Abstract
Aldehyde dehydrogenase (ALDH) enzymes are critical in the detoxification of aldehydes. The human genome contains 19 ALDH genes, mutations in which are the basis of several diseases. The expression, subcellular localization, enzyme kinetics and role of ALDH3B1 against aldehyde- and oxidant-induced cytotoxicity were investigated. ALDH3B1 was purified from Sf9 cells using chromatographic methods and enzyme kinetics were determined spectrophotometrically. ALDH3B1 demonstrated high affinity for hexanal (Km 62 μM), octanal (Km 8 μM), 4-hydroxy-2-nonenal (4HNE) (Km 52 μM) and benzaldehyde (Km 46 μM). Low affinity was seen towards acetaldehyde (Km 23.3 mM), malondialdehyde (Km 152 mM) and the ester p-nitrophenylacetate (Km 3.6 mM). ALDH3B1 mRNA was abundant in testis, lung, kidney and ovary. ALDH3B1 protein was highly expressed in these tissues and the liver. Immunofluorescence microscopy of ALDH3B1-transfected human embryonic kidney (HEK293) cells and subcellular fractionation of mouse kidney and liver revealed a cytosolic protein localization. ALDH3B1-transfected HEK293 cells were significantly protected from the lipid peroxidation-derived aldehydes trans-2-octenal, 4HNE and hexanal, and the oxidants H2O2 and menadione. In addition, ALDH3B1 protein expression was up-regulated by 4HNE in ARPE-19 cells. The results detailed in this study support a pathophysiological role for ALDH3B1 in protecting cells from the damaging effects of oxidative stress.
Keywords: aldehyde dehydrogenase 3B1 (ALDH3B1), 4-hydroxy-2-nonenal, lipid peroxidation, mRNA, protein expression, enzyme kinetics, aldehyde toxicity
Introduction
Aldehydes are highly reactive compounds oxidatively-generated from numerous precursors including lipids, alcohols, neurotransmitters and xenobiotics [1, 2]. Aldehydes and their associated toxicity have been implicated in the etiology and progression of human pathologies such as neurodegenerative diseases, alcoholic liver disease, cancer and male infertility [3-6]. Oxidative stress and resulting lipid peroxidation (LPO) can lead to the generation of more than 200 reactive aldehydes, including the highly toxic 4HNE [7]. While normal cellular levels of LPO-derived aldehydes, such as 4HNE, are in the micromolar range, millimolar concentrations can be reached in pathological states [7].
The oxidation of aldehydes to carboxylic acids, catalyzed by the aldehyde dehydrogenase (ALDH) enzyme superfamily, represents a significant metabolic route of aldehyde detoxification [8]. The clinical importance of ALDHs is underscored by the fact that mutations in several ALDH genes are the molecular basis of diseases including Sjögren-Larsson syndrome, type II hyperprolinemia, γ-hydroxybutyric aciduria and pyridoxine-dependent epilepsy [9, 10]. In addition, ALDH enzymes contribute to other pathological conditions such as cancer, in which ALDH expression is a factor in drug resistance, oxidative stress response and patient prognosis and outcome [11-14]. In addition to aldehyde metabolism, several ALDHs possess esterase and nitrate reductase activity [15, 16] and some are hormone- and/or xenobiotic-binding proteins [9, 17].
The ALDH3B1 gene encodes a protein of 468 amino acids (52 kDa), which belongs to the ALDH3 family of proteins [18-20]. Members of the ALDH3 family (ALDH3A1, ALDH3A2, ALDH3B1 and ALDH3B2) appear to have unique roles in the cellular defense against oxidative stress and aldehyde toxicity. ALDH3A1, one of the most abundant proteins in the cornea, efficiently metabolizes LPO-derived aldehydes and protects the cornea against ultraviolet radiation- and 4HNE-induced oxidative damage [21, 22]. ALDH3A2 plays a critical role in the oxidation of long-chain fatty aldehydes and mutations in ALDH3A2 result in Sjögren-Larsson syndrome, an inherited neurocutaneous disorder [23, 24]. The physiological significance of the ALDH3B proteins, ALDH3B1 and ALDH3B2, are only now being elucidated. Using crude cellular lysates, we have previously shown that ALDH3B1 has enzymatic activity directed towards various aldehyde substrates including 4HNE [25], one of the most reactive and cytotoxic aldehydes formed during LPO [26]. As such, we postulated that ALDH3B1 may play an important physiological role against oxidative stress processes. The present study was designed to characterize the expression and subcellular localization of ALDH3B1 and determine the enzymatic properties of the purified enzyme. In addition, the role of ALDH3B1 in the cellular defense against aldehyde-and oxidant-induced cytotoxicity was investigated. This study represents the most complete characterization of ALDH3B1 to date and the results described herein provide direct evidence that ALDH3B1 has an important role in the defense against oxidative stress.
Materials and Methods
Baculovirus expression
The previously cloned human ALDH3B1 cDNA [25] was used to obtain the coding region of ALDH3B1, which was subcloned into the pBluebac 4.5 baculovirus expression vector (Invitrogen, Carlsbad, CA, USA). The insert did not contain any native 5′ or 3′-untranslated sequence, but the leader was modified to contain an added ‘Kozak’ sequence motif (GCCACC) at the 5′-end (just in front of the ATG start methionine codon) for correct initiation of translation in eukaryotic cells and to increase protein expression, as previously described [27]. Sequence analyses verified the correct construction. Viruses were plaque-purified and amplified in Sf9 insect cells (Spodoptera frugiperda), as previously described [28]. Plaques were tested for ALDH3B1 protein expression by Western blot analyses. More than 50% of the viral plaques produced a single protein band at 52 kDa that immuno-reacted with the anti-human ALDH3B1 antibody. No reactive bands were detected in cell extracts derived from uninfected Sf9 cells. Sf9 cells were infected with baculoviruses encoding the human ALDH3B1 protein at a multiplicity of 1 for 48 h. Infected cells (500 ml culture) were harvested by centrifugation at 1000 g for 5 min and washed with PBS. Recombinant human ALDH3B1 was purified from cell pellets.
Purification of ALDH3B1
ALDH3B1 was purified from Sf9 cell pellets using a combination of ammonium sulfate precipitation and chromatography techniques. All procedures were conducted at 4°C, unless otherwise noted. ALDH3B1-expressing Sf9 cell pellets from two 500 ml cell cultures were suspended in 12 ml homogenization buffer (100 mM potassium phosphate, pH 7, 1 mM EDTA, 1 mM β-mercaptoethanol, 0.02% Triton X-100, 0.2% sodium deoxycholate, 30 mM KCl) containing 0.5 ml Complete Protease Inhibitor Cocktail (Roche Applied Science, Indianapolis, IN, USA). Cells were sonicated and subsequently centrifuged at 100,000 g for 1 h, as described previously [29]. Cell supernatant was collected, diluted 10:1 with 0.5 M Tris-HCl (pH 8.5) and titrated with saturated ammonium sulfate drop-by-drop to 35%. The protein solution was stirred on ice for 30 min to ensure complete precipitation of contaminating proteins, after which it was subjected to centrifugation for 20 min at 10,000 g. The supernatant was collected and used to screen 7 different hydrophobic interaction columns under a variety of purification conditions to determine maximum specific binding and elution of ALDH3B1. The least hydrophobic media, Butyl-S Fast Flow (GE Healthcare, Piscataway, NJ, USA), was found to have the highest selectivity for ALDH3B1. Ammonium sulfate precipitation supernatant was diluted 1:1 with hydrophobic interaction chromatography binding buffer (50 mM sodium phosphate, pH 7, 0.8 M ammonium sulfate) and applied drop-by-drop to two 4.5 ml aliquots of Butyl-S Fast Flow chromatography media packed into two Econo Columns (Bio-Rad, Hercules, CA, USA). Columns were washed with 10 ml of binding buffer and ALDH3B1 protein was eluted with 20 ml elution buffer (50 mM sodium phosphate, pH 7, 1 mM EDTA, 1 mM β-mercaptoethanol, 0.01% Triton-X 100). Butyl-S eluate was concentrated to 1 ml using Amicon Ultra-15 centrifugal filter tubes (Millipore, Billerica, MA, USA) and subjected to centrifugation at 3000 rpm for 24 min. The resulting concentrated protein solution was used to screen 7 different ion exchange columns under a variety of purification conditions. The weak anion exchanger, HiTrap DEAE FF (GE Healthcare), was determined to be optimal in its selectivity of ALDH3B1. Concentrated protein solution was diluted 1:4 in ion exchange chromatography binding buffer (20 mM Tris-HCl, pH 8.5, 1 mM EDTA, 1 mM β-mercaptoethanol, 50 mM NaCl), filtered through a 0.22 μm filter and injected onto a HiTrap DEAE FF ion exchange chromatography column connected to an Akta FPLC system (GE Healthcare). ALDH3B1 protein solution was eluted using a linear gradient of DEAE elution buffer (20 mM Tris-HCl, pH 8.5, 1 mM EDTA, 1 mM β-mercaptoethanol, 1 M NaCl). Eluate fractions were checked for ALDH3B1 enzymatic activity with NAD+ and octanal; highest activity fractions were pooled and concentrated. Concentrated DEAE eluate (500 μl) was filtered, applied to a Superdex 200 10/300 GL (GE Healthcare) gel filtration chromatography column and purified ALDH3B1 protein was eluted in gel filtration buffer (50 mM sodium phosphate pH 7, 1 mM EDTA, 1 mM β-mercaptoethanol and 0.01% Triton X-100). Purified ALDH3B1 was concentrated and desalted using Protein Desalting Spin Columns (Pierce, Rockford, IL, USA). Protein concentrations were estimated by BCA kit (Pierce) according to the manufacturer's instructions.
Enzyme kinetic assays
The enzymatic activity of purified recombinant ALDH3B1 was measured spectrophotometrically (using a Beckman DU-640 instrument) by monitoring the production of NADH (340 nm) during the enzymatic oxidation of aldehyde substrates, as described previously [29]. Assays were conducted at 25°C in a 1 ml reaction volume of 75 mM sodium pyrophosphate (pH 8), 1 mM NAD+, 1 mM pyrazole and 1-5 μg of ALDH3B1 recombinant protein. The enzymatic reaction was initiated by the addition of 100 μl of various concentrations of aldehyde substrates (unless otherwise specified, purchased from Sigma, St. Louis, MO, USA). Stock concentrations of aldehydes were prepared in 20% methanol, with the final concentration in the reaction mixture being less than 1%. 4HNE was purchased from Cayman Chemical Company (Ann Arbor, MI, USA). Malondialdehyde (MDA) was synthesized as described previously [7]. The ester p-nitrophenylacetate (NPA) was dissolved in acetone and ester hydrolysis was determined by measuring p-nitrophenol formation at 400 nm in 0.1M sodium phosphate (pH 7.4), as described previously [30]. To determine the apparent Km and Vmax values, ALDH3B1 enzymatic activities were measured using various substrate concentrations ranging from one-tenth of the apparent Km to 10 times the Km for each substrate, as substrate solubility allowed. All enzyme assays were performed in triplicate and rates were fitted to the Michaelis-Menten equation using SigmaPlot software for enzyme kinetics (Sigma Plot Version 9.0, 2004). Enzyme specific activities are expressed as nmol (or μmol) of NADH/min per mg protein).
Protein sequence analyses
The human ALDH3B1 protein sequence was analyzed for predicted subcellular localization and targeting peptides using iPSORT [31], WOLF PSORT [32], Target P [33], MitoProII [34], Predotar [35] and SOSUI programs [36]. Potential phosphorylation sites in the human ALDH3B1 sequence were identified using NetPhos and NetPhosK programs [37].
Animals
C57BL/6J mice were euthanized by CO2 inhalation followed by cervical dislocation. Animals were maintained in full compliance with all regulations stipulated by the Institutional Animal Care and Use Committee of the University of Colorado Denver and all published National Institutes of Health guidelines.
Western blot analyses
Mouse tissues were processed and subjected to Western blot analyses using anti-mouse ALDH3B1 (1:500) antibody [25]. For the detection of human ALDH3B1 protein, anti-human ALDH3B1 (1:200) antibody was used [25]. Antibody binding was detected using peroxidase-conjugated goat anti-rabbit IgG (1:5000) (Calbiochem, San Diego, CA, USA) and proteins were visualized using chemiluminescence (NEN Life Science Products, Boston, MA, USA) and hyperfilm (GE Healthcare). Equivalent loading of protein samples was confirmed by re-probing membranes with mouse monoclonal anti-β-actin IgG (1:10,000; Sigma) followed by peroxidase-conjugated rabbit anti-mouse IgG (1:5000; Sigma).
mRNA expression of ALDH3B1 in mouse tissues
Mouse tissues were harvested from male and female C57BL/6J mice (60-90 d old) and stored in RNA Later solution (Applied Biosystems, Foster City, CA, USA) at −80°C. Total RNA was isolated according to tissue type using Qiagen RNeasy Mini Kits according to the manufacturer's protocols (Qiagen, Valencia, CA, USA). RNA was treated with Turbo DNA-free DNase (Ambion, Austin, TX, USA). cDNA was synthesized using High Capacity cDNA RT-PCR Kit (Ambion). Real-time PCR of the cDNA (50 ng) was performed using ALDH3B1 (assay ID Mm00550698_m1) and β-actin (endogenous control) Taqman Gene Expression Assays and Universal PCR Master Mix (Applied Biosystems). Relative quantification of mRNA expression was determined using a Real-Time PCR 7500 SDS System and Software (Applied Biosystems). RNA purity and integrity were verified by measuring the ratio of absorbance at 260 and 280 nm (A260/A280) and by denaturing agarose gel electrophoresis, respectively.
Generation of HEK293 cell lines stably transfected with ALDH3B1
HEK293 cells were grown in Dulbecco's Modified Eagle Medium with high glucose (Invitrogen, Carlsbad, CA, USA) and 7% FBS (Gemini, West Sacramento, CA, USA) at 37°C in a 5% CO2 incubator. For transfection, the human ALDH3B1 cDNA (coding region only, with added Kozak sequence) was digested from the pBlueBac4.5/hALDH3B1-Kozak plasmid and ligated into the pcDNA3.1(-) plasmid. HEK293 cells were transfected with either pcDNA3.1(-) (vector only) or pcDNA3.1(-)/hALDH3B1-Kozak using a standard calcium phosphate precipitation method, as described previously [38]. Stable cell populations were selected using geneticin (Invitrogen). Following limiting dilution, cells were allowed to form colonies, which were further expanded, analyzed for ALDH3B1 expression by Western blot analyses and cloned a second time to ensure 100% cellular expression of ALDH3B1. ALDH3B1/HEK293 clones 5 and 32 were used for subsequent experiments in this study.
Immunofluorescence microscopy and image analysis of HEK293 cell lines
Vector only- and ALDH3B1-transfected HEK293 cells were grown on glass coverslips in culture dishes for at least 3 d prior to experiments, with media being changed every 2 d. Cells were plated sparsely and harvested before confluence to ensure adequate space between cells in order to accurately identify potential antigens present at the cell edge. At the time of harvest, media was removed and replaced immediately with 3.7% formaldehyde for 10 min to fix the cells, and then with 50% ethanol for 4 min to permeabilize the cells. A final wash with PBS was performed for 30 min. Blocking solution (2% BSA in PBS) was applied for 20 min followed by exposure to anti-human ALDH3B1 antibody [1:75] in blocking buffer for 25 min. Coverslips were rinsed with PBS, re-blocked for 5 min and then incubated with Alex Fluor® 488 labeled goat anti-rabbit IgG (Invitrogen) for 25 min. The coverslips were rinsed again with PBS, incubated with DAPI (0.1 μg/ml) for 20 min, rinsed with PBS and mounted on glass slides using Aquamount (ScyTek, Logan, UT, USA). Immunofluorescent images were captured at room temperature on a Nikon Diaphot fluorescence microscope equipped with a Cooke SensiCam CCD camera (Tonawand, NY, USA) using Slidebook software (Intelligent Imaging Innovations Inc., Denver, CO, USA). All images were digitally deconvolved using the No Neighbors algorithm (Slidebook), converted to TIFF files and processed using Photoshop (Adobe Systems Inc., Mountain View, CA, USA).
Subcellular fractionation of mouse liver and kidney
Fresh liver and kidney were harvested from C57BL/6J mice and processed to obtain subcellular fractions, as previously described [39]. Fifteen μg total protein from each fraction was separated by SDS-PAGE and ALDH3B1 was identified by Western blotting. Purity was assessed by probing the blots with glutamate cysteine ligase modifier subunit (GCLM), Na+/K+ ATPase β1 (sc-16053; Santa Cruz Biotechnology), voltage-dependent anion channel 1 (VDAC1) (ab15895; Abcam), superoxide dismutase 2 (SOD2) (06-984; Millipore) and histone H1 (sc-10806; Santa Cruz Biotechnology) antibodies for cytosolic, membranes, mitochondrial membranes, mitochondrial soluble and nuclear fractions, respectively.
Cytotoxicity assays
Vector- and ALDH3B1-transfected HEK293 cells were seeded into 24-well plates at 2 × 105 cells per well in 1 ml complete growth media overnight in a tissue culture incubator (37°C, 5% CO2). The following day (≈ 80% confluence), the media was carefully removed and replaced with various concentrations of aldehydes (4HNE, benzaldehyde, T2O, hexanal) or oxidants (H2O2, menadione, paraquat) in 1 ml phenol red-free DMEM containing 1% fetal bovine serum (Gemini). Treated cells were incubated for 16 h followed by subsequent analysis with MTT (3-(4, 5-dimethylthiazolyl-2)-2, 5-diphenyltetrazolium bromide) or sulforhodamine B (SRB) assays, as previously described [21, 40]. Cell images were taken using a Nikon Eclipse TS100 inverted microscope equipped with a Nikon DS-Fi1 digital camera (Nikon, Melville, NY, USA). For MTT assays, 200 ml MTT solution (5 mg/ml in PBS) was added per well to the existing media and allowed to incubate for 3 h. The media was then carefully removed and replaced with 1.5 ml MTT solvent (4 mM HCl, 0.1% (vol/vol) NP40 in isopropanol). Plates were then placed in the dark and incubated for 2 h at room temperature to dissolve precipitant. Cell viability was determined from the cellular reduction of MTT to formazan by measuring the absorbance at 570 nm using a Spectramax Plus384 plate reader (Molecular Devices, Sunnyvale, CA, USA). For the SRB assay, vector- and ALDH3B1-transfected HEK293 cells were seeded and treated with aldehydes as described for the MTT assay. Following aldehyde treatment, 1 ml cold 10% trichloroacetic acid was added to the existing media and the cells were incubated for an additional 1 h at 4°C. The plates were gently washed four times with water and allowed to dry overnight at room temperature. After drying, 0.5 ml 0.057% (wt/vol) SRB (Sigma) in 1% (vol/vol) acetic acid was added to each well and incubated at room temperature for 30 min. Plates were subsequently washed four times with 1% (vol/vol) acetic acid to remove any unbound stain and left overnight at room temperature to dry completely. One ml 10 mM Tris Base (pH 10.5) was added to each well and the plates were shaken at room temperature for 15 min to dissolve the bound dye. Cell survival was measured by taking fluorescence measurements at an excitation and emission wavelength of 488 nm and 570 nm, respectively, using a Spectramax Gemini EM plate reader (Molecular Devices). Using the SRB and MTT assay data, linear interpolation of log concentration survival and viability curves were constructed and acute EC50 values were calculated using regression analysis and the equation of four-parameter logistic curve (SigmaPlot software, version 9.0, 2004), as previously described [21].
Cell lines and culture conditions
Human embryonic kidney (HEK293) cells, human alveolar basal epithelial (A549) cells and human hepatocellular carcinoma (HepG2) cells were grown in Dulbecco's Modified Eagle Medium (DMEM) with high glucose supplemented with 10% heat inactivated fetal bovine serum (Gemini, West Sacramento, CA, USA) and 1% of a stock solution containing 10,000 IU/mL penicillin and 10 mg/mL streptomycin (Sigma-Aldrich, St. Louis, MO, USA). Human renal proximal tubular epithelial (HK-2) cells and human retinal pigment epithelial (ARPE-19) cells were grown in DMEM/F12 supplemented with 5% heat inactivated FBS and 1% of a stock solution containing 10,000 IU/mL penicillin and 10 mg/mL streptomycin. All cell lines were cultured at 37°C in a 5% CO2 incubator.
ALDH3B1 gene regulation in vitro
Gene regulation of ALDH3B1 by various compounds was studied in vitro using cultured cell lines. First, to determine putative transcription factor binding sites that may be involved in gene regulation, the 5Kb upstream (of the translation initiation site) promoter sequence for both human and mouse ALDH3B1 were screened with PROMO version 3.02 [41, 42] utilizing version 8.3 of the TRANSFAC transcription factor binding site database [43]. Putative conserved binding sites that were identified in homologous regions of human and mouse ALDH3B1 sequences included Nrf2, ER-alpha & -beta, PPAR-alpha and NF-kB. The Nrf2 pathway is activated during oxidative or electrophilic stress. Upon activation, Nrf2 transactivates antioxidant response elements (ARE) in promoter regions of target genes. A number of compounds, including hydrogen peroxide (H2O2) [44] and 4HNE [45], are known to activate the Nrf2 pathway. The presence of multiple ER2-alpha and -beta binding sites indicated possible ALDH3BB1 gene regulation by estrogen. Clofibrate, a potent PPAR-alpha activator, was used to address possible regulation by PPAR transcription factors. Lipopolysaccharide (LPS) activates pro-inflammatory cytokines resulting in the up-regulation of a number of downstream genes. The NF-kB pathway has been shown to be a target of both LPS [46] and H2O2 [47]. To determine if the ALDH3B1 gene is regulated by these compounds, A549, ARPE-19, HepG2 and HK-2 cells were grown in 100mm culture dishes in complete media until 50% confluent. The media was then replaced with antibiotic-free media containing reduced 1% heat inactivated FBS and supplemented with 100 μM clofibrate, 100 nM estradiol (E2), 50 μM H2O2, 10 μM 4HNE or 2 μg/mL LPS. All media was removed and replaced with fresh treatment after 18 h and allowed to incubate for an additional 18 h, for a total treatment period of 36 h. The cells were then harvested and processed in RIPA buffer. Fifteen μg lysate was separated by SDS-PAGE and ALDH3B1 expression identified by Western blotting. All blots were re-probed with β-actin antibody to confirm equal sample loading. As an added control, samples were also probed for the expression of an additional ALDH enzyme (ALDH7A1), which is not believed to be regulated by the above-mentioned compounds.
Statistical analysis
All values are expressed as mean ± SEM. Groups were compared by Student's unpaired t-test (SigmaPlot, version 9.0, 2004). P<0.05 was considered to be significant.
Results
Purification of recombinant human ALDH3B1
Purification of recombinant human ALDH3B1 was achieved from ALDH3B1-infected Sf9 cells in a multi-step process involving ammonium sulfate precipitation followed by hydrophobic interaction, ion-exchange and gel filtration chromatography (Table 1). A purification of approximately 30-fold, as evidenced by the 30-fold increase in ALDH3B1 specific activity was achieved. This method resulted in a major protein band at 52 kDa by Coomassie staining (Figure 1A, lane 6), which was also immunostained as the purified ALDH3B1 protein by Western blot (Figure 1B, lane 2). High molecular weight protein bands were also visible in the final purified samples of ALDH3B1 processed for SDS PAGE; these may represent un-denatured ALDH3B1 protein. We have previously shown that non-infected Sf9 cells have negligible enzymatic activity towards aldehyde substrates [25]; thus, if contaminating Sf9 cell proteins are present in purified ALDH3B1 samples, they are not believed to contribute to aldehyde oxidation as measured in this study.
Table 1. Purification yield of recombinant human ALDH3B1 expressed in Sf9 cells.
| Purification step | Total protein (mg) | Specific activity a (μmol NADH/min per mg) | Yield (%) | Purification b (-fold) |
|---|---|---|---|---|
| cell lysate | 370 | 0.11 | 100 | - |
| 100,000 g supernatant | 195 | 0.25 | 53 | 2 |
| 35% ammonium sulfate | 50 | 0.55 | 14 | 5 |
| Butyl-S FF eluate | 28 | 1.74 | 8 | 16 |
| DEAE FF eluate | 2 | 2.17 | 0.62 | 20 |
| Superdex 200 eluate | 0.28 | 3.16 | 0.08 | 30 |
Specific activity was determined using octanal as an aldehyde substrate
Purification is expressed as -fold of specific activity in cell lysate
Figure 1.

Purification of human ALDH3B1 from baculovirus-infected Sf9 cells. Purified recombinant human ALDH3B1 as analyzed by Coomassie blue staining (A) and immunoblot analysis (B). Lane 1 (A), cell lysate; lane 2 (A), 35% ammonium sulfate precipitation; lane 3 (A), Butyl-S FF eluate; lane 4 (A), DEAE FF eluate; lane 5 (A), Superdex 200 pooled eluate; lane 6 (A), Superdex 200 fractionated eluate. Lane 1 (B), cell lysate; lane 2 (B), purified ALDH3B1.
Kinetic properties of human ALDH3B1
The purified ALDH3B1 protein was subsequently used for kinetic studies. Apparent Km and Vmax values of recombinant human ALDH3B1 were determined for various aldehyde substrates and the ester p-nitrophenylacetate (Table 2). The recombinant enzyme was capable of oxidizing medium-chain saturated and unsaturated aldehydes including hexanal (Km = 62 μM), 4HNE (Km = 52 μM) and octanal (Km = 8 μM), as well as the aromatic aldehyde benzaldehyde (Km = 46 μM). Both the highest affinity and efficiency (Vmax/Km = 155) was directed towards octanal, a C8 aliphatic saturated aldehyde. Short-chain aldehydes (acetaldehyde and malondialdehyde) appeared to be poor substrates, with Vmax/Km being less than 0.1. ALDH3B1 was active towards the ester p-nitrophenylacetate with an affinity constant (Km) of 3.6 mM. The addition of NAD+ to the reaction mixture inhibited esterase activity by 21% (data not shown). The Kms for NAD+ and NADP+ were determined to be 316 μM and 1041 μM, respectively. Both affinity and catalytic efficiency were higher for NAD+ as opposed to NADP+, which has also been observed with other ALDH3 family members [48]. Similar to that reported for ALDH3A1 [49], no inhibition of ALDH3B1 by disulfiram was demonstrated in this study. However, 2 mM cyanamide produced a mild inhibitory effect (17% decrease) on ALDH3B1 enzymatic activity (Table 2).
Table 2. Kinetic and inhibitor properties of recombinant human ALDH3B1.
| Substrate | Vmax (nmol NADH/min/mg)a | Km (μM) | Vmax/Kmb |
|---|---|---|---|
| Acetaldehyde | 315 ± 8 | 23300 ± 1500 | 0.01 |
| Malondialdehyde | 3399 ± 377 | 152000 ± 17110 | 0.02 |
| Hexanal | 679 ± 27 | 62 ± 8 | 11.0 |
| Octanal | 1239 ± 59 | 8 ± 2 | 155.0 |
| 4HNE | 653 ± 18 | 52 ± 5 | 12.6 |
| Benzaldehyde | 129 ± 6 | 46 ± 11 | 2.8 |
| p-Nitrophenylacetate Cofactorc | 1319 ± 80 | 3600 ± 250 | 0.4 |
| NAD+ | 2987 ± 218 | 316 ± 54 | 9.5 |
| NADP+ | 5121 ± 740 | 1041 ± 311 | 4.9 |
|
| |||
| Inhibitorc | Concentration | % Uninhibited ALDH3B1 activity | P value |
|
| |||
| None | 100 ± 3 | ||
| Disulfiram | 100 μM | 100 ± 3 | no effect |
| 500 μM | 103 ± 1 | no effect | |
| Cyanamided | 400 μM | 106 ± 4 | no effect |
| 2000 μM | 83 ± 1 | *P<0.05 | |
Apparent Vmax and Km values were determined by fitting the data to the Michaelis-Menten equation using SigmaPlot.
Vmax/Km represents aldehyde (or esterase, in the case of p-nitrophenylacetate) oxidizing capacity (expressed as nmol NADH produced/min per mg protein per nmol substrate per ml).
Cofactor and inhibitor kinetics were determined using octanal as substrate
Cyanamide requires metabolic activation; thus, ALDH3B1-infected Sf9 cell lysates as opposed to purified protein were used to determine cyanamide-mediated enzyme inhibition.
Data represent means ± SEM from triplicate experiments.
ALDH3B1 protein and mRNA expression in mouse tissues
ALDH3B1 protein was found to be expressed in a number of mouse tissues including kidney, liver, stomach, lung, spleen, tongue, pancreas (Figure 2A), eye, ovary and testis (Figure 2B). ALDH3B1 mRNA expression was found in nearly all tissues examined (Table 3). Relative to kidney (100%), values for the quantities of mRNA ranged from 4% (stomach) to 1396% (testis). In addition to testis, tissues with relatively high expression included lung (254%), ovary (215%), adrenal gland (180%), femur bone (168%), skull bone (166%), peripheral leukocytes (105%) and trachea (105%). In addition to the stomach (4%), tissues with detectable but relatively low expression of ALDH3B1 mRNA included lacrimal gland (7%), eye lens (10%), large intestine (13%), submandibular gland (16%), seminal vesicle (17%), tongue (17%), thymus (17%), bladder (18%) and skin (19%).
Figure 2.
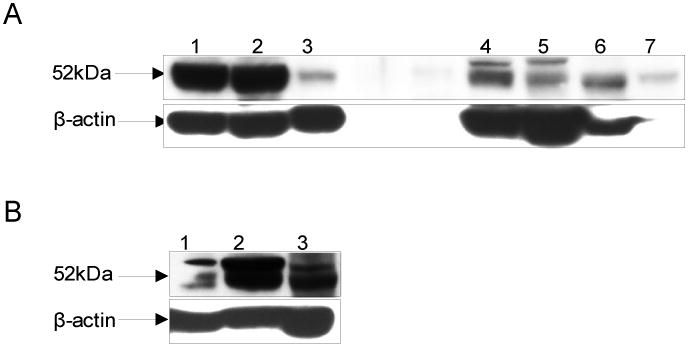
Expression of ALDH3B1 protein in mouse tissues. Immunoblot analysis of ALDH3B1 expression in C57BL/6J mice. A: kidney (lane 1), liver (lane 2), stomach (lane 3), lung (lane 4), spleen (lane 5), tongue (lane 6), pancreas (lane 7); B: eye (lane 1), ovary (lane 2), testis (lane 3).
Table 3. Relative ALDH3B1 mRNA expression in mouse tissues a.
| Male Reproductive | Brain | Digestive | Urinary |
|---|---|---|---|
| Testis (1396) | Spinal Cord (63) | Esophagus (92) | Kidney (100) |
| Vas Deferens (63) | Hypothalamus (57) | Pancreas (53) | Bladder (18) |
| Epididymus (25) | Thalamus (51) | Rectum (35) | |
| Seminal Vesicle (17) | Midbrain (49) | Small Intestine (34) | Immune |
| Frontal Cortex (47) | Parotid Gland (28) | Spleen (27) | |
| Female Reproductive | Whole Brain (46) | Liver (24) | Thymus (17) |
| Ovary (215) | Cortex (45) | Tongue (17) | |
| Uterine Horn (124) | Striatum (39) | Submandibular Gland (16) | Bone |
| Female Mammary (79) | Cerebellum (37) | Large Intestine (13) | Femur (168) |
| Fallopian Tubes (63) | Hippocampus (31) | Stomach (4) | Skull (166) |
| Skin and Adipose | Muscle | Respiratory | Eye |
| Skin (19) | Mandible Muscle (69) | Lung (254) | Total Eye (47) |
| Inguinal Fat Pad (46) | Abdominal Muscle (64) | Trachea (105) | Cornea (37) |
| Skeletal Muscle (54) | Lens (10) | ||
| Endocrine | Diaphragm (47) | Blood | Lacrimal Gland (7) |
| Adrenal Gland (180) | Heart (34) | Peripheral Leukocytes (105) | |
| Thyroid (40) |
Numbers in parentheses denote percent expression relative to kidney, which was empirically defined as being 100%. Tissues are in descending order of expression level within physiological systems.
Expression of ALDH3B1 in transfected HEK293 cells
To investigate the subcellular expression and protective role of ALDH3B1 against aldehyde toxicity, Stably-transfected ALDH3B1-expressing HEK293 cell clones were created and isolated using a pcDNA3.1(-) plasmid encoding the human ALDH3B1 cDNA. HEK293 cells stably transfected with the pcDNA3.1(-) vector alone were used as controls. Expression of the 52kDa ALDH3B1 was seen in approximately 70% of the clones (Figure 3A, lanes 2-7, 3B, lane 2). ALDH3B1 protein was undetectable in vector-only control cells (Figure 3A, lane 1, 3B, lane 1).
Figure 3.
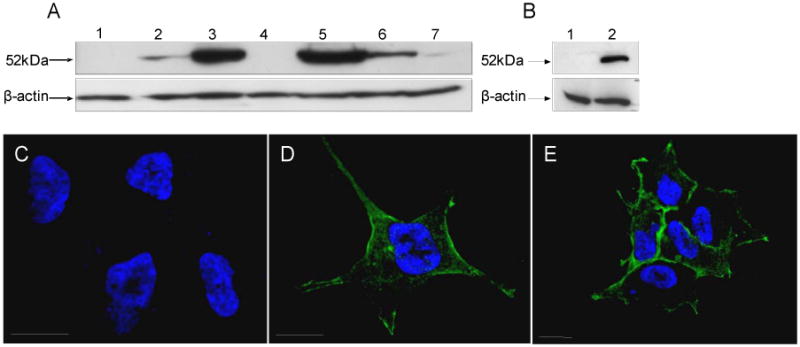
Human ALDH3B1 expression and subcellular localization in HEK293 cells. HEK293 clones stably transfected with human ALDH3B1 (A, lanes 2-7; B, lane 2) or transfected with vector alone (A, lane 1; B, lane 1). Clones 32 (A, lane 5) and 5 (B, lane 2) were used for immunofluorescence microscopy (C-E). Vector only-transfected HEK293 cells (C) and human ALDH3B1-transfected HEK293 clones 32 (D) and 5 (E), were stained using anti-human ALDH3B1 primary antibody [1:75] and secondary goat anti-rabbit antibody. Expression is indicated by green color. Cells were counterstained with DAPI to show nuclei. Bars = 20 microns.
Immunofluorescence microscopy of transfected HEK293 cells
Immunofluorescent confocal microscopy was used to determine the subcellular localization of ALDH3B1 in transfected HEK293 cell clones (Figures 3D, E), ALDH3B1 appears to be cytoplasmic but possibly membrane-associated or plasma membrane-bound. Both ALDH3B1-transfected HEK293 cell clones 32 (Figure 3D) and 5 (Figure 3E) were investigated and displayed similar localizations. No nuclear, mitochondrial or microsomal localization was demonstrated within either clonal cell line. Neither parental HEK293 cells, vector only-transfected HEK293 control cells nor non-expressing ALDH3B1-transfected clones (non-expressing as determined by Western blot analyses) showed specific immunohistochemical staining.
Analysis of ALDH3B1 protein sequence
Analysis of the amino acid sequence of human ALDH3B1 using protein subcellular localization prediction programs revealed no signal peptide or specific subcellular targeting sequence. However, the SOSUI program identified two potential transmembrane regions (amino acids 110-132 and 151-173) and the WOLF PSORT program gave the highest probability for ALDH3B1 being a plasma membrane protein (12.5%), as compared to other subcellular regions, indicating ALDH3B1 may theoretically be membrane-associated.
Subcellular fractionation of kidney and liver
To further determine the subcellular localization of the ALDH3B1 protein, subcellular compartments were isolated from mouse kidney and liver and analyzed for ALDH3B1 expression using an anti-mouse ALDH3B1 antibody. A cytosolic subcellular localization of mouse ALDH3B1 was demonstrated (Figures 4A, B). Fraction purity was confirmed by probing the isolated subcellular samples for the expression of enzymes with known localizations (GCLM - cytosol; Na+/K+ ATPase β1 -membranes; VDAC1 - mitochondrial membranes; SOD2 - mitochondrial soluble fraction; histone H1 -nuclear fraction (Figures 4A, B).
Figure 4.

Subcellular localization of mouse ALDH3B1. Individual organs were isolated from adult C57BL/6J mice. ALDH3B1 expression in subcellular fractions from kidney (A) and liver (B) was determined via Western blot analysis using anti-mouse ALDH3B1 antibody (as described in Materials and Methods). Fifteen μg whole cell or subcellular fraction lysate was loaded into each lane. Blots were also probed with GCLM (cytosol), Na+/K+ ATPase β1 (membranes), VDAC1 (VDAC, mitochondrial membranes), superoxide dismutase 2 (SOD2, mitochondria soluble), and histone H1 (nuclei) to assess fraction purity.
Protective role of ALDH3B1 against aldehyde cytotoxicity
The ability of ALDH3B1 to protect cells from the cytotoxic effects of various aldehydes was examined in vector-only (control) and ALDH3B1-transfected HEK293 cells (clone 5) by both MTT (cell viability) and SRB (cell survival) assays (Figure 5). The MTT assay analyzes cell viability by measuring the cellular reduction of MTT to formazan. The SRB assay is a sensitive measure of cytotoxicity that detects the binding of SRB to basic amino acids of cellular proteins and is an estimate of cell survival [50]. ALDH3B1-expressing cells were significantly less sensitive to the toxic effects of 4HNE (Figure 5A, B), hexanal (Figure 5C, D), benzaldehyde (Figure 5E, F) and T2O (Figure 5G), when compared to the stably-transfected vector-only cells. In the case of 4HNE, cell survival in vector only-transfected cells, as measured by the SRB assay, significantly decreased relative to ALDH3B1-expressing cells at concentrations exceeding 10 μM (Figure 5B). However, cell viability of vector only-transfected cells treated with 4HNE, as measured by the MTT assay, started to be significantly reduced relative to ALDH3B1-expressing cells at the lowest concentration of 4HNE tested, viz. 5 μM (Figure 5A). At 100 μM 4HNE, cell survival and viability of vector only-transfected cells was nominal (2% and 4%, respectively) while cell survival and viability of ALDH3B1-expressing cells remained fairly robust at 54% and 43%, respectively. EC50 values for 4HNE in vector only-transfected cells were 35.9 ± 1.7 μM and 18.0 ± 0.4 μM, as measured by SRB and MTT assays, respectively. In contrast, EC50 values for 4HNE in ALDH3B1-transfected cells were 113.0 ± 17.6 μM and 97.6 ± 3.4 μM, respectively. In cells treated with 4HNE, which is administered in an ethanol vehicle, the toxicity of the ethanol vehicle itself was examined relative to untreated cells. No toxicity from ethanol vehicle was demonstrated, either in vector-only or ALDH3B1-transfected cells (data not shown).
Figure 5.
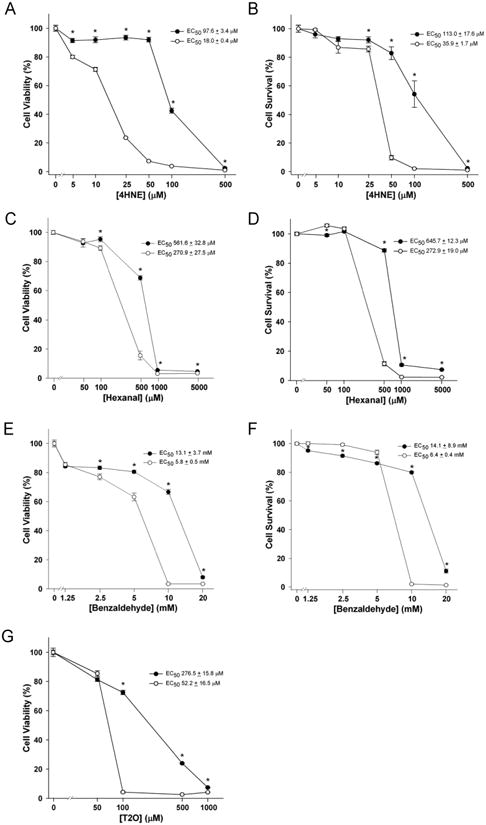
ALDH3B1 protection against aldehyde cytotoxicity. HEK293 cells stably-transfected with human ALDH3B1 (clone 5) (filled circles) or vector-alone (open circles) were treated with 4HNE (0-0.5mM; A, B), hexanal (0-5mM; C, D), benzaldehyde (0-20mM; E, F) or trans-2-octenal (T2O) (0-1mM; G) and analyzed for cell viability by the MTT assay (A, C, E, G) and cell survival by the SRB assay (B, D, F). Data represent means ± SEM from triplicates in at least three separate experiments. * Statistically significant differences between cytotoxic dose responses (P<0.05).
Protection from hexanal in ALDH3B1-expressing cells was seen beginning at 100 μM, as measured by the MTT assay, and at 500 μM, as measured by the SRB assay (Figures 5C, D). EC50 values for hexanal in vector only-transfected cells were 272.9 ± 19.0 μM and 270.9 ± 27.5 μM, as measured by SRB and MTT assays, respectively. In contrast, EC50 values for hexanal in ALDH3B1-transfected cells were 645.7 ± 12.3 μM and 561.6 ± 32.8 μM, as measured by SRB and MTT assays, respectively. The aromatic aldehyde benzaldehyde significantly reduced cell viability (MTT assay) and cell survival (SRB assay) of vector only-transfected cells beginning at 2.5 mM and 10 mM (respectively) relative to ALDH3B1-expressing cells (Figure 5E, F). EC50 values for benzaldehyde in vector only-transfected cells were 6.4 ± 0.4 mM and 5.8 ± 0.5 mM, as measured by SRB and MTT assays, respectively. In contrast, EC50 values for benzaldehyde in ALDH3B1-transfected cells were 14.1 ± 8.9 mM and 13.1 ± 3.7 mM, respectively. Cell viability (MTT assay) of vector only-transfected HEK293 cells treated with T2O was markedly reduced (to 4%) beginning at 100 μM T2O, as compared to ALDH3B1-transfected cells (to 72%) at the same concentration (Figure 5G). EC50 values for T2O were 52.2 ± 16.5 μM and 276.5 ± 15.8 μM in vector only-transfected and ALDH3B1-transfected cells, respectively.
Cell survival (as measured by SRB assay) was significantly decreased in ALDH3B1-expressing cells at benzaldehyde concentrations of 1.25, 2.5 and 5 mM and hexanal at 50 μM, when compared to vector-only transfected cells. Measures of cell viability using the MTT assay did not display this trend. It can be speculated that benzaldehyde and hexanal may have an initial mild proliferative effect that is mitigated in ALDH3B1-expressing cells. Nonetheless, EC50 values demonstrated that ALDH3B1-expressing cells were significantly less susceptible to aldehyde-induced cytotoxicity, compared to vector-only transfected cells (Figure 5, A-G, curve legends). Both SRB and MTT assays gave comparable EC50 values, confirming the cytotoxic validity of these assays. Cell images of vector-transfected and ALDH3B1-transfected HEK293 cells were consistent with the results of cytotoxicity assays (data not shown).
Protective role of ALDH3B1 against oxidant cytotoxicity
The ability of ALDH3B1 to also protect cells from the cytotoxic effects of the oxidants H2O2, menadione and paraquat was examined in vector-only (control) and ALDH3B1-transfected HEK293 cells (clone 5) by MTT assay (Figure 6). ALDH3B1-expressing cells were significantly less sensitive to the toxic effects of H2O2 (Figure 6A), menadione (Figure 6B) and paraquat (Figure 6C) at various concentrations, when compared to vector-only cells. In the case of H2O2, cell viability in vector only-transfected cells significantly decreased relative to ALDH3B1-expressing cells at all concentrations studied (0.1 - 2.5 mM) (Figure 6A). At 250 μM H2O2, cell viability of vector only-transfected cells decreased to 14% while cell viability of ALDH3B1-expressing cells remained at 39%. The EC50 value for H2O2 in vector only-transfected cells was estimated to be 0.02 ± 0.01 mM compared to 0.8 ± 0.1 mM in ALDH3B1-expressing cells. Protection from the oxidant menadione in ALDH3B1-expressing cells was also seen at all concentrations studied (5-30 μM; Figure 6B). The EC50 value for menadione in vector only-transfected cells was 10.8 ± 0.2 μM while in ALDH3B1-transfected cells it was 12.0 ± 0.4 μM. The redox-cycling agent paraquat significantly reduced cell viability of vector only-transfected cells only at 500 μM relative to ALDH3B1-expressing cells (Figure 6C). Lower concentrations of paraquat produced no change in cell viability between ALDH3B1-expressing and vector-only cells.
Figure 6.
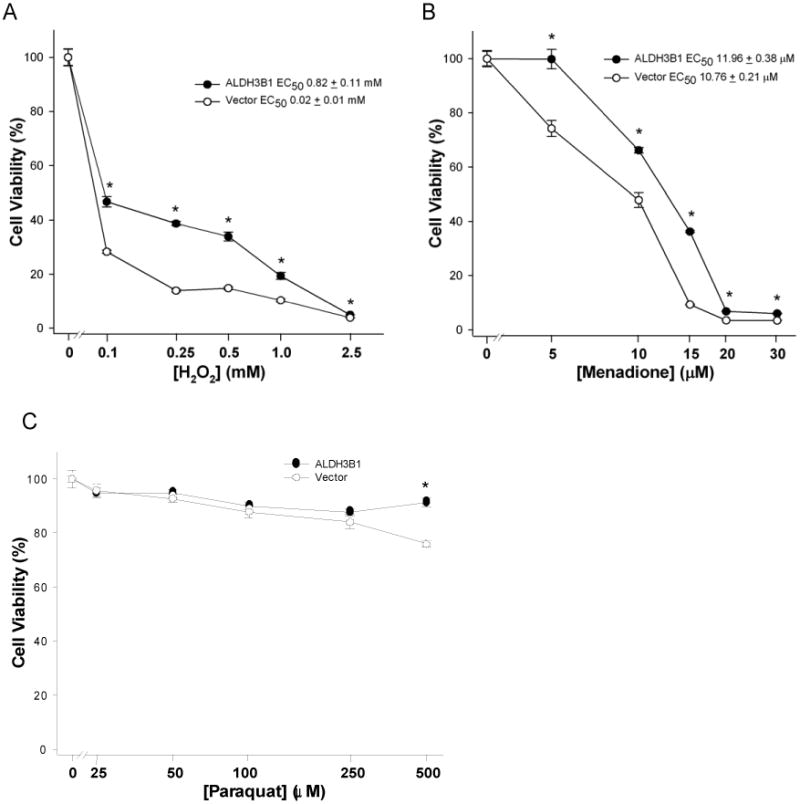
ALDH3B1 protection against oxidant cytotoxicity. HEK293 cells stably-transfected with human ALDH3B1 (clone 5) (filled circles) or vector-alone (open circles) were treated with H2O2 (0-2.5mM; A), menadione (0-30 μM; B), or paraquat (0-0.5mM; C) and analyzed for cell viability by MTT assay. Data represent means ± SEM from triplicates in at least three separate experiments. *Statistically significant differences between cytotoxic dose responses (P<0.05).
Induction of ALDH3B1 protein expression
To investigate potential compounds that may be involved in ALDH3B1 gene regulation, ALDH3B1 protein expression was determined in multiple cell lines following treatment with various compounds. Clofibrate is a PPAR alpha ligand that has been shown to decrease ALDH3A1 expression in hepatoma cell lines [51]. Estradiol (E2) has been shown to directly induce expression of various ALDH enzymes, particularly those involved in retinoic acid biosynthesis pathways [52]. Cells were also treated with the oxidative stress-inducing compounds H2O2, 4HNE and LPS. No change in ALDH3B1 expression was found in A549, HepG2 or HK-2 cell lines from any treatment when compared to untreated control cells. However, treatment with 10 μM 4HNE in ARPE-19 cells resulted in a significant increase in ALDH3B1 protein expression over that of control cells, indicating the ALDH3B1 substrate and LPO-derived aldehyde may be involved in ALDH3B1 gene regulation (Figure 7). As validation of these findings, no change in ALDH7A1 expression was found in any cell line following any treatment studied (data not shown).
Figure 7.

Induction of ALDH3B1 gene expression in vitro. ARPE-19 cells were untreated (control, C) or treated with 100 μM clofibrate (Clof), 100 nM estradiol (E2), 10 μM 4-hydroxy-2-nonenal (4HNE) or 2 μg/mL lipopolysaccharide (LPS) for 36 h (as described in Materials and Methods). ALDH3B1 induction in response to the treatments was determined by Western blot. ALDH3B1 protein bands were normalized relative to their respective β-actin expression. Density is displayed as means ± SEM from triplicate experiments. * Statistically significant difference compared to untreated control cells (P < 0.05).
Discussion
The ALDH3 family has unique roles in a variety of tissues. In the cornea, ALDH3A1 protects ocular structures against UV-induced damage by metabolizing LPO-derived aldehydes [21], absorbing ultraviolet light [22] and scavenging ROS [40]. Mutations in ALDH3A2 result in Sjögren-Larsson syndrome, a disorder characterized by congenital ichthyosis, mental retardation and ocular abnormalities [24]. The ALDH3B1 protein shares 53% and 55% sequence homology with ALDH3A1 and ALDH3A2, respectively. While little is known about the physiological role of the ALDH3B1 protein, our previous findings suggested that ALDH3B1 may defend against oxidative stress processes [25]. The characterization of ALDH3B1 detailed in this report, including the biochemical characterization and role as a cellular defense system against aldehyde and oxidant toxicity, lends further support for an important physiological role of ALDH3B1 against oxidative stress.
We and others have reported isolating ALDH enzymes to 95% or higher purity by one-step affinity chromatography using NAD+-analog (5′-AMP) columns [28, 29, 53]. However, ALDH3B1 binds very poorly to 5′-AMP and other NAD+-analog columns. In addition, ALDH3B1 binds essentially irreversibly to 2′,5′-ADP (NADP+-analog) columns. In this regard, human ALDH3B1 appears to have distinct biochemical properties from other ALDH enzymes including sequence-related ALDH3 members. The Rossman fold cofactor-binding turn region (GxTxxG) is highly conserved in ALDH sequences and is considered to be essential for interacting with the nicotinamide ring [54]. However, in human ALDH3B1, the threonine is replaced with a proline to give the sequence 188GxPxxG193. Similar to other ALDH3 family members, ALDH3B1 is able to use either NAD+ or NADP+ as cofactors; however, the affinity and catalytic efficiency for both cofactors was found to be markedly lower than other ALDH3 enzymes [48, 55-57]. ALDH3A1 demonstrates an affinity constant for NAD+ as low as 3 μM, indicating the enzyme has approximately 100-fold higher affinity for the cofactor than ALDH3B1. Likewise, catalytic efficiency values of NAD+ reported for ALDH3A1 are approximately 400-fold higher than the present findings for ALDH3B1. These findings may also explain the distinct differences seen in ALDH3B1 affinity for the cofactor-analogs 5′-AMP and 2′,5′-ADP when compared with other ALDH enzymes, particularly those that are easily purified using cofactor-analog chromatography columns, such as ALDH3A1.
Multiple expression and purification systems were utilized in an attempt to purify human ALDH3B1 to homogeneity while also preserving enzymatic activity for biochemical analysis. These included the expression of the ALDH3B1 protein as a GST- and hexahistidine (His6)-tagged fusion protein in E. Coli and Sf9 cells. No detectable enzymatic activity was found in E. Coli-expressing cell lysates or purified samples. Recent data in the literature [58] and protein phosphorylation prediction programs (NetPhos, NetPhosK) suggest that ALDH3B1 may be phosphorylated in its native state, particularly at serine residues 368 and 464. Phosphorylation has been reported to activate ALDH2 activity [59]; its potential effect on ALDH3B1 enzymatic activity remains to be clarified. Eukaryotic Sf9 cell lysates expressing ALDH3B1-GST were enzymatically active but protein binding to the GSH resin was negligible, indicating that the GSH binding region was obscured. Solubility was also a significant obstacle when expressing ALDH3B1 as a fusion protein. Denaturing conditions produced pure protein; however, enzymatic activity could not be recovered following refolding attempts. To achieve high expression levels of ALDH3B1 while maintaining enzymatic activity, a baculovirus Sf9 expression vector was created in which only the coding region of the ALDH3B1 cDNA was inserted and an added Kozak (GCCACC) sequence put in place before the first methionine, as described previously [27, 28]. These changes increased ALDH3B1 expression approximately 40-fold and allowed the protein to be purified using a multi-step process while both maintaining enzymatic activity and sufficient yield for enzyme kinetic analyses.
The affinities and catalytic efficiencies of human ALDH3B1 for 4HNE, hexanal, octanal and benzaldehyde reported here are consistent with those demonstrated for other ALDH enzymes believed to metabolize these aldehydes in vivo [29]. Substrate specificity, as determined here with pure protein by kinetic constants, is in agreement with previous findings comparing enzymatic activity of cell lysates [25]. In addition, 4HNE, hexanal, T2O and benzaldehyde cytotoxicity was significantly diminished in ALDH3B1-transfected HEK293 cells. Similar results have been reported for ALDH3A1, which is believed to play a major role in the cellular protection against LPO-derived aldehydes in vivo [29]. Short-chain aldehydes (acetaldehyde and malondialdehyde) were found to be poor substrates, which is consistent with that reported for other ALDH3 family members [29]. The binding of NAD+ to ALDH3B1 inhibited esterase activity, which is in contrast to ALDH2 but consistent with ALDH3A1, indicating that ALDH2 and ALDH3 family members may display distinct effects of NAD+-binding to their active site regions [60].
An intronic SNP (rs3751082 A) in ALDH3B1 has been linked to paranoid schizophrenia [61]. Intronic SNPs may play a role in schizophrenia as a result of altered pre-mRNA splicing and subsequent exon deletion [62]. Increased oxidative stress occurs in schizophrenics [63] and LPO-derived aldehydes including 4HNE have been implicated to play major roles in other pathological disease states of the brain including Alzheimer's and Parkinson's diseases [3, 64]. Of particular interest is the putative link between LPO-derived aldehydes and dopamine, which is directly metabolized by ALDH enzymes to the neurotoxic 3,4-dihydroxyphenylacetaldehyde (DOPAL) [65]. Neuronal accumulation of DOPAL can occur when this pathway is impaired [65, 66]. Recent reports indicate that LPO-derived aldehydes including 4HNE are responsible for inhibiting the ALDH-mediated oxidation of DOPAL [66, 67]. While DOPAL is a poor substrate for ALDH3B1 [25], our data collectively suggest that ALDH3B1 may prevent the accumulation of the DOPAL neurotoxin by metabolizing LPO-derived aldehydes. In this regard, ALDH3B1 may prove to have a protective role in a variety of disease states of the brain in which oxidative stress is implicated.
Several ALDH isozymes, including members of the ALDH3 family, have been found to be localized to multiple subcellular compartments where they perform distinct functions [1]. ALDH3A1, primarily found in the cytosol, is also present in the nucleus where it may have a role in cell cycle control. In addition, a splice variant of the microsomal ALDH3A2 is localized in peroxisomal membranes where it is involved in phytanic acid metabolism. As such, an important aspect of the present study was the identification of the subcellular localization of human ALDH3B1. In transfected HEK293 cells, ALDH3B1 appeared cytosolic, albeit with a higher concentration at the cell membrane. Analysis of the ALDH3B1 protein sequence revealed no signal or targeting peptides, indicating the protein may be cytosolic but associating with the membrane through other unknown proteins. Fractionation of mouse kidney and liver supported the finding that ALDH3B1 is a cytosolic protein.
The tissue expression of ALDH3B1 protein and mRNA in tissues is supportive of a role in oxidative stress. ALDH3B1 protein appeared high in the liver where ethanol metabolism and other oxidative processes lead to increased oxidative stress, LPO-derived aldehydes, free radicals and DNA damage [68]. In contrast, ALDH3A1 was negligibly expressed in liver [69]. ALDH3B1 was also highly expressed in the kidney where various disease states are associated with increased production of ROS and cell death [70]. High levels of ALDH3B1 were also found in the lung; direct contact with the atmosphere makes the lung vulnerable to inhaled aldehydes, such as those present in environmental pollutants. Ovary and testis, both of which show high ALDH3B1 protein and mRNA expression, are also sensitive to oxidative stress processes. In the testis, oxidative stress and the production of 4HNE impair spermatogenesis [5] and environmental pollutants cause increased aldehydes, aldehyde-protein adducts and DNA damage [71]. Expression of ALDH3B1 in the eye is also interesting as the sequence-related ALDH3A1 plays a critical role in protecting ocular structures from oxidative damage [22]. Similar to other ALDH3 enzymes [72], ALDH3B1 can use NADP+ as a cofactor [25]. Thus, ALDH3B1 may have an additional antioxidant role in tissues through the production of NAD(P)H and the regeneration of reduced GSH. Gene regulation (or control of the mRNA by miRNA) of ALDH3B1 may explain observed differences between ALDH3B1 protein and mRNA expression.
ALDH3B1 protein expression was induced in ARPE-19 cells by a sub-lethal concentration of 4HNE. We have previously shown that ALDH3B1 is up-regulated in various human tumors, which may be the result of oxidative stress processes [73]. In breast cancer cell lines, ALDH3A1 is induced by transient exposure to electrophiles which activate ARE [74]. The central regulator of the ARE response is the transcription factor Nrf2 which, on activation, dissociates from its inhibitor Keap1, translocates to the nucleus and activates ARE-dependent genes. 4HNE has been shown to modify Keap1 and activate ARE [75]. Our findings indicate that 4HNE may mediate redox cell signaling and the transcriptional regulation of the ALDH3B1 antioxidant enzyme.
In summary, kinetic and cytotoxicity studies reveal that ALDH3B1 efficiently metabolizes and protects cells from LPO-derived aldehydes and oxidant compounds, indicating the enzyme has an important role in the cellular defense against both oxidative stress and downstream aldehydes. In addition, this study also reveals the cytosolic subcellular localization of ADH3B1 and the putative 4HNE-mediated regulation of the ALDH3B1 gene. We believe the results presented here will be useful in providing further insight into the physiological significance of ALDH3B1.
Acknowledgments
The authors thank Dr. David Thompson for critical reading and discussion of this manuscript. The work described in this report was supported by NIH Grants EY11490 and EY17963 (VV) and NIH/NIAAA Predoctoral Fellowship AA016875 (SAM).
List of Abbreviations
- ALDH
aldehyde dehydrogenase
- 4HNE
4-hydroxy-2-nonenal
- ALDH3B1
aldehyde dehydrogenase 3B1
- ROS
reactive oxygen species
- LPO
lipid peroxidation
- HEK293
human embryonic kidney 293 cells
- Sf9
Spodoptera frugiperda
- MDA
malondialdehyde
- MTT
3-(4, 5-dimethylthiazolyl-2)-2, 5-diphenyltetrazolium bromide
- SRB
sulforhodamine B
- T2O
trans-2-octenal
- DOPAL
3,4-dihydroxyphenylacetaldehyde
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Marchitti SA, Brocker C, Stagos D, Vasiliou V. Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert Opin Drug Metab Toxicol. 2008;4:697–720. doi: 10.1517/17425250802102627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lindahl R. Aldehyde dehydrogenases and their role in carcinogenesis. Crit Rev Biochem Mol Biol. 1992;27:283–335. doi: 10.3109/10409239209082565. [DOI] [PubMed] [Google Scholar]
- 3.Yoritaka A, Hattori N, Uchida K, Tanaka M, Stadtman ER, Mizuno Y. Immunohistochemical detection of 4-hydroxynonenal protein adducts in Parkinson disease. Proc Natl Acad Sci U S A. 1996;93:2696–701. doi: 10.1073/pnas.93.7.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brooks PJ, Theruvathu JA. DNA adducts from acetaldehyde: implications for alcohol-related carcinogenesis. Alcohol. 2005;35:187–93. doi: 10.1016/j.alcohol.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 5.Shiraishi K, Naito K. Effects of 4-hydroxy-2-nonenal, a marker of oxidative stress, on spermatogenesis and expression of p53 protein in male infertility. J Urol. 2007;178:1012–7. doi: 10.1016/j.juro.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 6.Fuchs P, Loeseken C, Schubert JK, Miekisch W. Breath gas aldehydes as biomarkers of lung cancer. Int J Cancer. 2009 doi: 10.1002/ijc.24970. [DOI] [PubMed] [Google Scholar]
- 7.Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 8.Brichac J, Ho KK, Honzatko A, Wang R, Lu X, Weiner H, et al. Enantioselective oxidation of trans-4-hydroxy-2-nonenal is aldehyde dehydrogenase isozyme and Mg2+ dependent. Chem Res Toxicol. 2007;20:887–95. doi: 10.1021/tx7000509. [DOI] [PubMed] [Google Scholar]
- 9.Vasiliou V, Pappa A, Estey T. Role of human aldehyde dehydrogenases in endobiotic and xenobiotic metabolism. Drug Metab Rev. 2004;36:279–99. doi: 10.1081/dmr-120034001. [DOI] [PubMed] [Google Scholar]
- 10.Mills PB, Struys E, Jakobs C, Plecko B, Baxter P, Baumgartner M, et al. Mutations in antiquitin in individuals with pyridoxine-dependent seizures. Nat Med. 2006;12:307–9. doi: 10.1038/nm1366. [DOI] [PubMed] [Google Scholar]
- 11.Patel M, Lu L, Zander DS, Sreerama L, Coco D, Moreb JS. ALDH1A1 and ALDH3A1 expression in lung cancers: correlation with histologic type and potential precursors. Lung Cancer. 2008;59:340–9. doi: 10.1016/j.lungcan.2007.08.033. [DOI] [PubMed] [Google Scholar]
- 12.Sreerama L, Sladek NE. Identification and characterization of a novel class 3 aldehyde dehydrogenase overexpressed in a human breast adenocarcinoma cell line exhibiting oxazaphosphorine-specific acquired resistance. Biochem Pharmacol. 1993;45:2487–505. doi: 10.1016/0006-2952(93)90231-k. [DOI] [PubMed] [Google Scholar]
- 13.Moreb JS, Baker HV, Chang LJ, Amaya M, Lopez MC, Ostmark B, et al. ALDH isozymes downregulation affects cell growth, cell motility and gene expression in lung cancer cells. Mol Cancer. 2008;7:87. doi: 10.1186/1476-4598-7-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang B, Liu G, Xue F, Rosen DG, Xiao L, Wang X, et al. ALDH1 expression correlates with favorable prognosis in ovarian cancers. Mod Pathol. 2009;22:817–23. doi: 10.1038/modpathol.2009.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sladek NE. Human aldehyde dehydrogenases: potential pathological, pharmacological, and toxicological impact. J Biochem Mol Toxicol. 2003;17:7–23. doi: 10.1002/jbt.10057. [DOI] [PubMed] [Google Scholar]
- 16.Vasiliou V, Nebert DW. Analysis and update of the human aldehyde dehydrogenase (ALDH) gene family. Hum Genomics. 2005;2:138–43. doi: 10.1186/1479-7364-2-2-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graves PR, Kwiek JJ, Fadden P, Ray R, Hardeman K, Coley AM, et al. Discovery of novel targets of quinoline drugs in the human purine binding proteome. Mol Pharmacol. 2002;62:1364–72. doi: 10.1124/mol.62.6.1364. [DOI] [PubMed] [Google Scholar]
- 18.Hsu LC, Chang WC, Yoshida A. Cloning of a cDNA encoding human ALDH7, a new member of the aldehyde dehydrogenase family. Gene. 1994;151:285–9. doi: 10.1016/0378-1119(94)90672-6. [DOI] [PubMed] [Google Scholar]
- 19.Hsu LC, Chang WC, Yoshida A. Human aldehyde dehydrogenase genes, ALDH7 and ALDH8: genomic organization and gene structure comparison. Gene. 1997;189:89–94. doi: 10.1016/s0378-1119(96)00839-6. [DOI] [PubMed] [Google Scholar]
- 20.Vasiliou V, Bairoch A, Tipton KF, Nebert DW. Eukaryotic aldehyde dehydrogenase (ALDH) genes: human polymorphisms, and recommended nomenclature based on divergent evolution and chromosomal mapping. Pharmacogenetics. 1999;9:421–34. [PubMed] [Google Scholar]
- 21.Pappa A, Chen C, Koutalos Y, Townsend AJ, Vasiliou V. Aldh3a1 protects human corneal epithelial cells from ultraviolet- and 4-hydroxy-2-nonenal-induced oxidative damage. Free Radic Biol Med. 2003;34:1178–89. doi: 10.1016/s0891-5849(03)00070-4. [DOI] [PubMed] [Google Scholar]
- 22.Lassen N, Bateman JB, Estey T, Kuszak JR, Nees DW, Piatigorsky J, et al. Multiple and Additive Functions of ALDH3A1 and ALDH1A1: Cataract phenotype and ocular oxidative damage in Aldh3a1(-/-)/Aldh1a1(-/-) knock-out mice. J Biol Chem. 2007;282:25668–76. doi: 10.1074/jbc.M702076200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rizzo WB. Sjogren-Larsson syndrome: Molecular genetics and biochemical pathogenesis of fatty aldehyde dehydrogenase deficiency. Mol Genet Metab. 2007;90:1–9. doi: 10.1016/j.ymgme.2006.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rizzo WB, Carney G. Sjogren-Larsson syndrome: diversity of mutations and polymorphisms in the fatty aldehyde dehydrogenase gene (ALDH3A2) Hum Mutat. 2005;26:1–10. doi: 10.1002/humu.20181. [DOI] [PubMed] [Google Scholar]
- 25.Marchitti SA, Orlicky DJ, Vasiliou V. Expression and initial characterization of human ALDH3B1. Biochem Biophys Res Commun. 2007;356:792–8. doi: 10.1016/j.bbrc.2007.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benedetti A, Comporti M, Esterbauer H. Identification of 4-hydroxynonenal as a cytotoxic product originating from the peroxidation of liver microsomal lipids. Biochim Biophys Acta. 1980;620:281–96. doi: 10.1016/0005-2760(80)90209-x. [DOI] [PubMed] [Google Scholar]
- 27.Bunting KD, Townsend AJ. Protection by transfected rat or human class 3 aldehyde dehydrogenase against the cytotoxic effects of oxazaphosphorine alkylating agents in hamster V79 cell lines. Demonstration of aldophosphamide metabolism by the human cytosolic class 3 isozyme. J Biol Chem. 1996;271:11891–6. doi: 10.1074/jbc.271.20.11891. [DOI] [PubMed] [Google Scholar]
- 28.Manzer R, Qamar L, Estey T, Pappa A, Petersen DR, Vasiliou V. Molecular cloning and baculovirus expression of the rabbit corneal aldehyde dehydrogenase (ALDH1A1) cDNA. DNA Cell Biol. 2003;22:329–38. doi: 10.1089/104454903322216671. [DOI] [PubMed] [Google Scholar]
- 29.Pappa A, Estey T, Manzer R, Brown D, Vasiliou V. Human aldehyde dehydrogenase 3A1 (ALDH3A1): biochemical characterization and immunohistochemical localization in the cornea. Biochem J. 2003;376:615–23. doi: 10.1042/BJ20030810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheikh S, Ni L, Hurley TD, Weiner H. The potential roles of the conserved amino acids in human liver mitochondrial aldehyde dehydrogenase. J Biol Chem. 1997;272:18817–22. doi: 10.1074/jbc.272.30.18817. [DOI] [PubMed] [Google Scholar]
- 31.Bannai H, Tamada Y, Maruyama O, Nakai K, Miyano S. Extensive feature detection of N-terminal protein sorting signals. Bioinformatics. 2002;18:298–305. doi: 10.1093/bioinformatics/18.2.298. [DOI] [PubMed] [Google Scholar]
- 32.Horten P, Park KJ, Obayashi T, Nakai K. Protein subcellular localization prediction with WoLF PSORT (abstract). Proceedings of Asian Pacific Bioinformatics Conference; 2006. [Google Scholar]
- 33.Emanuelsson O, Nielsen H, Brunak S, von HG. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–16. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 34.Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241:779–86. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 35.Small I, Peeters N, Legeai F, Lurin C. Predotar: A tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics. 2004;4:1581–90. doi: 10.1002/pmic.200300776. [DOI] [PubMed] [Google Scholar]
- 36.Hirokawa T, Boon-Chieng S, Mitaku S. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics. 1998;14:378–9. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- 37.Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294:1351–62. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 38.Jordan M, Schallhorn A, Wurm FM. Transfecting mammalian cells: optimization of critical parameters affecting calcium-phosphate precipitate formation. Nucleic Acids Res. 1996;24:596–601. doi: 10.1093/nar/24.4.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cox B, Emili A. Tissue subcellular fractionation and protein extraction for use in mass-spectrometry-based proteomics. Nat Protoc. 2006;1:1872–8. doi: 10.1038/nprot.2006.273. [DOI] [PubMed] [Google Scholar]
- 40.Lassen N, Pappa A, Black WJ, Jester JV, Day BJ, Min E, et al. Antioxidant function of corneal ALDH3A1 in cultured stromal fibroblasts. Free Radic Biol Med. 2006;41:1459–69. doi: 10.1016/j.freeradbiomed.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 41.Messeguer X, Escudero R, Farre D, Nunez O, Martinez J, Alba MM. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002;18:333–4. doi: 10.1093/bioinformatics/18.2.333. [DOI] [PubMed] [Google Scholar]
- 42.Farre D, Roset R, Huerta M, Adsuara JE, Rosello L, Alba MM, et al. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003;31:3651–3. doi: 10.1093/nar/gkg605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wingender E, Chen X, Fricke E, Geffers R, Hehl R, Liebich I, et al. The TRANSFAC system on gene expression regulation. Nucleic Acids Res. 2001;29:281–3. doi: 10.1093/nar/29.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seng S, Avraham HK, Birrane G, Jiang S, Avraham S. Nuclear matrix protein (NRP/B) modulates the nuclear factor (erythroid-derived 2)-like 2 (NRF2)-dependent oxidative stress response. J Biol Chem. 2010 doi: 10.1074/jbc.M109.095786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siow RC, Ishii T, Mann GE. Modulation of antioxidant gene expression by 4-hydroxynonenal: atheroprotective role of the Nrf2/ARE transcription pathway. Redox Rep. 2007;12:11–5. doi: 10.1179/135100007X162167. [DOI] [PubMed] [Google Scholar]
- 46.Beinke S, Ley SC. Functions of NF-kappaB1 and NF-kappaB2 in immune cell biology. Biochem J. 2004;382:393–409. doi: 10.1042/BJ20040544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gloire G, Legrand-Poels S, Piette J. NF-kappaB activation by reactive oxygen species: fifteen years later. Biochem Pharmacol. 2006;72:1493–505. doi: 10.1016/j.bcp.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 48.Lindahl R, Petersen DR. Lipid aldehyde oxidation as a physiological role for class 3 aldehyde dehydrogenases. Biochem Pharmacol. 1991;41:1583–7. doi: 10.1016/0006-2952(91)90157-z. [DOI] [PubMed] [Google Scholar]
- 49.King G, Holmes RS. Human corneal aldehyde dehydrogenase: purification, kinetic characterisation and phenotypic variation. Biochem Mol Biol Int. 1993;31:49–63. [PubMed] [Google Scholar]
- 50.Vichai V, Kirtikara K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat Protoc. 2006;1:1112–6. doi: 10.1038/nprot.2006.179. [DOI] [PubMed] [Google Scholar]
- 51.Canuto RA, Maggiora M, Trombetta A, Martinasso G, Muzio G. Aldehyde dehydrogenase 3 expression is decreased by clofibrate via PPAR gamma induction in JM2 rat hepatoma cell line. Chem Biol Interact. 2003;143-144:29–35. doi: 10.1016/s0009-2797(02)00169-2. [DOI] [PubMed] [Google Scholar]
- 52.Li XH, Kakkad B, Ong DE. Estrogen directly induces expression of retinoic acid biosynthetic enzymes, compartmentalized between the epithelium and underlying stromal cells in rat uterus. Endocrinology. 2004;145:4756–62. doi: 10.1210/en.2004-0514. [DOI] [PubMed] [Google Scholar]
- 53.Lassen N, Estey T, Tanguay RL, Pappa A, Reimers MJ, Vasiliou V. Molecular cloning, baculovirus expression, and tissue distribution of the zebrafish aldehyde dehydrogenase 2. Drug Metab Dispos. 2005;33:649–56. doi: 10.1124/dmd.104.002964. [DOI] [PubMed] [Google Scholar]
- 54.Liu ZJ, Sun YJ, Rose J, Chung YJ, Hsiao CD, Chang WR, et al. The first structure of an aldehyde dehydrogenase reveals novel interactions between NAD and the Rossmann fold. Nat Struct Biol. 1997;4:317–26. doi: 10.1038/nsb0497-317. [DOI] [PubMed] [Google Scholar]
- 55.Kelson TL, Secor M, Jr, Rizzo WB. Human liver fatty aldehyde dehydrogenase: microsomal localization, purification, and biochemical characterization. Biochim Biophys Acta. 1997;1335:99–110. doi: 10.1016/s0304-4165(96)00126-2. [DOI] [PubMed] [Google Scholar]
- 56.Mitchell DY, Petersen DR. Oxidation of aldehydic products of lipid peroxidation by rat liver microsomal aldehyde dehydrogenase. Arch Biochem Biophys. 1989;269:11–7. doi: 10.1016/0003-9861(89)90081-7. [DOI] [PubMed] [Google Scholar]
- 57.Evces S, Lindahl R. Characterization of rat cornea aldehyde dehydrogenase. Arch Biochem Biophys. 1989;274:518–24. doi: 10.1016/0003-9861(89)90465-7. [DOI] [PubMed] [Google Scholar]
- 58.Imami K, Sugiyama N, Kyono Y, Tomita M, Ishihama Y. Automated phosphoproteome analysis for cultured cancer cells by two-dimensional nanoLC-MS using a calcined titania/C18 biphasic column. Anal Sci. 2008;24:161–6. doi: 10.2116/analsci.24.161. [DOI] [PubMed] [Google Scholar]
- 59.Chen CH, Budas GR, Churchill EN, Disatnik MH, Hurley TD, Mochly-Rosen D. Activation of aldehyde dehydrogenase-2 reduces ischemic damage to the heart. Science. 2008;321:1493–5. doi: 10.1126/science.1158554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mann CJ, Weiner H. Differences in the roles of conserved glutamic acid residues in the active site of human class 3 and class 2 aldehyde dehydrogenases. Protein Sci. 1999;8:1922–9. doi: 10.1110/ps.8.10.1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Y, Hu Y, Fang Y, Zhang K, Yang H, Ma J, et al. Evidence of epistasis between the catechol-O-methyltransferase and aldehyde dehydrogenase 3B1 genes in paranoid schizophrenia. Biol Psychiatry. 2009;65:1048–54. doi: 10.1016/j.biopsych.2008.11.027. [DOI] [PubMed] [Google Scholar]
- 62.Law AJ, Kleinman JE, Weinberger DR, Weickert CS. Disease-associated intronic variants in the ErbB4 gene are related to altered ErbB4 splice-variant expression in the brain in schizophrenia. Hum Mol Genet. 2007;16:129–41. doi: 10.1093/hmg/ddl449. [DOI] [PubMed] [Google Scholar]
- 63.Wang JF, Shao L, Sun X, Young LT. Increased oxidative stress in the anterior cingulate cortex of subjects with bipolar disorder and schizophrenia. Bipolar Disord. 2009;11:523–9. doi: 10.1111/j.1399-5618.2009.00717.x. [DOI] [PubMed] [Google Scholar]
- 64.Fukuda M, Kanou F, Shimada N, Sawabe M, Saito Y, Murayama S, et al. Elevated levels of 4-hydroxynonenal-histidine Michael adduct in the hippocampi of patients with Alzheimer's disease. Biomed Res. 2009;30:227–33. doi: 10.2220/biomedres.30.227. [DOI] [PubMed] [Google Scholar]
- 65.Marchitti SA, Deitrich RA, Vasiliou V. Neurotoxicity and metabolism of the catecholamine-derived 3,4-dihydroxyphenylacetaldehyde and 3,4-dihydroxyphenylglycolaldehyde: the role of aldehyde dehydrogenase. Pharmacol Rev. 2007;59:125–50. doi: 10.1124/pr.59.2.1. [DOI] [PubMed] [Google Scholar]
- 66.Jinsmaa Y, Florang VR, Rees JN, Anderson DG, Strack S, Doorn JA. Products of oxidative stress inhibit aldehyde oxidation and reduction pathways in dopamine catabolism yielding elevated levels of a reactive intermediate. Chem Res Toxicol. 2009;22:835–41. doi: 10.1021/tx800405v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Florang VR, Rees JN, Brogden NK, Anderson DG, Hurley TD, Doorn JA. Inhibition of the oxidative metabolism of 3,4-dihydroxyphenylacetaldehyde, a reactive intermediate of dopamine metabolism, by 4-hydroxy-2-nonenal. Neurotoxicology. 2006 doi: 10.1016/j.neuro.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 68.Seitz HK, Becker P. Alcohol metabolism and cancer risk. Alcohol Res Health. 2007;30:38–7. [PMC free article] [PubMed] [Google Scholar]
- 69.Vasiliou V, Puga A, Nebert DW. Negative regulation of the murine cytosolic aldehyde dehydrogenase-3 (Aldh-3c) gene by functional CYP1A1 and CYP1A2 proteins. Biochem Biophys Res Commun. 1992;187:413–9. doi: 10.1016/s0006-291x(05)81508-6. [DOI] [PubMed] [Google Scholar]
- 70.Han HJ, Lee YJ, Park SH, Lee JH, Taub M. High glucose-induced oxidative stress inhibits Na+/glucose cotransporter activity in renal proximal tubule cells. Am J Physiol Renal Physiol. 2005;288:F988–F996. doi: 10.1152/ajprenal.00327.2004. [DOI] [PubMed] [Google Scholar]
- 71.Che W, Qiu H, Liu G, Ran Y, Zhang H, Zhang L, et al. Oxidative damage of the extracts of condensate, particulate and semivolatile organic compounds from gasoline engine exhausts on testicles of rats. Bull Environ Contam Toxicol. 2009;83:42–7. doi: 10.1007/s00128-009-9757-x. [DOI] [PubMed] [Google Scholar]
- 72.Hempel J, Kuo I, Perozich J, Wang BC, Lindahl R, Nicholas H. Aldehyde dehydrogenase. Maintaining critical active site geometry at motif 8 in the class 3 enzyme. Eur J Biochem. 2001;268:722–6. doi: 10.1046/j.1432-1327.2001.01926.x. [DOI] [PubMed] [Google Scholar]
- 73.Marchitti SA, Orlicky DJ, Brocker C, Vasiliou V. Aldehyde Dehydrogenase 3B1 (ALDH3B1): Immunohistochemical Tissue Distribution and Cellular-specific Localization in Normal and Cancerous Human Tissues. J Histochem Cytochem. 2010 doi: 10.1369/jhc.2010.955773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sladek NE. Transient induction of increased aldehyde dehydrogenase 3A1 levels in cultured human breast (adeno)carcinoma cell lines via 5′-upstream xenobiotic, and electrophile, responsive elements is, respectively, estrogen receptor-dependent and -independent. Chem Biol Interact. 2003;143-144:63–74. doi: 10.1016/s0009-2797(02)00174-6. [DOI] [PubMed] [Google Scholar]
- 75.Levonen AL, Landar A, Ramachandran A, Ceaser EK, Dickinson DA, Zanoni G, et al. Cellular mechanisms of redox cell signalling: role of cysteine modification in controlling antioxidant defences in response to electrophilic lipid oxidation products. Biochem J. 2004;378:373–82. doi: 10.1042/BJ20031049. [DOI] [PMC free article] [PubMed] [Google Scholar]


