertical axis) onto all possible mutational combinations in a population. Fitter genotypes correspond to higher elevations in the topography, and a population located anywhere on the map ascends the local slope by natural selection to the nearest peak, much as a mountaineer ascends a mountain. Although the adaptive landscape metaphor is a useful way of thinking about evolution, it has a glaring practical problem: namely, the near impossibility of obtaining the real map relating all possible mutational combinations to fitness in an organism (or cancer) for any given environment (individual), let alone across environments (individuals).
In contrast to the adaptive landscape approach, our recent theoretical work focuses on the mountaineer rather than the mountains. Just as a mountaineer’s gait and pace may reveal something about the terrain underfoot, an evolving population’s composition can carry information about the underlying adaptive landscape and even about what lies a few paces ahead. We have shown that by sampling members of a population in real time and characterising the population’s fitness distribution at multiple time points, evolution can successfully be predicted tens of generations into the future in simulations [12], experimental E. coli populations (Sprouffskeet al., manuscript in preparation) and, potentially, cancer lineages. This approach might be applicable to the analysis and prediction of cancer progression within individual patients, though we caution that its practicability and scope require considerable further research. Nonetheless, the potential of this approach is, we think, illustrative of the main point of our review: namely, that evolution experiments with microbes may have new things to tell us about cancer.
Acknowledgments
We thank Aaron Shaver, Yevgeniy Raynes, Gail Kienitz and two anonymous reviewers for comments that improved the manuscript. K.S., P.J.G. and P.S. were supported by US National Institutes of Health grants R01 GM079843-01 and ARRA PDS#35063; in addition, P.J.G. was supported by European Commission (ec.europa.eu) grant FP7231807. C.C.M. was supported by Research Scholar Grant #117209-RSG-09-163-01-CNE from the American Cancer Society, the Addario Lung Cancer Medical Institute, and US National Institutes of Health grants P01 CA91955, R01 CA149566 and R01 CA140657.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nowell P. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 2.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Merlo L, Pepper J, Reid B, Maley C. Cancer as an evolutionary and ecological process. Nat. Rev. Cancer. 2006;6:924–935. doi: 10.1038/nrc2013. [DOI] [PubMed] [Google Scholar]
- 4.Crespi B, Summers K. Evolutionary biology of cancer. Trends Ecol. Evol. 2005;20:545–552. doi: 10.1016/j.tree.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 5.Frank SA. Dynamics of Cancer: Incidence, Inheritance, and Evolution. Princeton University Press; Princeton: 2007. [PubMed] [Google Scholar]
- 6.Elena SF, Lenski RE. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat. Rev. Genet. 2003;4:457–469. doi: 10.1038/nrg1088. [DOI] [PubMed] [Google Scholar]
- 7.Sniegowski PD, Gerrish PJ, Lenski RE. Evolution of high mutation rates in experimental populations of E. coli. Nature. 1997;387:703–705. doi: 10.1038/42701. [DOI] [PubMed] [Google Scholar]
- 8.Sniegowski P, Gerrish P, Johnson T, Shaver A. The evolution of mutation rates: separating causes from consequences. BioEssays. 2000;22:1057–1066. doi: 10.1002/1521-1878(200012)22:12<1057::AID-BIES3>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 9.Martens E, Kostadinov R, Maley CC, Hallatschek O. Spatial structure increases the waiting time for cancer. New J. Phys. 2011;13:115014. doi: 10.1088/1367-2630/13/11/115014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martens EA, Hallatschek O. Interfering waves of adaptation promote spatial mixing. Genetics. 2011;189:1045–1060. doi: 10.1534/genetics.111.130112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Q, Lambert G, Liao D, Kim H, Robin K, Tung C.-k., Pourmand N, Austin RH. Acceleration of emergence of bacterial antibiotic resistance in connected microenvironments. Science. 2011;333:1764–1767. doi: 10.1126/science.1208747. [DOI] [PubMed] [Google Scholar]
- 12.Gerrish PJ, Sniegowski PD. Real time forecasting of nearfuture evolution. J. Roy. Soc. Interface. 2012:1742–5662. doi: 10.1098/rsif.2012.0119. http://dx.doi.org/ 10.1098/rsif.2012.0119, Advance Online Publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campos PRA, Neto PSCA, de Oliveira VM, Gordo I. Environmental heterogeneity enhances clonal interference. Evolution. 2008;62:1390–1399. doi: 10.1111/j.1558-5646.2008.00380.x. [DOI] [PubMed] [Google Scholar]
- 14.Velicer GJ. Social strife in the microbial world. Trends Microbiol. 2003;11:330–337. doi: 10.1016/s0966-842x(03)00152-5. [DOI] [PubMed] [Google Scholar]
- 15.Dunham MJ, Badrane H, Ferea T, Adams J, Brown PO, Rosenzweig F, Botstein D. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2002;99:16144–16149. doi: 10.1073/pnas.242624799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gresham D, Desai MM, Tucker CM, Jenq HT, Pai DA, Ward A, De-Sevo CG, Botstein D, Dunham MJ. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet. 2008;4:e1000303. doi: 10.1371/journal.pgen.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rancati G, Pavelka N, Fleharty B, Noll A, Trimble R, Walton K, Perera A, Staehling-Hampton K, Seidel CW, Li R. Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell. 2008;135:879–893. doi: 10.1016/j.cell.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Darwin C. The Origin of Species. John Murray; London: 1859. [Google Scholar]
- 19.Coyne JA. Why Evolution is True (Viking Adult) 2009 [Google Scholar]
- 20.Grant P. Ecology and Evolution of Darwin’s Finches. Princeton University Press; Princeton: 1999. [Google Scholar]
- 21.Atwood KC, Schneider LK, Ryan FJ. Periodic selection in Escherichia coli. Proc. Natl. Acad. Sci. USA. 1951;37:146–155. doi: 10.1073/pnas.37.3.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ryan F. Evolution observed. Sci. Am. 1953;189:78–82. [Google Scholar]
- 23.Paquin C, Adams J. Frequency of fixation of adaptive mutations is higher in evolving diploid than haploid yeast populations. Nature. 1983;302:495–500. doi: 10.1038/302495a0. [DOI] [PubMed] [Google Scholar]
- 24.Paquin C, Adams J. Relative fitness can decrease in evolving asexual populations of S. cerevisiae. Nature. 1983;306:368–371. doi: 10.1038/306368a0. [DOI] [PubMed] [Google Scholar]
- 25.Hall BG. Evolution of new metabolic functions in laboratory organisms. In: Nei M, Koehn RK, editors. Evolution of Genes and Proteins. Sinauer; Sunderland, MA: 1983. pp. 234–257. [Google Scholar]
- 26.Mortlock RR. Microorganisms as Model Systems for Studying Evolution. Springer; Plenum, NY: 1984. [Google Scholar]
- 27.Bell G. Selection: the Mechanism of Evolution. 2nd Edition Oxford University Press; USA: 2009. [Google Scholar]
- 28.Garland TG, Rose MR. Experimental Evolution: Concepts, Methods and Applications of Selection Experiments. University of California Press; Berkeley and Los Angeles, CA: 2009. [Google Scholar]
- 29.Schiffels S, Szollosi GJ, Mustonen V, Lässig M. Emergent neutrality in adaptive asexual evolution. Genetics. 2011;189:1361–1375. doi: 10.1534/genetics.111.132027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Desai MM, Fisher DS. The balance between mutators and nonmutators in asexual populations. Genetics. 2011;188:997–1014. doi: 10.1534/genetics.111.128116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerrish PJ, Colato A, Perelson AS, Sniegowski PD. Complete genetic linkage can subvert natural selection. Proc. Natl. Acad. Sci. USA. 2007;104:6266–6271. doi: 10.1073/pnas.0607280104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campos PRA, Wahl LM. The adaptation rate of asexuals: deleterious mutations, clonal interference and population bottlenecks. Evolution. 2010;64:1973–1983. doi: 10.1111/j.1558-5646.2010.00981.x. [DOI] [PubMed] [Google Scholar]
- 33.Sniegowski PD, Gerrish PJ. Beneficial mutations and the dynamics of adaptation in asexual populations. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010;365:1255–1263. doi: 10.1098/rstb.2009.0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fogle CA, Nagle JL, Desai MM. Clonal interference, multiple mutations and adaptation in large asexual populations. Genetics. 2008;180:2163–2173. doi: 10.1534/genetics.108.090019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gerrish P. The rhythm of microbial adaptation. Nature. 2001;413:299–302. doi: 10.1038/35095046. [DOI] [PubMed] [Google Scholar]
- 36.Kim Y, Orr HA. Adaptation in sexuals vs. asexuals: clonal interference and the Fisher-Muller model. Genetics. 2005;171:1377–1386. doi: 10.1534/genetics.105.045252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park S-C, Krug J. Clonal interference in large populations. Proc. Natl. Acad. Sci. USA. 2007;104:18135–18140. doi: 10.1073/pnas.0705778104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rouzine IM, Wakeley J, Coffin JM. The solitary wave of asexual evolution. Proc. Natl. Acad. Sci. USA. 2003;100:587–592. doi: 10.1073/pnas.242719299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rouzine IM, Brunet E, Wilke CO. The traveling-wave approach to asexual evolution: Muller’s ratchet and speed of adaptation. Theor. Popul. Biol. 2008;73:24–46. doi: 10.1016/j.tpb.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilke CO. The speed of adaptation in large asexual populations. Genetics. 2004;167:2045–2053. doi: 10.1534/genetics.104.027136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.André J, Godelle B. The evolution of mutation rate in finite asexual populations. Genetics. 2006;172:611–626. doi: 10.1534/genetics.105.046680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caulin AF, Maley CC. Peto’s paradox: evolution’s prescription for cancer prevention. Trends Ecol. Evol. 2011;26:175–182. doi: 10.1016/j.tree.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Otto SP, Orive ME. Evolutionary consequences of mutation and selection within an individual. Genetics. 1995;141:1173–1187. doi: 10.1093/genetics/141.3.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Michod RE, Roze D. Cooperation and conflict in the evolution of multicellularity. Heredity. 2001;86:1–7. doi: 10.1046/j.1365-2540.2001.00808.x. [DOI] [PubMed] [Google Scholar]
- 45.Nunney L. Lineage selection and the evolution of multistage carcinogenesis. Proc. Biol. Sci. 1999;266:493–498. doi: 10.1098/rspb.1999.0664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nunney L. The population genetics of multistage carcinogenesis. Proc. Biol. Sci. 2003;270:1183–1191. doi: 10.1098/rspb.2003.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lenski RE. Evolution in action: a 50,000 generation salute to Charles Darwin. Microbe. 2011;6:30–33. [Google Scholar]
- 48.Shibata D. Mutation and epigenetic molecular clocks in cancer. Carcinogenesis. 2011;32:123–128. doi: 10.1093/carcin/bgq239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat. Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 50.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Edkins S, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bozic I, Antal T, Ohtsuki H, Carter H, Kim D, Chen S, Karchin R, Kinzler KW, Vogelstein B, Nowak MA. Accumulation of driver and passenger mutations during tumor progression. Proc. Natl. Acad. Sci. USA. 2010;107:18545–18550. doi: 10.1073/pnas.1010978107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fisher RA. The Genetical Theory of Natural Selection. Oxford University Press; Oxford, UK: 1930. [Google Scholar]
- 53.Muller H. Some genetic aspects of sex. Am. Nat. 1932;66:118–138. [Google Scholar]
- 54.Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri M, Dick J. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 56.O’Brien C, Pollett A, Gallinger S, Dick J. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 57.Gupta PB, Chaffer CL, Weinberg RA. Cancer stem cells: mirage or reality? Nat. Med. 2009;15:1010–1012. doi: 10.1038/nm0909-1010. [DOI] [PubMed] [Google Scholar]
- 58.Rosen JM, Jordan CT. The increasing complexity of the cancer stem cell paradigm. Science. 2009;324:1670–1673. doi: 10.1126/science.1171837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shipitsin M, Campbell LL, Argani P, Weremowicz S, Bloushtain-Qimron N, Yao J, Nikolskaya T, Serebryiskaya T, Beroukhim R, Hu M, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11:259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 60.Park SY, Lee HE, Li H, Shipitsin M, Gelman R, Polyak K. Heterogeneity for stem cell-related markers according to tumor subtype and histologic stage in breast cancer. Clin. Cancer Res. 2010;16:876–887. doi: 10.1158/1078-0432.CCR-09-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Merlo LMF, Maley CC. The role of genetic diversity in cancer. J. Clin. Invest. 2010;120:401–403. doi: 10.1172/JCI42088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morrison S, Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 2006;441:1068–1074. doi: 10.1038/nature04956. [DOI] [PubMed] [Google Scholar]
- 63.Michor F, Nowak MA, Frank SA, Iwasa Y. Stochastic elimination of cancer cells. Proc. Biol. Sci. 2003;270:2017–2024. doi: 10.1098/rspb.2003.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dingli D, Traulsen A, Michor F. (A)symmetric stem cell replication and cancer. PLoS Comput. Biol. 2007;3:e53. doi: 10.1371/journal.pcbi.0030053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Salmon SE, Smith BA. Immunoglobulin synthesis and total body tumor cell number in IgG multiple myeloma. J. Clin. Invest. 1970;49:1114–1121. doi: 10.1172/JCI106327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hayden EJ, Ferrada E, Wagner A. Cryptic genetic variation promotes rapid evolutionary adaptation in an RNA enzyme. Nature. 2011;474:92–95. doi: 10.1038/nature10083. [DOI] [PubMed] [Google Scholar]
- 67.Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, Olivier M. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum. Mutat. 2007;28:622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 68.Bull JJ, Badgett MR, Wichman HA, Huelsenbeck JP, Hillis DM, Gulati A, Ho C, Molineux IJ. Exceptional convergent evolution in a virus. Genetics. 1997;147:1497–1507. doi: 10.1093/genetics/147.4.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Woods R, Schneider D, Winkworth CL, Riley MA, Lenski RE. Tests of parallel molecular evolution in a long-term experiment with Escherichia coli. Proc. Natl. Acad. Sci. USA. 2006;103:9107–9112. doi: 10.1073/pnas.0602917103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tenaillon O, Rodríguez-Verdugo A, Gaut RL, McDonald P, Bennett AF, Long AD, Gaut BS. The molecular diversity of adaptive convergence. Science. 2012;335:457–461. doi: 10.1126/science.1212986. [DOI] [PubMed] [Google Scholar]
- 71.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hou Y, Song L, Zhu P, Zhang B, Tao Y, Xu X, Li F, Wu K, Liang J, Shao D, et al. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell. 2012;148:873–885. doi: 10.1016/j.cell.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 73.Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012 doi: 10.1038/nature10933. http:// dx.doi.org/10.1038/nature10933, Advance Online Publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu X, Hou Y, Yin X, Bao L, Tang A, Song L, Li F, Tsang S, Wu K, Wu H, et al. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell. 2012;148:886–895. doi: 10.1016/j.cell.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Walter MJ, Shen D, Ding L, Shao J, Koboldt DC, Chen K, Larson DE, McLellan MD, Dooling D, Abbott R, et al. Clonal architecture of secondary acute myeloid leukemia. N. Engl. J. Med. 2012;366:1090–1098. doi: 10.1056/NEJMoa1106968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Freudenreich CH. Chromosome fragility: molecular mechanisms and cellular consequences. Front. Biosci. 2007;12:4911–4924. doi: 10.2741/2437. [DOI] [PubMed] [Google Scholar]
- 78.Bignell GR, Greenman CD, Davies H, Butler AP, Edkins S, Andrews JM, Buck G, Chen L, Beare D, Latimer C, et al. Signatures of mutation and selection in the cancer genome. Nature. 2010;463:893–898. doi: 10.1038/nature08768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lai LA, Kostadinov R, Barrett MT, Peiffer DA, Pokholok D, Odze R, Sanchez CA, Maley CC, Reid BJ, Gunderson KL, et al. Deletion at fragile sites is a common and early event in Barrett’s esophagus. Mol. Cancer Res. 2010;8:1084–1094. doi: 10.1158/1541-7786.MCR-09-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 81.Hughes AL. Looking for Darwin in all the wrong places: the misguided quest for positive selection at the nucleotide sequence level. Heredity. 2007;99:364–373. doi: 10.1038/sj.hdy.6801031. [DOI] [PubMed] [Google Scholar]
- 82.Fraser HB. Genome-wide approaches to the study of adaptive gene expression evolution. BioEssays. 2011;33:469–477. doi: 10.1002/bies.201000094. [DOI] [PubMed] [Google Scholar]
- 83.Kryazhimskiy S, Plotkin JB. The population genetics of dN/dS. PLoS Genet. 2008;4:e1000304. doi: 10.1371/journal.pgen.1000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Charlesworth J, Eyre-Walker A. The McDonald-Kreitman test and slightly deleterious mutations. Mol. Biol. Evol. 2008;25:1007–1015. doi: 10.1093/molbev/msn005. [DOI] [PubMed] [Google Scholar]
- 85.Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin M-L, Ordóñez GR, Bignell GR, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Illingworth CJR, Mustonen V. Distinguishing driver and passenger mutations in an evolutionary history categorized by interference. Genetics. 2011;189:989–1000. doi: 10.1534/genetics.111.133975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dean AM. Selection and neutrality in lactose operons of Escherichia coli. Genetics. 1989;123:441–454. doi: 10.1093/genetics/123.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Suiter AM, Bänziger O, Dean AM. Fitness consequences of a regulatory polymorphism in a seasonal environment. Proc. Natl. Acad. Sci. USA. 2003;100:12782–12786. doi: 10.1073/pnas.2134994100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lenski RE, Travisano M. Dynamics of adaptation and diversification: a 10,000-generation experiment with bacterial populations. Proc. Natl. Acad. Sci. USA. 1994;91:6808–6814. doi: 10.1073/pnas.91.15.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lunzer M, Natarajan A, Dykhuizen DE, Dean AM. Enzyme kinetics, substitutable resources and competition: from biochemistry to frequency-dependent selection in lac. Genetics. 2002;162:485–499. doi: 10.1093/genetics/162.1.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Weinreich DM, Delaney NF, Depristo MA, Hartl DL. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science. 2006;312:111–114. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- 92.Sanjuán R, Cuevas JM, Moya A, Elena SF. Epistasis and the adaptability of an RNA virus. Genetics. 2005;170:1001–1008. doi: 10.1534/genetics.105.040741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chou H-H, Chiu H-C, Delaney NF, Segre D, Marx CJ. Diminishing returns epistasis among beneficial mutations decelerates adaptation. Science. 2011;332:1190–1192. doi: 10.1126/science.1203799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Khan AI, Dinh DM, Schneider D, Lenski RE, Cooper TF. Negative epistasis between beneficial mutations in an evolving bacterial population. Science. 2011;332:1193–1196. doi: 10.1126/science.1203801. [DOI] [PubMed] [Google Scholar]
- 95.Lunzer M, Golding GB, Dean AM. Pervasive cryptic epistasis in molecular evolution. PLoS Genet. 2010;6:e1001162. doi: 10.1371/journal.pgen.1001162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Reid BJ, Li X, Galipeau PC, Vaughan TL. Barrett’s oesophagus and oesophageal adenocarcinoma: time for a new synthesis. Nat. Rev. Cancer. 2010;10:87–101. doi: 10.1038/nrc2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Maley C, Galipeau P, Li X, Sanchez C, Paulson T, Reid B. Selectively advantageous mutations and hitchhikers in neoplasms: p16 lesions are selected in Barrett’s esophagus. Cancer Res. 2004;64:3414–3427. doi: 10.1158/0008-5472.CAN-03-3249. [DOI] [PubMed] [Google Scholar]
- 98.Patel JP, Gönen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J, Van Vlierberghe P, Dolgalev I, Thomas S, Aminova O, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N. Engl. J. Med. 2012;366:1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Weinstein IB, Joe A. Oncogene addiction. Cancer Res. 2008;68:3077–3080. doi: 10.1158/0008-5472.CAN-07-3293. [DOI] [PubMed] [Google Scholar]
- 100.Maisnier-Patin S, Andersson D. Adaptation to the deleterious effects of antimicrobial drug resistance mutations by compensatory evolution. Res. Microbiol. 2004;155:360–369. doi: 10.1016/j.resmic.2004.01.019. [DOI] [PubMed] [Google Scholar]
- 101.Fearon E, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 102.Sprouffske K, Pepper JW, Maley CC. Accurate reconstruction of the temporal order of mutations in neoplastic progression. Cancer Prev. Res. 2011;4:1135–1144. doi: 10.1158/1940-6207.CAPR-10-0374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Smith G, Carey F, Beattie J, Wilkie M, Lightfoot T, Coxhead J, Garner R, Steele R, Wolf C. Mutations in APC, Kirsten-ras, and p53-alternative genetic pathways to colorectal cancer. Proc. Natl. Acad. Sci. USA. 2002;99:9433–9438. doi: 10.1073/pnas.122612899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Campbell PJ, Stephens PJ, Pleasance ED, O’Meara S, Li H, Santarius T, Stebbings LA, Leroy C, Edkins S, Hardy C, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat. Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Stephens PJ, McBride DJ, Lin M-L, Varela I, Pleasance ED, Simpson JT, Stebbings LA, Leroy C, Edkins S, Mudie LJ, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Campbell P, Yachida S, Mudie L, Stephens P, Pleasance E, Stebbings L, Morsberger L, Latimer C, McLaren S, Lin M-L, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Stratton MR. Exploring the genomes of cancer cells: progress and promise. Science. 2011;331:1553–1558. doi: 10.1126/science.1204040. [DOI] [PubMed] [Google Scholar]
- 108.Lynch M. Rate, molecular spectrum, and consequences of human mutation. Proc. Natl. Acad. Sci. USA. 2010;107:961–968. doi: 10.1073/pnas.0912629107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shibata D, Lieber MR. Is there any genetic instability in human cancer? DNA Repair. 2010;9:858. doi: 10.1016/j.dnarep.2010.04.011. discussion 859-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Loeb LA, Loeb KR, Anderson JP. Multiple mutations and cancer. Proc. Natl. Acad. Sci. USA. 2003;100:776–781. doi: 10.1073/pnas.0334858100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Frank SA. Genetic predisposition to cancer — insights from population genetics. Nat. Rev. Genet. 2004;5:764–772. doi: 10.1038/nrg1450. [DOI] [PubMed] [Google Scholar]
- 112.Jones S, Chen W-D, Parmigiani G, Diehl F, Beerenwinkel N, Antal T, Traulsen A, Nowak MA, Siegel C, Velculescu VE, et al. Comparative lesion sequencing provides insights into tumor evolution. Proc. Natl. Acad. Sci. USA. 2008;105:4283–4288. doi: 10.1073/pnas.0712345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tomlinson IP, Novelli MR, Bodmer WF. The mutation rate and cancer. Proc. Natl. Acad. Sci. USA. 1996;93:14800–14803. doi: 10.1073/pnas.93.25.14800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mao EF, Lane L, Lee J, Miller JH. Proliferation of mutators in a cell population. J. Bacteriol. 1997;179:417–422. doi: 10.1128/jb.179.2.417-422.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shaver AC, Dombrowski PG, Sweeney JY, Treis T, Zappala RM, Sniegowski PD. Fitness evolution and the rise of mutator alleles in experimental Escherichia coli populations. Genetics. 2002;162:557–566. doi: 10.1093/genetics/162.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Notley-McRobb L, Seeto S, Ferenci T. Enrichment and elimination of mutY mutators in Escherichia coli populations. Genetics. 2002;162:1055–1062. doi: 10.1093/genetics/162.3.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wylie CS, Ghim C-M, Kessler D, Levine H. The fixation probability of rare mutators in finite asexual populations. Genetics. 2009;181:1595–1612. doi: 10.1534/genetics.108.094532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Taddei F, Radman M, Maynard-Smith J, Toupance B, Gouyon PH, Godelle B. Role of mutator alleles in adaptive evolution. Nature. 1997;387:700–702. doi: 10.1038/42696. [DOI] [PubMed] [Google Scholar]
- 119.Tenaillon O, Toupance B, Le Nagard H, Taddei F, Godelle B. Mutators, population size, adaptive landscape and the adaptation of asexual populations of bacteria. Genetics. 1999;152:485–493. doi: 10.1093/genetics/152.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gerrish PJ, Lenski RE. The fate of competing beneficial mutations in an asexual population. Genetica. 1998:102–103. 127–144. [PubMed] [Google Scholar]
- 121.Desai MM, Fisher DS, Murray AW. The speed of evolution and maintenance of variation in asexual populations. Curr. Biol. 2007;17:385–394. doi: 10.1016/j.cub.2007.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Merlo LMF, Howard TC, Gardiner KL, Caulin AF, Blum SM, Sprouffske K, Evans P, Bedalov A, Sniegowski PD, Maley CC. Application of simultaneous selective pressures retards adaptation to single selective pressures in Saccharomyces cerevisiae. Submitted. 2012 doi: 10.1111/eva.13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bull JJ, Wilke CO. Lethal mutagenesis of bacteria. Genetics. 2008;180:1061–1070. doi: 10.1534/genetics.108.091413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Eigen M. Selforganization of matter and the evolution of biological macromolecules. Naturwissenschaften. 1971;58:465–523. doi: 10.1007/BF00623322. [DOI] [PubMed] [Google Scholar]
- 125.Eigen M, Schuster P. The hypercycle. A principle of natural self-organization. Part A: Emergence of the hypercycle. Naturwissenschaften. 1977;64:541–565. doi: 10.1007/BF00450633. [DOI] [PubMed] [Google Scholar]
- 126.Haigh J. The accumulation of deleterious genes in a population-Muller’s Ratchet. Theor. Popul. Biol. 1978;14:251–267. doi: 10.1016/0040-5809(78)90027-8. [DOI] [PubMed] [Google Scholar]
- 127.Lynch M, Gabriel W. Mutation load and the survival of small populations. Evolution. 1990;44:1725–1737. doi: 10.1111/j.1558-5646.1990.tb05244.x. [DOI] [PubMed] [Google Scholar]
- 128.Lynch M, Bürger R, Butcher D, Gabriel W. The mutational meltdown in asexual populations. J. Hered. 1993;84:339–344. doi: 10.1093/oxfordjournals.jhered.a111354. [DOI] [PubMed] [Google Scholar]
- 129.Muller HJ. The relation of recombination to mutational advance. Mutat. Res. 1964;106:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- 130.Crotty S, Cameron C, Andino R. Ribavirin’s antiviral mechanism of action: lethal mutagenesis? J. Mol. Med. 2002;80:86–95. doi: 10.1007/s00109-001-0308-0. [DOI] [PubMed] [Google Scholar]
- 131.Solé RV, Deisboeck TS. An error catastrophe in cancer? J. Theor. Biol. 2004;228:47–54. doi: 10.1016/j.jtbi.2003.08.018. [DOI] [PubMed] [Google Scholar]
- 132.Fox EJ, Loeb LA. Lethal mutagenesis: targeting the mutator phenotype in cancer. Semin. Cancer Biol. 2010;20:353–359. doi: 10.1016/j.semcancer.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Frank SA, Nowak MA. Problems of somatic mutation and cancer. BioEssays. 2004;26:291–299. doi: 10.1002/bies.20000. [DOI] [PubMed] [Google Scholar]
- 134.Cairns J, Overbaugh J, Miller S. The origin of mutants. Nature. 1988;335:142–145. doi: 10.1038/335142a0. [DOI] [PubMed] [Google Scholar]
- 135.Sniegowski P, Lenski R. Mutation and adaptation: the directed mutation controversy in evolutionary perspective. Annu. Rev. Ecol. Syst. 1995;26:553–578. [Google Scholar]
- 136.van Delft FW, Horsley S, Colman S, Anderson K, Bateman C, Kempski H, Zuna J, Eckert C, Saha V, Kearney L, et al. Clonal origins of relapse in ETV6-RUNX1 acute lymphoblastic leukemia. Blood. 2011;117:6247–6254. doi: 10.1182/blood-2010-10-314674. [DOI] [PubMed] [Google Scholar]
- 137.Turke AB, Zejnullahu K, Wu Y-L, Song Y, Dias-Santagata D, Lifshits E, Toschi L, Rogers A, Mok T, Sequist L, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, Inserra E, Diederichs S, Iafrate AJ, Bell DW, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N. Engl. J. Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Roche-Lestienne C, Preudhomme C. Mutations in the ABL kinase domain pre-exist the onset of imatinib treatment. Semin. Hematol. 2003;40:80–82. doi: 10.1053/shem.2003.50046. [DOI] [PubMed] [Google Scholar]
- 140.Inukai M, Toyooka S, Ito S, Asano H, Ichihara S, Soh J, Suehisa H, Ouchida M, Aoe K, Aoe M, et al. Presence of epidermal growth factor receptor gene T790M mutation as a minor clone in non-small cell lung cancer. Cancer Res. 2006;66:7854–7858. doi: 10.1158/0008-5472.CAN-06-1951. [DOI] [PubMed] [Google Scholar]
- 141.Bonhoeffer S, May RM, Shaw GM. Virus dynamics and drug therapy. Proc. Natl. Acad. Sci. USA. 1997;94:6971–6976. doi: 10.1073/pnas.94.13.6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.zur Wiesch PA, Kouyos R, Engelstädter J, Regoes RR, Bonhoeffer S. Population biological principles of drug-resistance evolution in infectious diseases. Lancet Infect. Dis. 2011;11:236–247. doi: 10.1016/S1473-3099(10)70264-4. [DOI] [PubMed] [Google Scholar]
- 143.Björkholm B, Sjölund M, Falk PG, Berg OG, Engstrand L, Andersson DI. Mutation frequency and biological cost of antibiotic resistance in Helicobacter pylori. Proc. Natl. Acad. Sci. USA. 2001;98:14607–14612. doi: 10.1073/pnas.241517298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ribeiro RM, Bonhoeffer S. Production of resistant HIV mutants during antiretroviral therapy. Proc. Natl. Acad. Sci. USA. 2000;97:7681–7686. doi: 10.1073/pnas.97.14.7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Olofsson SK, Cars O. Optimizing drug exposure to minimize selection of antibiotic resistance. Clin. Infect. Dis. 2007;45(Suppl. 2):S129–136. doi: 10.1086/519256. [DOI] [PubMed] [Google Scholar]
- 146.Marchbanks CR, McKiel JR, Gilbert DH, Robillard NJ, Painter B, Zinner SH, Dudley MN. Dose ranging and fractionation of intravenous ciprofloxacin against Pseudomonas aeruginosa and Staphylo-coccus aureus in an in vitro model of infection. Antimicrob. Agents Chemother. 1993;37:1756–1763. doi: 10.1128/aac.37.9.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Read AF, Day T, Huijben S. The evolution of drug resistance and the curious orthodoxy of aggressive chemotherapy. Proc. Natl. Acad. Sci. USA. 2011;108(Suppl 2):10871–10877. doi: 10.1073/pnas.1100299108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Foo J, Michor F. Evolution of resistance to targeted anti-cancer therapies during continuous and pulsed administration strategies. PLoS Comput. Biol. 2009;5:e1000557. doi: 10.1371/journal.pcbi.1000557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Komarova NL, Wodarz D. Drug resistance in cancer: principles of emergence and prevention. Proc. Natl. Acad. Sci. USA. 2005;102:9714–9719. doi: 10.1073/pnas.0501870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J. Clin. Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 152.Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, Bell GW, Richardson AL, Polyak K, Tubo R, Weinberg RA. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- 153.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 154.Maley C, Galipeau P, Finley J, Wongsurawat V, Li X, Sanchez C, Paulson T, Blount P, Risques R-A, Rabinovitch P, et al. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat. Genet. 2006;38:468–473. doi: 10.1038/ng1768. [DOI] [PubMed] [Google Scholar]
- 155.Campbell PJ, Pleasance ED, Stephens PJ, Dicks E, Rance R, Good-head I, Follows GA, Green AR, Futreal PA, Stratton MR. Subclonal phylogenetic structures in cancer revealed by ultra-deep sequencing. Proc. Natl. Acad. Sci. USA. 2008;105:13081–13086. doi: 10.1073/pnas.0801523105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Park SY, Gönen M, Kim HJ, Michor F, Polyak K. Cellular and genetic diversity in the progression of in situ human breast carcinomas to an invasive phenotype. J. Clin. Invest. 2010;120:636–644. doi: 10.1172/JCI40724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Notta F, Mullighan CG, Wang JCY, Poeppl A, Doulatov S, Phillips LA, Ma J, Minden MD, Downing JR, Dick JE. Evolution of human BCR-ABL1 lymphoblastic leukaemia-initiating cells. Nature. 2011;469:362–367. doi: 10.1038/nature09733. [DOI] [PubMed] [Google Scholar]
- 158.Losi L, Baisse B, Bouzourene H, Benhattar J. Evolution of intratumoral genetic heterogeneity during colorectal cancer progression. Carcinogenesis. 2005;26:916–922. doi: 10.1093/carcin/bgi044. [DOI] [PubMed] [Google Scholar]
- 159.Kerr B, Riley MA, Feldman MW, Bohannan BJM. Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature. 2002;418:171–174. doi: 10.1038/nature00823. [DOI] [PubMed] [Google Scholar]
- 160.Rainey PB, Travisano M. Adaptive radiation in a heterogeneous environment. Nature. 1998;394:69–72. doi: 10.1038/27900. [DOI] [PubMed] [Google Scholar]
- 161.Turner P, Souza V, Lenski R. Tests of ecological mechanisms promoting the stable coexistence of two bacterial genotypes. Ecology. 1996;77:2119–2129. [Google Scholar]
- 162.Treves D, Manning S, Adams J. Repeated evolution of an acetate-crossfeeding polymorphism in long-term populations of Escherichia coli. Mol. Biol. Evol. 1998;15:789–797. doi: 10.1093/oxfordjournals.molbev.a025984. [DOI] [PubMed] [Google Scholar]
- 163.Rozen D, Lenski R. Long-term experimental evolution in Escherichia coli. VIII. Dynamics of a balanced polymorphism. Am. Nat. 2000;155:24–35. doi: 10.1086/303299. [DOI] [PubMed] [Google Scholar]
- 164.Rosenzweig RF, Sharp RR, Treves DS, Adams J. Microbial evolution in a simple unstructured environment: genetic differentiation in Escherichia coli. Genetics. 1994;137:903–917. doi: 10.1093/genetics/137.4.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.West SA, Diggle SP, Buckling A, Gardner A, Griffins AS. The social lives of microbes. Annu. Rev. Ecol. Evol. Syst. 2007;38:53–77. [Google Scholar]
- 166.Boles BR, Thoendel M, Singh PK. Self-generated diversity produces “insurance effects” in biofilm communities. Proc. Natl. Acad. Sci. USA. 2004;101:16630–16635. doi: 10.1073/pnas.0407460101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 168.Pepper J, Sprouffske K, Maley C. Animal cell differentiation patterns suppress somatic evolution. PLoS Comput. Biol. 2007;3:e250. doi: 10.1371/journal.pcbi.0030250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Weinreich D, Watson R, Chao L. Perspective: sign epistasis and genetic constraint on evolutionary trajectories. Evolution. 2005;59:1165–1174. [PubMed] [Google Scholar]
- 170.Dykhuizen DE, Dean AM. Predicted fitness changes along an environmental gradient. Evol. Ecol. 1994;8:524–541. [Google Scholar]
- 171.Crow JF, Kimura M. Evolution in sexual and asexual populations. Am. Nat. 1965;99:439–450. [Google Scholar]