Abstract
BACKGROUND
Prostate cancer (PrCa) has a high incidence in Western countries and at present, there is no cure for hormone refractory prostate cancer. Transgenic mouse models have proven useful for understanding mechanisms of prostate carcinogenesis. The characterization of genetically modified mouse PrCa models using high-throughput genomic analyses provides important information to guide appropriate experiment applications for such model.
METHODS
We have analyzed the transcriptome of the hormone refractory and highly metastatic Fetal Globin-SV40/T-antigen (Gγ-globin-Tag) transgenic mouse model for PrCa compared to normal mouse prostate tissue. Gene expression patterns found in Gγ-globin-Tag mouse prostate tumors were compared with publicly available human localized and metastatic prostate tumors (GEO accession # GSE3325) through hierarchical cluster analysis, Pearson’s rank correlation coefficient, and Self Organizing Feature Maps (SOM) analyses.
RESULTS
Gγ-globin-Tag tumors clustered closely with human metastatic tumors and gene expression patterns had a significant correlation (P < 0.01), unlike human localized primary tumors (P > 0.6). Bioinformatic analyses identified deregulated genetic pathways and networks in Gγ-globin-Tag tumors, which displayed similarities to alterations in human PrCa. Changes in the expression of genes involved in DNA replication and repair (Rb1, p53, Myc, PCNA, DNMT3A) and growth factor signaling pathways (TGFβ2, ERK1/2, NRas, and Notch1) are deregulated in the Gγ-globin-Tag tumors, suggesting their key role in the oncogenic process. Identification of an enrichment of putative binding sites for transcription factors revealed eight transcription factors that may be important in Gγ-globin-Tag carcinogenesis, including SP1, NF-Y, CREB, Elk1, and E2F. Novel genes related to microtubule regulation were also identified in Gγ-globin-Tag tumors as potentially important candidate targets for PrCa. Overexpression of stathmin-1, whose expression was increased in human metastatic prostate tumors, was validated in Gγ-globin-Tag tumors by immunohistochemistry. This protein belongs to the SV40/T-antigen cancer signature identified in previous studies in prostate, breast, and lung cancer mouse models.
CONCLUSIONS
Our results show that the Gγ-globin-Tag model for hormone refractory PrCa shares important features with aggressive, metastatic human PrCa. Given the role of stathmin-1 in the destabilization of microtubles and taxane resistance, the Gγ-globin-Tag model and other SV40/T-antigen driven transgenic models may be useful for testing potential therapies directed at stathmin-1 in human prostate tumors.
Keywords: prostate cancer, transgenic mouse, gene expression, microarrays, stathmin-1
INTRODUCTION
Prostate cancer (PrCa) is a leading cause of cancer in men [1] where measurement of serum prostate-specific antigen (PSA) may lead to the early detection of prostatic tumors [2]. If the malignant tissue is localized within the prostate, PrCa may be effectively treated with radical surgery, radiotherapy, and androgen blockade. However, treatment options for androgen-independent metastatic PrCa are generally not curative as these tumors lack specific targeted therapies. While most prostate tumors exhibit variable amounts of neuroendocrine (NE) cell differentiation [3], only a small proportion of prostate tumors are classified as NE (or small cell type) and prognosis is very poor for these patients [4].
Gene expression profiling of PrCa have led to a better molecular characterization of the disease and to the identification of potential new therapeutic targets for advanced metastatic PrCa [5,6]. Since PrCa is a very heterogeneous disease, the development of novel therapies depends on the discovery of specific targets present in subtypes of human tumors. Many novel genes altered in human PrCa, like alpha-methylacyl-CoA racemase (AMACR) [7], hepsin [6], and polycomb group protein enhancer of zeste homolog 2 (EZH2) [8], have been recently identified by microarray technology. Techniques to determine methylation patterns have also demonstrated that many genes are silenced in a high percentage of prostate carcinomas, including GSTP-1 (up to 100% of tumors) [9]. In addition, the development of knockout and transgenic mouse models targeting specific genes, such as NKX3.1 and PTEN, have shown the key role of particular molecular pathways in prostate carcinogenesis [10].
Animal models of PrCa that resemble human prostate carcinoma have been valuable for understanding underlying mechanisms of the human disease. However, it is critical to identify in what specific ways different models mimic subtypes of the human pathology, particularly on a molecular level. This is critical for appropriately using the models for preclinicial testing of therapies. Each particular model has strengths and weaknesses, and it is generally accepted that not a single model can encompass the diversity of human PrCa. Among the models most widely used for PrCa, five groups can be distinguished: (a) Xenografts; (b) Transgenics; (c) Knockouts; (d) Chemically induced models; and (e) Reconstitution models [11–13]. Examples of transgenic models include the C3(1)-SV40-Tag, probasin-Tag (TRAMP) probasin-Large-Tag (LADY), Fetal Globin-Tag (Gγ-globin-Tag, or FG/Tag), C3(1)-polyoma middle-T, C3(1)-Bcl-2, Cryptidin-Tag, and Myc-driven transgenics (probasin-Myc and ARR2/probasin-Myc) [14]. Although many features of these models have been elucidated (i.e., natural history, androgen dependence, metastatic ability, response to some pharmacologic compounds, etc.), comprehensive analyses of gene expression in tumors from these models need to be determined to ascertain in what ways the transcriptomes of transgenic mouse models are similar to human PrCa and which deregulated genes and pathways might be potential targets for therapy. Molecular profiling of the TRAMP and Myc models have been reported with specific molecular features also found in human PrCa [14,15]. TRAMP tumors shared genes in common with human PrCa in multiple functional categories. Myc-induced mouse prostate tumors demonstrated dysregulation of Nkx3.1 and Pim-1, genes also deregulated in subsets of human PrCa.
The Gγ-globin-Tag transgenic mouse model (also called Gγ/T-15 in previous publications) expresses the SV40/Tag oncogene driven by the human fetal globin (FG) promoter, resulting in androgen-independent prostate tumors with NE features [16,17]. This is a unique transgenic mouse model because the T-antigen is expressed in a subset of p63 basal epithelial cells, possibly representing adult prostate stem cells [18]. Similar to human PrCa, prostate intraepithelial neoplasia (PIN) is the first alteration in Gγ-globin-Tag mice, which progresses to invasive carcinoma with epithelial and NE features and metastasis to the lymph nodes and other distant sites [17]. Thus, the Gγ-globin-Tag model shares important features with human androgen-independent metastatic prostate carcinoma.
In the present study, we have performed high-throughput gene expression analyses to characterize the transcriptome of Gγ-globin-Tag prostate tumors in order to further define its relationship to human PrCa at the transcriptome level. We have used cDNA microarray technology and bioinformatics to analyze transcriptional profiles and to identify novel genes involved in prostate carcinogenesis. We show here that the Gγ-globin-Tag model for PrCa is an accurate model to study aggressive and metastatic human prostate tumors. Moreover, this model may be very useful for evaluating particular proteins (such as stathmin-1) as biomarkers or targets for therapy.
MATERIALS AND METHODS
Transgenic Mice and Tissue Processing
Four Gγ-globin-Tag male transgenic mice (17–25 weeks of age) in the C57Bl/6 background with poorly differentiated prostate tumors and four control non-transgenic mice prostates in the same background were used. Transgenic mice were identified by slot blot, as described previously [17] and control non-transgenic mice in the same background were utilized in this study. The natural history of tumor development has previously been published [16,17]. Mice were palpated in the urogenital area to grossly monitor tumor size. Primary and metastatic (lymph node) prostate tumors were removed, cut into 1 mm pieces with a scalpel, and immediately frozen in dry ice or immersed in 10% formalin overnight for histological studies. For controls, non-transgenic healthy male mice were euthanized at a similar age and prostates were processed as described for the Gγ-globin-Tag mice. TRAMP transgenic mice (C57Bl/6 background) were purchased from The Jackson Laboratory (Bar Harbor, Maine) and prostate tumors were processed as described above. All animals were treated according to the guidelines of Animal Care and Use Committee (NIH publication No. 86–23, 1985) under an approved animal protocol.
Cell Culture
Human LNCaP, PC-3, and DU 145 (tumorigenic cells), and RWPE-1 (a non-tumorigenic human cell line immortalized with the papillomavirus) that was previously characterized [19], and mouse C2-TRAMP PrCa cell lines were cultured. Cells were cultured in RPMI 1640 medium-Glutamax supplemented with 10% fetal calf serum, 100 U/ml penicillin, and 100 µg/ml streptomycin (Invitrogen, Paisley, UK) except for RWPE-1 cells that were cultured in keratinocyte serum free medium (KSFM) supplemented with 25 µg/ml bovine pituitary extract and 5 ng/ml recombinant EGF (Invitrogen).
cDNA Microarray Analysis
RNA extraction, preparation of the cDNA-labeled probes and hybridization
The Incyte mouse GEM2 set of cDNA clones containing 8.7K features was arrayed on poly-lysine coated glass slides at the National Cancer Institute Advanced Technology Center. The gene list is available at http://nciarray.nci.nih.gov. The detailed protocol for the microarray analysis has been published elsewhere [20].
Total RNA was extracted from frozen prostate tumor and normal prostate tissues using the RNeasy mini kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol. Twenty micrograms of RNA from normal prostates (n = 4) were pooled and used as the reference RNA for the competitive hybridizations with tumor RNA in microarray experiments. Four Gγ-globin-Tag PrCa RNA samples were used. The preparation of the cDNA-labeled probes was performed using the MICROMAX™ system (NEN Life Science Products, Boston, MA), according to the manufacture’s protocol.
Reverse-transcription was conducted with Cyanine 3-dUTP (for the reference) or Cyanine 5-dUTP for tumor samples. The labeled cDNA mixture was filter-concentrated and hybridized onto the microarray slide for 16 hr at 42°C. After hybridization, slides were washed sequentially in a series of solutions with increasing stringency and scanned with an Axon 4000B fluorescence laser-scanning instrument (Axon Instruments, Foster City, CA).
Data filtering, normalization, and statistical analysis
After scanning, image analysis, and calculation of average foreground signal adjusted for local channel-specific background was performed using the GenePix Pro 3.0 software. Spots with signal intensities in both channels <100 were excluded from consideration. If at least one channel for a feature had an intensity above 100, the feature intensity under 100 was set to 100. Each array was globally normalized to make the median value of the log 2 ratio equal to zero, to correct for dye bias, PMT voltage imbalance and variations between channels. Hierarchical clustering of samples was based on a Pearson’s correlation similarity metric. Genes differentially expressed between normal and tumoral prostates were identified based on differences in geometric means of ratios as well as statistical significance based on Student’s t-tests. Genes with geometric means of ratios either >2 or <0.5 were first filtered and Student’s t-tests were performed to select genes whose expression were statistically different (P < 0.05).
Bioinformatics and data mining
Biological interpretation of the filtered genes was carried out by hierarchical clustering and Gene Ontology (GO) enrichment analysis using GARBAN [21], and network and signaling pathway analysis was performed using Ingenuity Pathway Analysis software (Ingenuity Systems, Redwood City, CA). Microarray data from human localized prostate tumors and androgen-independent metastatic samples were retrieved from Gene Expression Omnibus database (GEO accession # GSE3325). Normalization of this dataset using RMA algorithm was performed with Bioconductor [22]. Self Organizing Feature Maps (SOM) analysis was carried out to find gene expression profiles capable of differentiating between benign and pathological tissue. For cross-comparison with the human dataset, genes differentially expressed in mouse experiments were mapped to the Affymetrix HG-U133 Plus 2 microarray using homology data included in EnsMart database [23]. Pearson’s rank correlation coefficient was analyzed to assess correlation between gene expression profiles found in either human primary or metastastatic tumors, with Gγ-globin-Tag tumors.
To analyze transcription factors (TFs) that might be involved in the deregulation of Gγ-globin-Tag PrCa genes, several bioinformatic resources were used. Proximal promoter sequences of the murine genome were taken from the EnsMart database [24] and position weight matrixes (PWM) of known transcription factor binding sites (TFBSs) from the Jaspar database [25]. FactorY software [26] was used to compare the TFBS distribution in our set of selected genes with the distribution in the murine genome. A TFBS enrichment P-value was calculated using the hypergeometric distribution, and P values lower than 0.01 were considered as statistically significant for this analysis.
Real-Time RT-PCR
Real-time RT-PCR was performed to validate the quantification of stathmin-1 in mouse samples. Total RNA was extracted from frozen tissues using the RNeasy mini kit (Qiagen) including the DNase step according to the manufacturers’ protocols. The sequences of PCR primers were as follows: sense 5′–3′: GCTTTC CTTGCCAGTGGATT; antisense 5′–3′:TTG ACC GAG GGC TGA GAA TC. Quantitative analysis of gene expression was generated using SYBR Green master mix kit (Applied BioSystems) and a BioRad I-Cycler IQ Real-Time detection system machine. The level of gene expression was calculated after normalizing it to the 28S RNA level in each sample, and is given in relative units. Primers used for normalization to detect 28S RNA levels were as follows: Sense 5′ –3′: GGG TGG TAA ACT CCA TCT AA; antisense 5′–3′: AGT TCT TTT CAA CTT TCC CT.
Fluorescence In Situ Hybridization (FISH)
FISH analyses were performed to detect gene copy number of stathmin-1 in Gγ-globin-Tag tumors, normal prostates, and the PrCa cell line C2-TRAMP (a cell line isolated from a TRAMP tumor, which was previously characterized) [27]. Cells were exposed to Colcemid (0.02 µg/ml) for 4 hr, trypsinized and treated in a 0.06 M KCl hypotonic solution for 30 min. Then, cells were fixed in methanol: acetic acid (3:1), and dropped onto wet slides. Metaphase spreads from C2-TRAMP cells were simultaneously hybridized with a chromosome 4 painting probe labeled with Digoxigenin-dUTP and a probe prepared from the BAC clone RP23-356I15 (Research Genetics) labeled with Biotin-dUTP, which included the whole sequence of stathmin-1. Paraffin-embedded tissues on slides were hybridized following standard procedures. The digoxigenin and the biotin-labeled probes were detected with FITC-conjugated sheep anti-digoxigenin antibodies, and streptavidin conjugated with Texas Red, respectively. Specimens were examined with a Nikon Eclipse E800 microscope equipped with epifluorescence optics and a Photometrics MicroMax cooled CCD camera (1,300 × 1,300 array, 6.7 µm pixel size, 5MHz, image pixel size 80 nm). Images were generated and analyzed using Meta-Morph Imaging System™ 4.6 processor.
Immunohistochemistry
Slides were deparaffinized and incubated for 10 min with 3% H2O2 in water to quench the endogenous peroxidase activity. An antigen retrieval method was used for detection of the antibody against stathmin-1 (microwave treatment for 15 min at 750 W). Tissues were incubated with 5% normal rabbit serum in TBS (Tris–HCl 0.05 M, 0.5 M NaCl, pH 7.3) for 30 min at room temperature. Dilutions of the primary antibodies were as follows: 1:100 for anti-PCNA; and 1:1,500 for anti-stathmin-1. Primary antibodies were incubated at 4°C overnight or for 1 hr at RT. The anti-stathmin-1 antibody was kindly provided by Dr. Sobel (INSERM, Universite Pierre et Marie Curie, France). Tissues were washed in TBS and incubated with the appropriate secondary antibody. For the indirect avidin–biotin–peroxidase method, biotiny-lated rabbit–anti-mouse Ig anti-serum was added at a 1:200 dilution for 30 min at RT. Slides were then incubated for 30 min at RT with the avidin–biotin complex at a 1:100 dilution. For the EnVision™ signal enhancement system, the secondary polyclonal complex was applied for 30 min at RT. After washing the slides in TBS, development of peroxidase was performed with diaminobenzidine and H2O2. The indirect avidin–biotin–peroxidase method was employed for the analysis of PCNA (clone PC10, Dako, Barcelona, Spain), and the EnVision (Dako) signal enhancement system was used for the analysis of stathmin-1. Sections were counterstained with Harris hematoxylin, dehydrated, and mounted. Negative controls included incubation of the slides without primary antibody or with a non-specific antibody of the same isotype (Dako).
Western Blot
For Western blots, cell extracts were lysed in RIPA buffer (1% Triton X-100, 50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 10 mM Na3VO4) with protease inhibitors (Roche, Penzberg, Germany). Twenty micrograms protein were electrophoresed in 12% Tris–HCl 0.1% SDS–polyacrylamide gel, then electrotransferred to PVDF membranes. Blots were blocked in 5% Skim milk dissolved in TBS Tween 0.05% (TBST). Primary rabbit polyclonal antibody anti-Stathmin-1 (Sigma, St. Louis, MO) was diluted 1:2,000 in 5% milk in TBST and blots were incubated overnight at 4°C. Membranes were then washed three times with TBST and treated for 5 min with the peroxidase substrate LumiLight Plus (Roche). Blots were exposed to ECL films (Amersham) and developed. After immunodetection of stathmin-1, blots were stripped in 1 M glycine solution, pH 2, for 2 hr at RT, equilibrated in TBS and reprobed with anti-β-actin antibody diluted 1:20,000 (Sigma).
Statistical Analysis
The two-sided Student’s t-test was used to evaluate significant differences between mean values. A P-value <0.05 was considered statistically significant.
RESULTS
Gene Expression Profiles of Gγ-Globin-Tag Mouse Prostate Tumors reveal Similarities to Human Prostate Cancer Metastasis With Neuroendocrine Differentiation
Gene expression in Gγ-globin-Tag (FG/Tag) transgenic prostate tumors was compared to the expression profile of normal prostate using cDNA microarray analysis. We identified 902 genes with statistically different expression (P < 0.05) between normal and malignant prostates; 571 genes were up-regulated and 331 genes were down-regulated. Pearson’s correlation analyses (r > 0.89) showed that gene expression profiles were very similar within the tumor samples or within the normal prostate samples. Genes were functionally classified using GeneCards (http://bioinformatics.weizmann.ac.il), and GO. Table I summarizes the differential expression of a selected list of deregulated genes. The complete list of genes can be found at http://caarraydb.nci.nih.gov/caarray/. Many of the genes with an aberrant expression in the Gγ-globin-Tag model were similarly deregulated in the human disease (Table I). From the categories that we have established for classification, a group of highly up-regulated genes were related to microtubule polymerization (Table I). Stathmin-1 and tubulin-β5 were highly increased in tumors (19 ± 2.1- and 12.3 ± 1.6-fold, respectively). Another substantial group of up-regulated genes were related to NE differentiation, including secretogranin III (4.0 ± 0.6) and dopa decarboxylase (3.1 ± 0.5). This result shows that Gγ-globin-Tag tumors display NE differentiation features, which occurs in a large percentage of human prostate tumors [3]. The NE phenotype of Gγ-globin-Tag tumors has been previously reported [18].
TABLE I.
Selected List of Genes Whose Expression Is Changed in FG/Tag Tumors Compared to Normal Prostates
| Category and accession # | Gene name | Fold change (± SEM) | Human PrCa |
|---|---|---|---|
| FG/Tag | |||
| Microtubule-related proteins | |||
| NM 019641 | Stathmin-1 (Oncoprotein-18) | 19.4 ± 2.1 | ↑ in 30% tumors |
| NM 011655 | Tubulin β-5 | 12.3 ± 1.6 | ↑ |
| NM 009451 | Tubulin β-4 | 2.9 ± 0.4 | ↑ |
| NM 009447 | Tubulin α-4 | 3.7 ± 0.2 | Unknown |
| NM 019410 | Profilin 2 | 3.2 ± 0.4 | Unknown |
| NM 013758 | Aduccin 3 (Gamma) | −3.3 ± 0.1 | ↓ mRNA level |
| Neuroendocrine | |||
| NM 010875 | Neural cell adhesion molecule 1 (Ncam1) | 4.7 ± 0.3 | ↑ in PrCa nerves |
| NM 009130 | Secretogranin III | 4.0 ± 0.6 | ↑ in N.E.T. |
| NM 016672 | Dopa decarboxylase | 3.1 ± 0.5 | Unknown |
| NM 023119 | Enolase-1 (non-neuron) | 2.4 ± 0.3 | Unknown |
| Cell cycle | |||
| NM 009875 | Cyclin-dependent kinase inh.1B (p27) | 1.6 ± 0.4 | ↑ in N.E. differentiation |
| NM 011045 | PCNA | 5.0 ± 0.4 | ↑ |
| NM 009029 | Retinoblastoma 1 | 2.1 ± 0.1 | ↑ |
| NM 157569 | Cyclin G | 5.0 ± 0.4 | ↑ |
| Apoptosis | |||
| NM 007522 | BCL-associated death promoter (Bad) | 5.1 ± 0.3 | ↑ |
| NM 009761 | Bcl-2/adenov19Kda (BNIP-3) | 2.3 ± 0.1 | Unknown |
| NM 011997 | Caspase 8-associated protein | 1.7 ± 0.4 | Unknown |
| NM 010019 | Death-associated kinase 2 | 2.2 ± 0.2 | ↓ |
| Cell adhesion, angiogenesis | |||
| NM 007585 | Annexin A2 | −3.3 ± 0.1 | ↓ |
| NM 007616 | Caveolin 1 (Cav1) | −4.7 ± 0.4 | ↓ |
| NM 007543 | Ceacam1 | −4.1 ± 0.4 | ↓ |
| NM 011582 | Thrombospondin 4 | 3.4 ± 0.47 | Unknown |
| NM 009864 | E-Cadherin | −2.0 ± 0.1 | ↓ |
| NM 003873 | Neuropilin-1 | 2.9 ± 0.4 | ↑ |
| Ribonucleoproteins/splicing | |||
| NM 177301 | hnRNP L | 1.6 ± 0.3 | Unknown |
| NM 182650 | hnRNP A2/B1 | 3.3 ± 0.1 | Unknown |
| NM 019868 | hnRNP H2 | 2.9 ± 0.1 | Unknown |
| NM 003017 | Splicing factor Sfrs3 | 2.9 ± 0.4 | Unknown |
| Proteases | |||
| NM 008610 | MMP-2 | 2.2 ± 0.2 | ↑ |
| NM 011594 | Tissue inhibitor of metalloproteinase 2 | −2.4 ± 0.1 | Inconclusive |
| NM 080639 | Tissue inhibitor of metalloproteinase 4 | −2.5 ± 0.1 | Unknown |
| Cell signaling and transcription factors | |||
| NM 138473 | SP1 | 2.1 ± 0.1 | ↑ |
| NM 008538 | MARCKS | 11.6 ± 1.4 | Unknown |
| NM 016899 | Rab 25 | −2.0 ± 0.1 | Unknown |
| NM 013685 | Transcription factor-4 | 5.5 ± 1.2 | Unknown |
| NM 008854 | PRKACA | 1.9 ± 0.2 | Unknown |
| NM 010937 | NRas | 2.4 ± 0.1 | ↑ |
| Metabolism | |||
| NM 008084 | GAPDH | 3.3 ± 0.1 | “Housekeeping” gene |
| NM 009155 | Selenoprotein P, plasma 1 (SePP1) | −2.0 ± 0.1 | ↓ in 60% tumors |
| NM 013602 | Metallothionein 1 | −3.4 ± 0.4 | ↓ |
| NM 001512 | Glutathione S-transferase alpha 4 | −2.0 ± 0.1 | Unknown |
| NM 008509 | Lipoprotein lipase | −5.0 ± 0.7 | Unknown |
| Growth factors | |||
| NM 009367 | TGF-beta 2 | 2.3 ± 0.2 | ↑ |
| NM 019971 | PDGF-D | −2.2 ± 0.1 | Unknown |
| Histones/methylation | |||
| NM 008229 | Histone deacetylase 2 | 2.7 ± 0.2 | Unknown |
| NM 016750 | H2A Histone family member Z | 6.2 ± 0.7 | Unknown |
| NM 002107 | Histone H3, family 3A | 3.2 ± 0.1 | Unknown |
| NM 1003963 | DNA methyltransferase 3A | 2.5 ± 0.2 | Unknown |
| Calcium binding proteins | |||
| NM 011313 | S100a6 (Calcyclin) | −2.8 ± 0.1 | ↓ |
| NM 009037 | Reticulocalbin | −5.1 ± 0.4 | Unknown |
| NM 009813 | Calsequestrin | −2.5 ± 0.2 | Unknown |
Deregulation of cell cycle-related genes was also identified in Gγ-globin-Tag tumors, compared to normal prostates. Many of these changes were similar to alterations described for human PrCa, such as PCNA, cyclin G, and Bad (Table I). Expression of specific genes related to angiogenesis, adhesion, metastasis, and regulatory proteins of the extracellular matrix were also similar between Gγ-globin-Tag and human prostate tumors. The VEGF165 receptor neuropilin-1 was up-regulated (2.9 ± 0.4-fold), whereas caveolin-1, E-cadherin, and annexin A2 were down-regulated (Table I). Selenoprotein-P, a gene involved in removing reactive oxygen species (ROS), which is decreased in 60% of human prostate tumors [20], was also down-regulated in the Gγ-globin-Tag model (−2.0 ± 0.1-fold). Some genes related to cell signaling were overexpressed, such as the myristoylated alanine-rich C-kinase substrate (MARCKS) (11.6 ± 1.4) and the ERK1/2/MAPK-related protein PRKACA (1.9 ± 0.2). Another group of up-regulated genes was the family of heterogeneous nuclear ribonucleoprotein/splicing proteins (Table I).
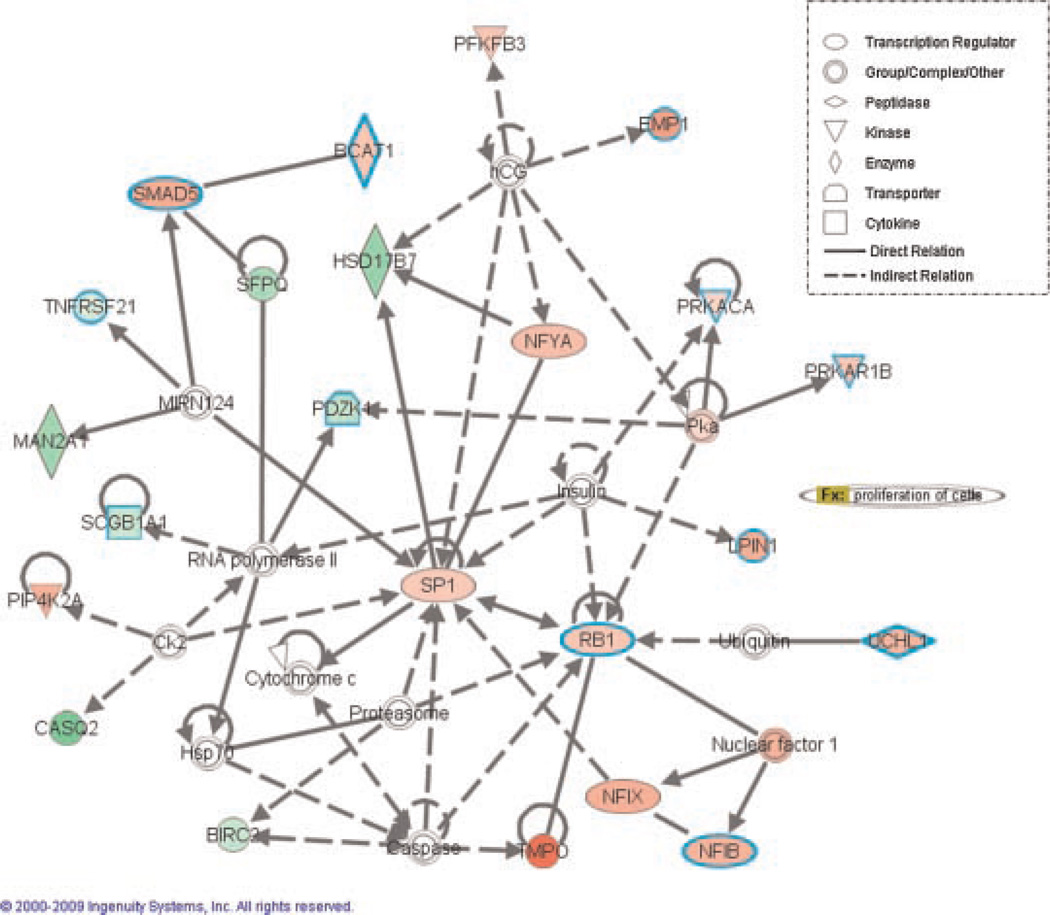
Gene network analysis using Ingenuity software identified nine networks with a high score for gene interactions. The complete set of networks and the list of genes involved in those networks can be found on line (http://caarraydb.nci.nih.gov/caarray/). The top-rated network of gene interaction was established by genes related to DNA replication, recombination repair, and cellular assembly and organization (Fig. 1). SP1 and Rb1 were the main nodes of this network (Fig. 1), which suggests that many altered pathways of Gγ-globin-Tag tumors are closely related to Rb1 dysfunction. Overexpression of Rb1 in this model is in keeping with the fact that T-antigen-transformed cells up-regulate Rb1 to compensate for loss of function [28]. The second and third highly scored networks had p53/HNF4A (Supplementary Fig. 1) and Myc (Supplementary Fig. 2) as main node genes. These results are in keeping with the alterations caused by T-antigen in Rb1 and p53 pathways in this model. A set of 10 genes were classified in the category “Metastasis” by Ingenuity, which included: MMP-2, Caveolin-1, Tubb4, Tuba4, N-CAM1, Fyn, ENPP2, POLE2, CNN1, and TXNIP. Altered expression of androgen receptor signaling regulators in Gγ-globin-Tag tumors, such as FKBP4, NCOA4, and calreticulin (see expression profiles at http://caarraydb.nci.nih.gov/caarray/) might be linked to the acquisition of androgen independence and aggressiveness in this model.
Fig. 1.
Gene network analysis using Ingenuity™. SP1 and Rb1 are the main “nodes” of the main top-ranked network of gene interactions. Genes in this network are related to DNA replication, recombination repair, and cellular assembly and organization.
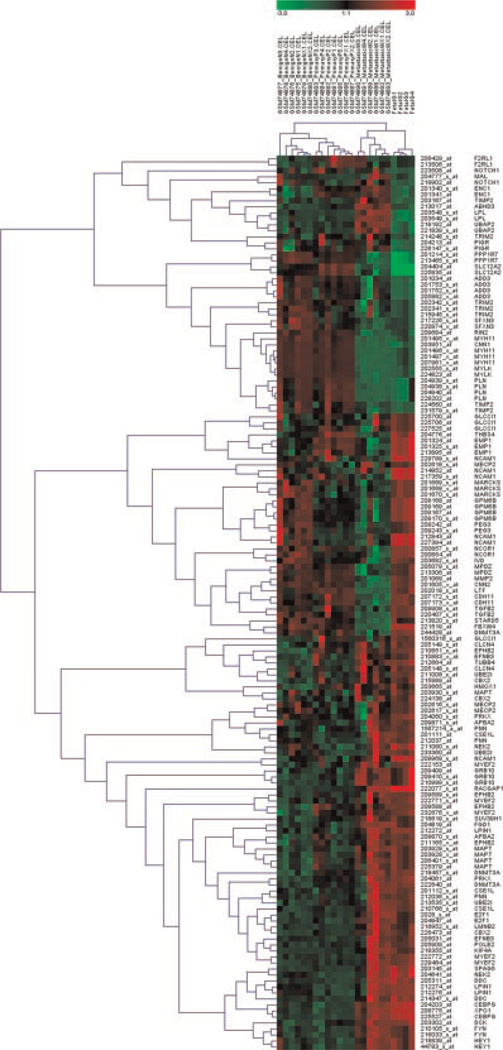
We next compared gene expression profiles between Gγ-globin-Tag and publicly available human PrCa samples (GSE3325).We identified 387 common probesets present in both human Affymetrix HG-U133 Plus 2 microarray and differentially expressed genes in the mouse dataset. Clustering analysis of these probes using SOM algorithm was used as a filtering method to identify deregulated genes that followed a similar pattern in most human tumors. The analyses of primary tumor and metastasis samples were carried out independently. Two clusters for each analysis including 46 genes whose expression is altered in primary tumors and 48 genes in metastasis, using benign prostate samples as reference in both contrasts, were found. Comparison of the expression patterns between human and Gγ-globin-Tag PrCa is shown in Table II. Concordant patterns were found in 27 out of 46 genes (59%) for primary tumors, and 36 out of 48 (75%) for metastasis. A two-way hierarchical clustering of the resulting 94-gene signature revealed that gene expression profiles from the Gγ-globin-Tag model were closely related with human metastatic prostate tumors (Fig. 2). The P-value from Pearson’s rank correlation coefficient for primary tumors was >0.6, whereas for metastasis samples the P-value was <0.01. This suggests that Gγ-globin-Tag tumors share important similarities with gene expression patterns of human PrCa metastasis, in keeping with the aggressive behavior of this model.
TABLE II.
Comparison of Gene Expression Patterns Between Human and Mouse FG/Tag Tumors
| Name | Description | Human | Mouse |
|---|---|---|---|
| Benign vs. primary | |||
| ADD3 | Adducin 3 (gamma) | ↓ | ↓ |
| APBA2 | Amyloid beta (A4) precursor protein-binding, family A, member 2 | ↓ | ↑ |
| CBX2 | Chromobox homolog 2 (Drosophila Pc class) | ↑ | ↑ |
| CDC2 | Cell cycle CDC2 Mm.4761 Cell division cycle control protein 2a | ↑ | ↑ |
| CLCN4 | Chloride channel 4–2 | ↑ | ↑ |
| CSE1L | Chromosome segregation 1-like (S. cerevisiae) | ↑ | ↑ |
| DNMT3A | DNA methyltransferase 3A | ↑ | ↑ |
| EMP1 | Epithelial membrane protein 1 | ↑ | ↑ |
| ENC1 | Ectodermal-neural cortex 1 | ↑ | ↑ |
| EPHB2 | Eph receptor B2 | ↑ | ↑ |
| F2RL1 | Coagulation factor II (thrombin) receptor-like 1 | ↑ | ↓ |
| FYN | Fyn proto-oncogene | ↓ | ↑ |
| GLCCI1 | Glucocorticoid-induced transcript 1 | ↑ | ↑ |
| GPM6B | Glycoprotein m6b | ↓ | ↑ |
| HEY1 | Hairy/enhancer-of-split related with YRPW motif 1 | ↓ | ↑ |
| HMOX1 | Heme oxygenase (decycling) 1 | ↑ | ↑ |
| IVD | UG5 isovaleryl coenzyme A dehydrogenase | ↓ | ↑ |
| KIF4A | Kinesin heavy chain member 4 | ↑ | ↑ |
| LMOD1 | Leiomodin 1 (smooth muscle) | ↓ | ↓ |
| LPIN1 | Lipin 1 | ↓ | ↑ |
| LTF | Lactotransferrin | ↑ | ↑ |
| MAL | Myelin and lymphocyte protein, T-cell differentiation protein | ↓ | ↓ |
| MAPT | Microtubule-associated protein tau | ↑ | ↑ |
| MARCKS | Myristoylated alanine rich protein kinase C substrate | ↓ | ↑ |
| MECP2 | Methyl CpG binding protein 2 | ↓ | ↑ |
| MGLL | Monoglyceride lipase | ↑ | ↓ |
| MYEF2 | Myelin basic protein expression factor 2, repressor | ↑ | ↑ |
| NCAM1 | Neural cell adhesion molecule 1 | ↓ | ↑ |
| NCOR1 | Nuclear receptor co-repressor 1 | ↓ | ↑ |
| NEK2 | NIMA (never in mitosis gene a)-related expressed kinase 2 | ↓ | ↑ |
| NOTCH1 | Notch gene homolog 1, (Drosophila) | ↓ | ↓ |
| PEG3 | Paternally expressed 3 | ↓ | ↑ |
| PIGR | Polymeric immunoglobulin receptor | ↑ | ↓ |
| PNN | Pinin | ↓ | ↑ |
| PPP1R7 | Protein phosphatase 1, regulatory (inhibitor) subunit 7 | ↓ | ↓ |
| PRKX | Putative serine/threonine kinase | ↑ | ↑ |
| RACGAP1 | Rac GTPase-activating protein 1 | ↑ | ↑ |
| RP13-15M17,2 | Ribosomal protein L13a | ↑ | ↑ |
| SFXN3 | Sideroflexin 3 | ↓ | ↓ |
| SLC12A2 | Solute carrier family 12, member 2 | ↑ | ↓ |
| SPAG5 | Sperm-associated antigen 5 | ↑ | ↑ |
| THBS4 | Thrombospondin 4 | ↑ | ↑ |
| TIMP2 | Tissue inhibitor of metalloproteinase 2 | ↓ | ↓ |
| TRIM2 | Tripartite motif protein 2 | ↓ | ↓ |
| TUBA1 | Tubulin, alpha 4 | ↓ | ↑ |
| UBE2I | Ubiquitin-conjugating enzyme E2I | ↓ | ↑ |
| Benign vs. metastasis | |||
| ABHD3 | Abhydrolase domain containing 3 | ↑ | ↑ |
| CBX2 | Chromobox homolog 2 (Drosophila Pc class) | ↑ | ↑ |
| CDC2 | Cell cycle CDC2 Mm.4761 Cell division cycle control protein 2a | ↑ | ↑ |
| CDH11 | Cadherin 11 | ↓ | ↑ |
| CEBPG | UG5 CCAAT/enhancer binding protein (C/EBP), gamma | ↑ | ↑ |
| CNN1 | Calponin 1 | ↓ | ↓ |
| CNN2 | UG5 calponin 2 | ↓ | ↑ |
| CSE1L | Chromosome segregation 1-like (S. cerevisiae) | ↑ | ↑ |
| DCK | Deoxycytidine kinase | ↑ | ↑ |
| DDC | Dopa decarboxylase | ↑ | ↑ |
| DNMT3A | DNA methyltransferase 3A | ↑ | ↑ |
| E2F1 | E2F transcription factor 1 | ↑ | ↑ |
| EFNB3 | Ephrin B3 | ↑ | ↑ |
| ENC1 | Ectodermal-neural cortex 1 | ↑ | ↓ |
| EPHB2 | UG5 Eph receptor B2 | ↑ | ↑ |
| FBXW4 | UG5 f-box and WD-40 domain protein 4 | ↓ | ↑ |
| FGD1 | Faciogenital dysplasia homolog (human) | ↑ | ↑ |
| FYN | Fyn proto-oncogene | ↑ | ↑ |
| GRB10 | Growth factor receptor bound protein 10 | ↑ | ↑ |
| HEY1 | Hairy/enhancer-of-split related with YRPW motif 1 | ↑ | ↑ |
| KIF4A | Kinesin heavy chain member 4 | ↑ | ↑ |
| LMNB2 | Lamin B2 | ↑ | ↑ |
| LMOD1 | Leiomodin 1 (smooth muscle) | ↓ | ↓ |
| LPL | Lipoprotein lipase | ↑ | ↓ |
| LTF | Lactotransferrin | ↓ | ↑ |
| MAPT | Microtubule-associated protein tau | ↑ | ↑ |
| MARCKS | Myristoylated alanine rich protein kinase C substrate | ↓ | ↑ |
| MMP2 | Matrix metalloproteinase 2 | ↓ | ↑ |
| MPDZ | Multiple PDZ domain protein | ↓ | ↑ |
| MYEF2 | Myelin basic protein expression factor 2, repressor | ↑ | ↑ |
| MYH11 | Myosin heavy chain 11, smooth muscle | ↓ | ↓ |
| MYLK | Myosin, light polypeptide kinase | ↓ | ↓ |
| NEK2 | NIMA (never in mitosis gene a)-related expressed kinase 2 | ↑ | ↑ |
| PLN | Phospholamban | ↓ | ↓ |
| POLE2 | Polymerase (DNA directed), epsilon 2 (p59 subunit) | ↑ | ↑ |
| PPP1R7 | Protein phosphatase 1, regulatory (inhibitor) subunit 7 | ↓ | ↓ |
| PRKX | Putative serine/threonine kinase | ↑ | ↑ |
| PYGGB | Brain glycogen phosphorylase | ↓ | ↓ |
| RACGAP1 | Rac GTPase-activating protein 1 | ↑ | ↑ |
| RIN2 | Ras and Rab interactor 2 | ↓ | ↓ |
| SFXN3 | Sideroflexin 3 | ↓ | ↓ |
| SLC12A2 | Solute carrier family 12, member 2 | ↓ | ↓ |
| SPAG5 | Sperm-associated antigen 5 | ↑ | ↑ |
| STARD5 | StAR-related lipid transfer (START) domain containing 5 | ↓ | ↑ |
| SUV39H1 | Suppressor of variegation 3–9 homolog 1 (Drosophila) | ↑ | ↑ |
| TGFB2 | Transforming growth factor, beta 2 | ↓ | ↑ |
| TUBB4 | Tubulin, beta 4 | ↑ | ↑ |
| UBAP2 | Ubiquitin-associated protein 2 | ↑ | ↓ |
| XPO1 | Exportin 1, CRM1 homolog (yeast) | ↑ | ↑ |
↑, up-regulation; ↓, down-regulation, in malignant tissues.
Human data were retrieved from the University of Michigan Comprehensive Cancer Center Affymetryx Core (GEO accession # GSE3325).
Fig. 2.
Hierarchical cluster analysis of human and mouse prostate samples. The dendogram shows that the Gγ-globin-Tag (FG/Tag) model clusters closely to metastatic human prostate cancer.
We further wished to identify TFs that may be involved in the coordinated deregulation of gene expression in Gγ-globin-Tag prostate tumors using FactorY. This analysis identified eight TFBSs with a very significant enrichment (P < 0.01) in the set of differentially expressed genes (Table III). This result suggests that the eight TFs are co-regulated and play a role in the altered expression of Gγ-globin-Tag tumor genes. The list of genes with TFBSs for these eight TFs can be found in Supplementary Table I, and the complete list of TFs, in Supplementary Table II. An analysis by Ingenuity was carried out as well to study gene interactions belonging to the category “transcription factors”. Supplementary Figure 3 shows that Rb1 and p53 may regulate the expression of the eight TFs in the Gγ-globin-Tag model, such as SP1, E2F1, NFYA, CREB1, and TFAP2A.
TABLE III.
Analysis of TFs With High Probability (P < 0.01) of Binding to TFBSs of Deregulated Genes (N = 902) Found in FG/Tag Tumors
| Motif # | Motif name | N total | N selection | P-value | Rank |
|---|---|---|---|---|---|
| MA0060 | NF-Y | 5,515 | 67 | 0.00000027643 | 1 |
| MA0006 | Ahr-ARNT | 7,343 | 81 | 0.00000052953 | 2 |
| MA0003 | AP2alpha | 5,273 | 58 | 0.00005313753 | 3 |
| MA0079 | SP1 | 8,768 | 85 | 0.00005631816 | 4 |
| MA0056 | MZF_1–4 | 5,765 | 58 | 0.00061676156 | 5 |
| MA0018 | CREB | 3,201 | 36 | 0.00122261734 | 6 |
| MA0028 | Elk-1 | 5,750 | 54 | 0.00476565119 | 7 |
| MA0024 | E2F | 1,132 | 15 | 0.00761377485 | 8 |
Motif # corresponds to the accession number for each TF in the Jaspar database. “N total” means the number of genes in the mouse genome with a specific TFBS. “N Selection” means the number of genes of our selected list with a specific TFBS. The “Rank” was based on “P” values.
The Gγ-Globin-Tag as a model to Study Stathmin-1 in Prostate Cancer
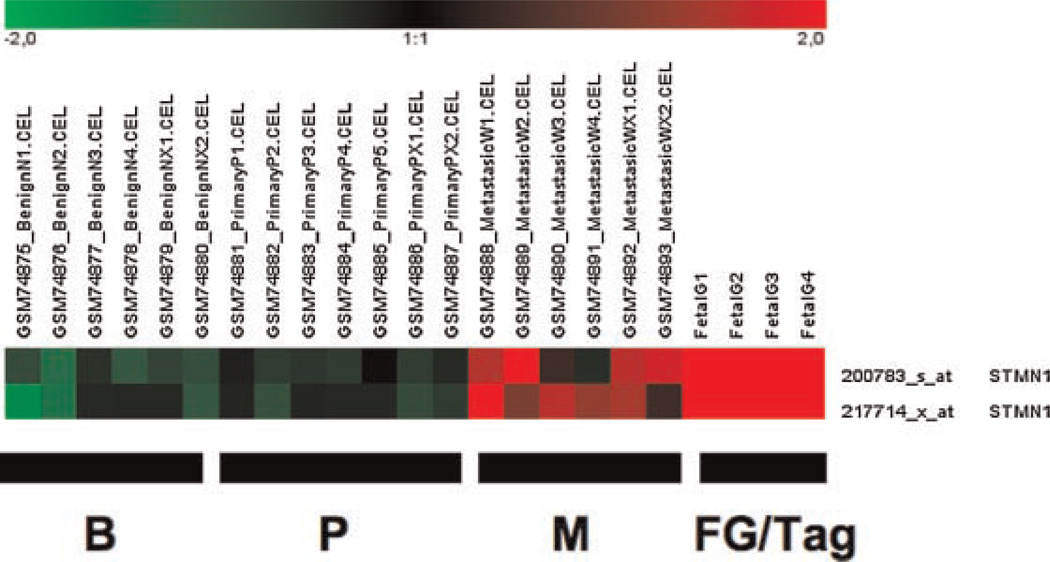
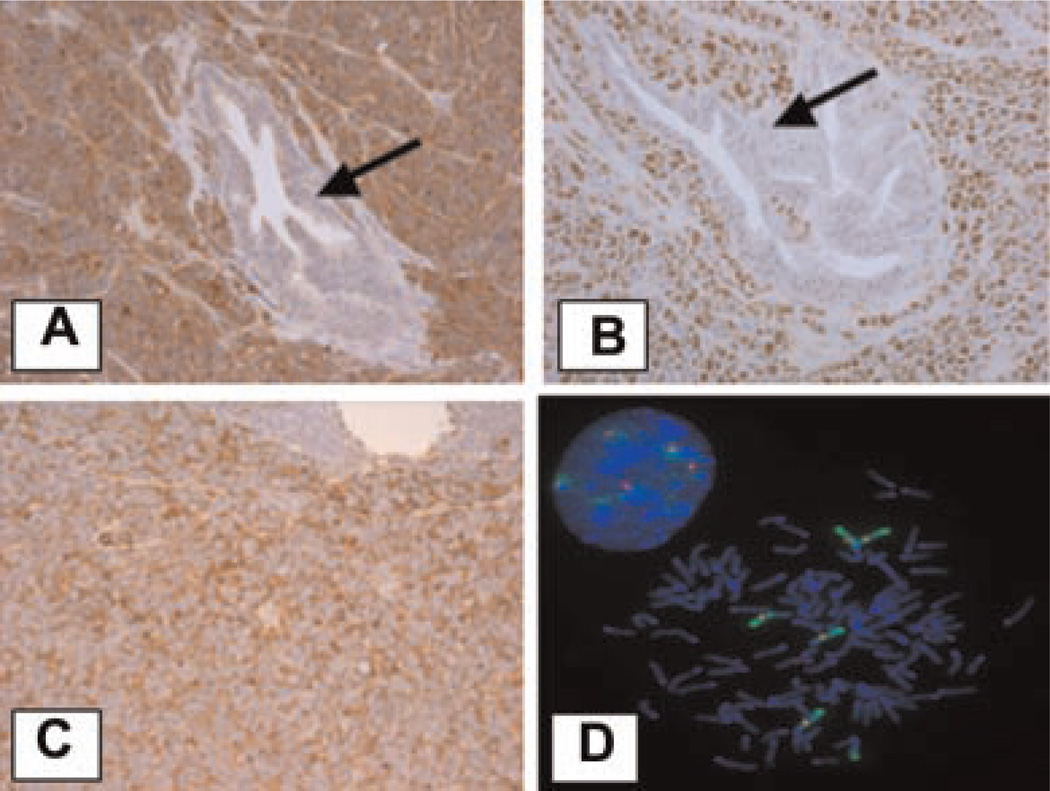
Stathmin-1 (STMN1), also called leukemia-associated gene and oncoprotein-18 (Op 18), was found to be highly overexpressed in Gγ-globin-Tag prostate tumors. In the human dataset, stathmin-1 expression was found up-regulated in metastatic but not in primary tumors (Fig. 3). Stathmin-1 is a p53-regulated gene that plays an important role in microtubule polymerization and intracellular signaling [29]. The up-regulation of stathmin-1 mRNA and protein levels in mouse prostate tumors was confirmed by real-time RT-PCR and immunohistochemistry. Quantitative real-time RT-PCR performed on Gγ-globin-Tag and TRAMP tumors and normal prostates (normalized with 28s RNA levels) confirmed a 20-fold higher level of expression in all tumors compared to normal prostates (results not shown). Protein levels were analyzed by immunohistochemistry in Gγ-globin-Tag tissues. A strong and widespread expression of stathmin-1 was observed in prostate tumors, but no expression in normal prostates or normal prostate glands embedded in tumors was found (Fig. 4A). High levels of stathmin-1 were also detected in metastasis (Fig. 4C). The expression of stathmin-1 correlated with the expression of PCNA-positive cells (Fig. 4B).
Fig. 3.
Expression patterns of stathmin-1 in human and mouse (FG/Tag) prostate samples. Stathmin-1 is highly expressed in human metastatic prostate cancer and mouse Gγ-globin-Tag tumors. B, benign; P, primary; M, metastasis.
Fig. 4.
Expression of stathmin-1, and PCNA in Gγ-globin-Tag mouse prostates. A: Stathmin-1 immunostaining showing strong expression in Gγ-globin-Tag prostate tumor but not in embedded normal prostate glands (arrow). B: PCNA is also highly expressed in tumor cells but not in normal glands embedded in the tumors (arrow).C: Staining for Stathmin-1in a lymph node metastatic foci. D: FISH analysis of stathmin-1 on C2-TR AMP tumor cells. Five copies of stathmin-1 are seen in these cells. Green color, chromosome 4; red color, stathmin-1; blue color, DAPI.
To test genetic amplification of the stathmin-1 gene in mouse PrCa we performed FISH analysis. Gγ-globin-Tag tumor samples showed heterogeneity in the number of copies of the stathmin-1 gene for all the cases analyzed (Table IV). Cells with 2, 3, and 4 signals for stathmin-1 were observed in Gγ-globin-Tag tumors. Two of the tumors exhibited a high percentage of cells with four copies (27% and 28%), and the other two exhibited a high percentage of cells with three copies (44% and 47%). These results demonstrate that the stathmin-1 gene is amplified in Gγ-globin-Tag tumors. We also analyzed gene copy number for stathmin-1 in TRAMP-C2 PrCa cells [30]. A complex hypertetraploid karyotype in the TRAMP-C2 cell line was found, and FISH analysis showed five copies of the stathmin-1 gene in these cells (Fig. 4D). Therefore, increased expression of stathmin-1 in mouse prostate tumors and cell lines may be in part due to gene amplification.
TABLE IV.
Quantification of the Number of FISH Signals for Stathmin-1 in FG/Tag Tumors and in Normal Prostates
| Number of cells (%) | ||||
|---|---|---|---|---|
| Sample | 1 FISH signal | 2 FISH signals | 3 FISH signals | 4 FISH signals |
| Normal prostate | 7 (7) | 93 (93) | ||
| PC-1 | 10 (4) | 97 (46) | 98 (47) | 6 (3) |
| PC-2 | 8 (5) | 67 (44) | 68 (44) | 10 (7) |
| PC-3 | 6 (4) | 101 (60) | 17 (10) | 46 (27) |
| PC-4 | 7 (3) | 127 (58) | 23 (10) | 61 (28) |
Numbers in parentheses indicate percentage of cells. PC: Prostate Cancer.
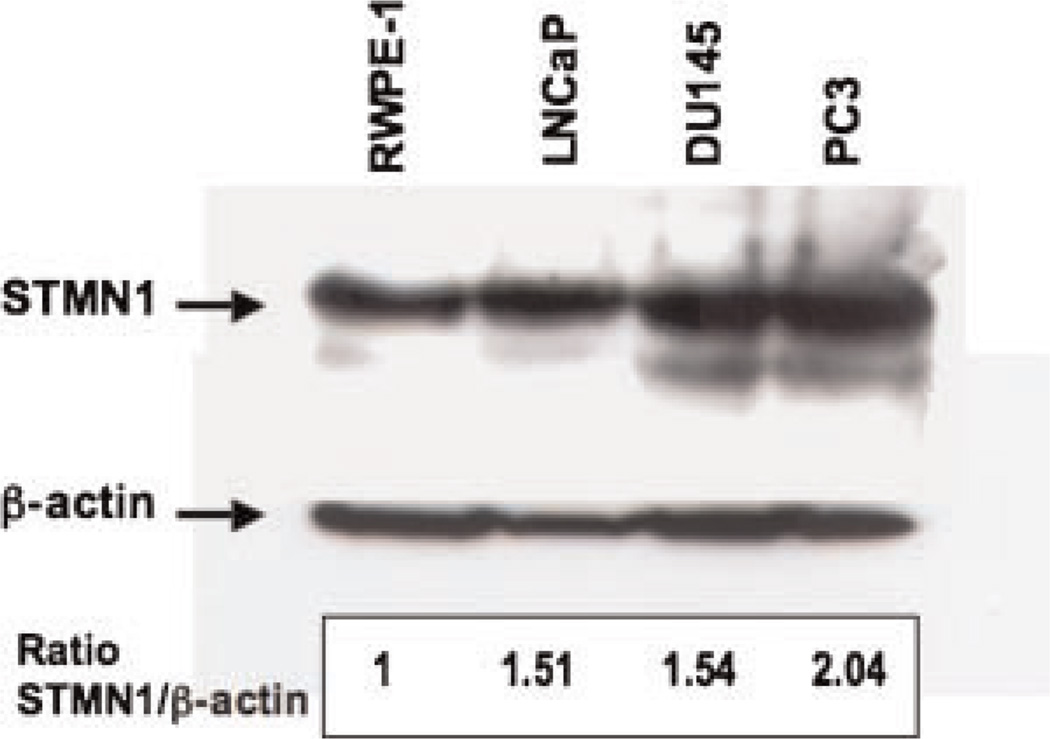
Western blot analysis showed a modest increase (~1.5-fold) in stathmin-1 in the tumor cell lines LNCaP and DU 145, and a ~2-fold increase in PC-3 PrCa cell line, compared to non-tumorigenic RWPE-1 cells (Fig. 5).
Fig. 5.
Representative Western blot analysis of stathmin-1 in the human prostate cell lines RWPE-1, LNCaP, DU 145, and PC-3. Densitometric analysis shows a ~1.5-fold increase in stathmin-1 in the tumor cell lines LNCaP and DU 145, and a ~2-fold increase in PC-3, compared to non-tumorigenic RWPE-1 cells.
DISCUSSION
Several transgenic animal models have been developed to represent aspects of human PrCa and to test targeted therapies. Despite their widespread use, xenograft models have significant shortcomings in the recapitulation of the natural history of the human disease. Pre-neoplastic lesions cannot be analyzed in these models, and cells have to be injected into immunocompromised mice. Transgenic models overcome these problems and are particularly useful for testing preventive therapies and targeted anti-tumor therapies. However, not many models have been examined by gene expression profiling. This approach provides a more comprehensive picture of their cancer transcriptome, which allows for the selection of potential targets that are shared by human PrCa, and for testing targeted therapies in such a model.
Cross-species comparisons of mouse and human cancers is a powerful means of identifying evolutionary conserved genes and genetic networks in cancer that may be especially important for therapeutic targeting. This study represents a first attempt to compare expression changes in the transcriptome of the Gγ-globin-Tag transgenic mouse model for PrCa with changes reported in human PrCa. However, the direct comparison of mouse and human microarray datasets remains challenging due to differences in the platforms used, reference samples and large variances in gene expression found in the human datasets. Different microarray studies comparing the transcriptome of benign human prostates with prostate carcinomas have found very heterogeneous results between patients [31–33]. Indeed, the variance in gene expression profiles between human tumors is a reason by which microarray data from the mouse can be powerful in sorting through the background noise in human microarray data. Unlike humans, the genetic background in transgenic mice is more homogeneous providing much less variance in gene expression of tumors from the same model. This suggests that identification of deregulated pathways in a specific transgenic mouse model may provide an important filter in helping to identify targets or pathways also important in sub-types of human PrCa.
The Gγ-globin-Tag is a unique model for PrCa. Unlike for other T-antigen based models, such as TRAMP, LADY, and C3(1)/Tag, the expression of T-antigen during malignant transformation of the prostate in the Gγ-globin-Tag is confined to a subset of p63+ cells located in the basal layer of the epithelium [18]. In the Gγ-globin-Tag mice, subsets of p63+ basal epithelial cells (possibly stem cells) are, therefore, the target cells of carcinogenesis [18]. Genetic studies in mice have shown that p63+ basal cells are required for normal prostate differentiation and that p63 is a key determinant of epithelial stem cells. However, whether p63+ basal cells are the target for human prostate carcinogenesis is currently unknown. Similar to human PrCa, the loss of p63+ basal cells is a common characteristic of prostate carcinogenesis in Gγ-globin-Tag transgenic mice [18]. A similar phenotype has also been observed in the PTEN KO model of PrCa [34].
In our study, many altered genes in the Gγ-globin-Tag model are deregulated in a manner similar to what has been reported for advanced human PrCa. Indeed, correlation analysis of human and Gγ-globin-Tag prostate tumor gene datasets shows a closely related pattern of expression profiles between the mouse tumors and hormone refractory and metastatic human tumors. This similarity suggests an aggressive behavior of Gγ-globin-Tag PrCa, which has been previously shown to frequently metastatize to lymph nodes, lungs, and bone [35]. Some of the genes identified in our study, such as MMP-2, Caveolin-1, Tubb4, Tuba4, N-CAM1, Fyn, ENPP2 (autotaxin), POLE2, CNN1, and TXNIP, are likely to play a role in the metastatic development of this model. MMP-2 has been found in different studies as a key mediator of the metastatic outbreak [36]. Other genes are emergent candidates to play a role in malignant spread. Such is the case of autotaxin (ENPP2), a metastasis promoting enzyme whose overexpression is associated with poor outcome. Collectively, our results show that this model may be particularly useful to study advanced, androgen-independent and NE-positive human PrCa.
Bioinformatic analysis of gene networks and signaling pathways have shown the deregulation of genes linked to p53, Rb, and Myc pathways in the Gγ-globin-Tag model. Since the Gγ-globin-Tag prostate tumors originate from a Tag-driven carcinogenesis process where p53 and Rb are known to be functionally inactivated, these results are consistent with loss of p53 and Rb function resulting in the alteration of numerous genes involved in cell cycle and apoptosis. Altered levels of Myc-related genes have also been found in C3(1)/Tag mice mammary tumors [37,38]. Interestingly, our previous study analyzing tumor-specific pathways has shown that MMTV–Myc mammary tumors share oncogenic signatures with Tag-based mammary models. These data, together with our results in PrCa suggests common altered pathways between Tag and Myc, which could be related to G1/G2 checkpoint disruption [38].
We have studied by computational analysis TFs with binding sites highly represented in promoter regions of deregulated genes in Gγ-globin-Tag tumors. Our results show that SP1, AP2alpha, MZF 1–4, E2F, ahr-ARNT, cyclic-AMP response element binding protein (CREB), Elk-1, and NY-F TFs may play an important role in prostate carcinogenesis in the Gγ-globin-Tag model. Some deregulated genes in this model involved in prostate carcinogenesis display TFBSs for several of such TFs, including Rb1, Hey1, Notch1, and vav3 oncogene (see Supplementary Table I). This suggests a tight regulation by these TFs of the carcinogenic process in Gγ-globin-Tag tumors. SP1 has been involved in PrCa development [39]. In addition, SP1 has been proposed as a molecular target for phytochemicals in PrCa therapy [40]. CREB is a TF that controls different pathways involved in proliferation, differentiation, and survival through induction of key target genes [41]. CREB phosphorylation increases by DHT and forskolin treatment in LNCaP cells, which correlates with enhancement of PSA transcription [42]. The acquisition of an androgen-independent phenotype is associated with a constitutive activation of ERK-1/2-CREB signaling pathways in LNCaP cells [43]. Elk-1 is another TF related to the ERK1/2-MAPK and p38 pathways, which mediates the early responses of c-fos promoter to growth factors. High levels of phosphorylated-Elk1 have been found in human PrCa in comparison to BPH [44]. Additional studies will further investigate the roles of these TFs in tumorigenesis of this model.
We have observed a strong up-regulation of markers for NE differentiation in Gγ-globin-Tag tumors. Some degree of NE differentiation appears to be a typical feature of the majority of human prostate tumors [3]. In fact, NE cells have been found in 30–100% of human PrCa, although with a variable proportion of NE cells [3]. Hirano et al. [45] quantified the percentage of NE cells in patients treated or not treated with hormone therapy. In the group of men with no therapy, the majority of tumors had <10% NE cells within the tumor. On the contrary, androgen deprivation increased significantly the number of NE cells, which represented 10–20% of total cells of the tumor. Similarly to hormone refractory human PrCa, Gγ-globin-Tag tumors show a NE-phenotype, with ~20% of cells positive for NE markers, as revealed previously by immunohistochemistry [18]. NE products stimulate prostate tumor cell proliferation in an autocrine–paracrine fashion, and may sustain tumor cell proliferation in the absence of androgens [3]. Gγ-globin-Tag tumors express typical NE markers, such as synaptophysin, secretogranin-III, and dopa decarboxylase. Interestingly, human microarray data also showed an increase in the expression of dopa decarboxylase for all metastatic samples analyzed. Dopa decarboxylase has been reported as a novel AR-interacting protein that enhances steroid receptor transactivation [46]. NE features have been demonstrated in TRAMP and LADY transgenic models for PrCa [47,48], whereas massive NE differentiation has been found in the Cryptidin-Tag model [49]. All these models could be used to investigate the role of NE cells in prostate carcinogenesis. Pure NE tumors are resistant to chemotherapy, and novel NE-based approaches, such as the use of somatostatin analogs are currently under investigation to block NE cell function in PrCa [50].
Many of the highly up-regulated genes in the Gγ-globin-Tag mouse model were related to microtubules. These include tubulin-β5, tubulin-β4, tubulin-α4, and stathmin-1/Op18. Stathmin-1, which we observed highly up-regulated in Gγ-globin-Tag mouse tumors and human metastatic tumors, plays a role in binding to tubulin at the microtubule ends, thus increasing hydrolysis. Stathmin-1 is highly expressed when cells proliferate and are exposed to mitogenic stimuli [29]. Overexpression of stathmin-1 was reported by immunohistochemistry in a subset of prostate and breast cancers [51,52]. Interestingly, a recently published transcriptomic analysis of TRAMP mice found stathmin-1 as one of the genes included in a gene signature associated with an aggressive tumor phenotype [15]. A poor prognosis signature associated with PI3K-activation/PTEN-loss recently identified in breast cancer, included stathmin-1 as well [53]. In fact, breast cancer patients with high stathmin-1 score had significantly worse disease-free survival [53]. Therefore, authors suggest that expression of stathmin-1 may be clinically relevant to stratify patients for anti-PI3K targeted therapy and monitoring therapeutic efficacy [53]. It is also worth noticing that stathmin-1 is one of the genes that belong to the SV40/T-antigen signature of prostate, breast, and lung mouse tumors, which predicts poor prognosis in human cancers [54]. Therefore, stathmin-1 could be considered as a metastatic biomarker and a therapeutic target for advanced prostate and breast cancers.
Overexpression of stathmin-1 in breast cancer cells decreased polymerization of microtubules and greatly reduced the binding of paclitaxel, while increasing the binding of vinblastin [55]. In lung carcinoma cells, expression of stathmin-1 is associated to sensitivity to vindesine, and it has been suggested that expression of this protein can serve as a surrogate marker for the sensitivity to Vinca alkaloids [56]. Therefore, inhibition of stathmin-1 may be a novel means of inhibiting drug resistence to certain drugs, such as taxanes or vinca derivatives. Mistry et al. [57] have suggested that stathmin-1 could be targeted for PrCa therapy. In their study, adenovirus-mediated transfer of anti-stathmin-1 ribozymes resulted in a dramatic cell growth inhibition, accumulation of cells in the G2/M phase, and increased apoptosis [57]. Interestingly, overexpression of stathmin-1 in PrCa cells is mediated predominantly through the E2F family of TFs [58], which is in keeping with the involvement of this TF we have identified in Gγ-globin-Tag carcinogenesis by predictive computational analysis. Thus, the Gγ-globin-Tag model for PrCa (or other T-antigen driven models) may be particularly useful for the preclinical targeting of stathmin-1.
In conclusion, our results show the potential use of the Gγ-globin-Tag model for PrCa to mimic human aggressive PrCa with NE characteristics and to target-specific signaling pathways that are altered during prostate carcinogenesis. Gγ-globin-Tag mice may serve as an accurate model to test the role of stathmin-1 as a therapeutic target with drugs such as taxanes, Vinca alkaloids, and PI3K inhibitors. Future comparative high-throughput genomic analyses between mouse models and human PrCa will provide further insights into similarities and distinctions between the models and sub-types of human cancer, leading to the most appropriate use of models for testing specific targeted therapies.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Sobel (INSERM, Universite Pierre et Marie Curie, France) for providing us the anti-stathmin-1 antibody. We also thank Dr. Jung-Im Huh for critical comments on our manuscript and Miriam Redrado for technical help. This work was supported in part by the Intramural Program of NCI, Center for Cancer Research of NIH (to J.E.G.) and by “UTE project CIMA,” ISCIII-RETIC RD06/0020 grant; Ministerio de Educacion y Ciencia grant SAF2007-64184 (to A.C.); and P.A.N. was supported by a Spanish Torres-Quevedo fellowship (PTQ05-01-01084).
Footnotes
Additional Supporting Information may be found in the online version of this article.
REFERENCES
- 1.Greenlee RT, Hill-Harmon MB, Murray T, Thun M. Cancer statistics, 2001. CA Cancer J Clin. 2001;51:15–36. doi: 10.3322/canjclin.51.1.15. [DOI] [PubMed] [Google Scholar]
- 2.Troyer DA, Mubiru J, Leach RJ, Naylor SL. Promise and challenge: Markers of prostate cancer detection, diagnosis and prognosis. Dis Markers. 2004;20:117–128. doi: 10.1155/2004/509276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abrahamsson PA. Neuroendocrine differentiation in prostatic carcinoma. Prostate. 1999;39:135–148. doi: 10.1002/(sici)1097-0045(19990501)39:2<135::aid-pros9>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 4.Cussenot O, Villette JM, Cochand-Priollet B, Berthon P. Evaluation and clinical value of neuroendocrine differentiation in human prostatic tumors. Prostate Suppl. 1998;8:43–51. [PubMed] [Google Scholar]
- 5.Glinsky GV, Glinskii AB, Stephenson AJ, Hoffman RM, Gerald WL. Gene expression profiling predicts clinical outcome of prostate cancer. J Clin Invest. 2004;113:913–923. doi: 10.1172/JCI20032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dhanasekaran SM, Barrette TR, Ghosh D, Shah R, Varambally S, Kurachi K, Pienta KJ, Rubin MA, Chinnaiyan AM. Delineation of prognostic biomarkers in prostate cancer. Nature. 2001;412:822–826. doi: 10.1038/35090585. [DOI] [PubMed] [Google Scholar]
- 7.Luo J, Zha S, Gage WR, Dunn TA, Hicks JL, Bennett CJ, Ewing CM, Platz EA, Ferdinandusse S, Wanders RJ, Trent JM, Isaacs WB, De Marzo AM. Alpha-methylacyl-CoA racemase: A new molecular marker for prostate cancer. Cancer Res. 2002;62:2220–2226. [PubMed] [Google Scholar]
- 8.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, Rubin MA, Chinnaiyan AM. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 9.Jeronimo C, Henrique R, Hoque MO, Mambo E, Ribeiro FR, Varzim G, Oliveira J, Teixeira MR, Lopes C, Sidransky D. A quantitative promoter methylation profile of prostate cancer. Clin Cancer Res. 2004;10:8472–8478. doi: 10.1158/1078-0432.CCR-04-0894. [DOI] [PubMed] [Google Scholar]
- 10.Kim MJ, Cardiff RD, Desai N, Banach-Petrosky WA, Parsons R, Shen MM, Abate-Shen C. Cooperativity of Nkx3.1 and Pten loss of function in a mouse model of prostate carcinogenesis. Proc Natl Acad Sci USA. 2002;99:2884–2889. doi: 10.1073/pnas.042688999. Epub 2002 Feb 2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Green JE, Greenberg NM, Ashendel CL, Barrett JC, Boone C, Getzenberg RH, Henkin J, Matusik R, Janus TJ, Scher HI. Workgroup 3: Transgenic and reconstitution models of prostate cancer. Prostate. 1998;36:59–63. doi: 10.1002/(sici)1097-0045(19980615)36:1<59::aid-pros11>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 12.Stearns ME, Ware JL, Agus DB, Chang CJ, Fidler IJ, Fife RS, Goode R, Holmes E, Kinch MS, Peehl DM, Pretlow TG, II, Thalmann GN. Workgroup 2: Human xenograft models of prostate cancer. Prostate. 1998;36:56–58. doi: 10.1002/(sici)1097-0045(19980615)36:1<56::aid-pros10>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 13.Lucia MS, Bostwick DG, Bosland M, Cockett AT, Knapp DW, Leav I, Pollard M, Rinker-Schaeffer C, Shirai T, Watkins BA. Workgroup I: Rodent models of prostate cancer. Prostate. 1998;36:49–55. doi: 10.1002/(sici)1097-0045(19980615)36:1<49::aid-pros9>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 14.Ellwood-Yen K, Graeber TG, Wongvipat J, Iruela-Arispe ML, Zhang J, Matusik R, Thomas GV, Sawyers CL. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell. 2003;4:223–238. doi: 10.1016/s1535-6108(03)00197-1. [DOI] [PubMed] [Google Scholar]
- 15.Kela I, Harmelin A, Waks T, Orr-Urtreger A, Domany E, Eshhar Z. Interspecies comparison of prostate cancer gene-expression profiles reveals genes associated with aggressive tumors. Prostate. 2009;69:1034–1044. doi: 10.1002/pros.20950. [DOI] [PubMed] [Google Scholar]
- 16.Perez-Stable C, Altman NH, Brown J, Harbison M, Cray C, Roos BA. Prostate, adrenocortical, and brown adipose tumors in fetal globin/T antigen transgenic mice. Lab Invest. 1996;74:363–373. [PubMed] [Google Scholar]
- 17.Perez-Stable C, Altman NH, Mehta PP, Deftos LJ, Roos BA. Prostate cancer progression, metastasis, and gene expression in transgenic mice. Cancer Res. 1997;57:900–906. [PubMed] [Google Scholar]
- 18.Reiner T, de Las Pozas A, Parrondo R, Perez-Stable C. Progression of prostate cancer from a subset of p63-positive basal epithelial cells in FG/Tag transgenic mice. Mol Cancer Res. 2007;5:1171–1179. doi: 10.1158/1541-7786.MCR-07-0024. [DOI] [PubMed] [Google Scholar]
- 19.Bello D, Webber MM, Kleinman HK, Wartinger DD, Rhim JS. Androgen responsive adult human prostatic epithelial cell lines immortalized by human papillomavirus 18. Carcinogenesis. 1997;18:1215–1223. doi: 10.1093/carcin/18.6.1215. [DOI] [PubMed] [Google Scholar]
- 20.Calvo A, Xiao N, Kang J, Best CJ, Leiva I, Emmert-Buck MR, Jorcyk C, Green JE. Alterations in gene expression profiles during prostate cancer progression: Functional correlations to tumorigenicity and down-regulation of selenoprotein-P in mouse and human tumors. Cancer Res. 2002;62:5325–5335. [PubMed] [Google Scholar]
- 21.Segura V, Podhorski A, Guruceaga E, Sevilla JL, Corrales FJ, Rubio A. GARBAN II: An integrative framework for extracting biological information from proteomic and genomic data. Proteomics. 2006;6(suppl 1):S12–S15. doi: 10.1002/pmic.200500313. [DOI] [PubMed] [Google Scholar]
- 22.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aerts S, Thijs G, Coessens B, Staes M, Moreau Y, De Moor B. Toucan: Deciphering the cis-regulatory logic of coregulated genes. Nucleic Acids Res. 2003;31:1753–1764. doi: 10.1093/nar/gkg268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kasprzyk A, Keefe D, Smedley D, London D, Spooner W, Melsopp C, Hammond M, Rocca-Serra P, Cox T, Birney E. EnsMart: A generic system for fast and flexible access to biological data. Genome Res. 2004;14:160–169. doi: 10.1101/gr.1645104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vlieghe D, Sandelin A, De Bleser PJ, Vleminckx K, Wasserman WW, van Roy F, Lenhard B. A new generation of JASPAR, the open-access repository for transcription factor binding site profiles. Nucleic Acids Res. 2006;34:D95–D97. doi: 10.1093/nar/gkj115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guruceaga E, Segura V, Corrales FJ, Rubio A. FactorY, a bioinformatic resource for genome-wide promoter analysis. Comput Biol Med. 2009;39:385–387. doi: 10.1016/j.compbiomed.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 27.Freeman KW, Gangula RD, Welm BE, Ozen M, Foster BA, Rosen JM, Ittmann M, Greenberg NM, Spencer DM. Conditional activation of fibroblast growth factor receptor (FGFR) 1, but not FGFR2, in prostate cancer cells leads to increased osteopontin induction, extracellular signal-regulated kinase activation, and in vivo proliferation. Cancer Res. 2003;63:6237–6243. [PubMed] [Google Scholar]
- 28.Cantalupo PG, Saenz-Robles MT, Rathi AV, Beerman RW, Patterson WH, Whitehead RH, Pipas JM. Cell-type specific regulation of gene expression by simian virus 40 T antigens. Virology. 2009;386:183–191. doi: 10.1016/j.virol.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rubin CI, Atweh GF. The role of stathmin in the regulation of the cell cycle. J Cell Biochem. 2004;93:242–250. doi: 10.1002/jcb.20187. [DOI] [PubMed] [Google Scholar]
- 30.Foster BA, Gingrich JR, Kwon ED, Madias C, Greenberg NM. Characterization of prostatic epithelial cell lines derived from transgenic adenocarcinoma of the mouse prostate (TRAMP) model. Cancer Res. 1997;57:3325–3330. [PubMed] [Google Scholar]
- 31.Glinsky GV, Berezovska O, Glinskii AB. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest. 2005;115:1503–1521. doi: 10.1172/JCI23412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Varambally S, Yu J, Laxman B, Rhodes DR, Mehra R, Tomlins SA, Shah RB, Chandran U, Monzon FA, Becich MJ, Wei JT, Pienta KJ, Ghosh D, Rubin MA, Chinnaiyan AM. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 33.Best CJ, Gillespie JW, Yi Y, Chandramouli GV, Perlmutter MA, Gathright Y, Erickson HS, Georgevich L, Tangrea MA, Duray PH, Gonzalez S, Velasco A, Linehan WM, Matusik RJ, Price DK, Figg WD, Emmert-Buck MR, Chuaqui RF. Molecular alterations in primary prostate cancer after androgen ablation therapy. Clin Cancer Res. 2005;11:6823–6834. doi: 10.1158/1078-0432.CCR-05-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang S, Garcia AJ, Wu M, Lawson DA, Witte ON, Wu H. Pten deletion leads to the expansion of a prostatic stem/progenitor cell subpopulation and tumor initiation. Proc Natl Acad Sci USA. 2006;103:1480–1485. doi: 10.1073/pnas.0510652103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perez-Stable C, Altman NH, Mehta PP, Deftos LJ, Roos BA. Prostate cancer progression, metastasis, and gene expression in transgenic mice. Cancer Res. 1997;57:900–906. [PubMed] [Google Scholar]
- 36.Gupta GP, Nguyen DX, Chiang AC, Bos PD, Kim JY, Nadal C, Gomis RR, Manova-Todorova K, Massague J. Mediators of vascular remodelling co-opted for sequential steps in lung metastasis. Nature. 2007;446:765–770. doi: 10.1038/nature05760. [DOI] [PubMed] [Google Scholar]
- 37.Desai KV, Xiao N, Wang W, Gangi L, Greene J, Powell JI, Dickson R, Furth P, Hunter K, Kucherlapati R, Simon R, Liu ET, Green JE. Initiating oncogenic event determines gene-expression patterns of human breast cancer models. Proc Natl Acad Sci USA. 2002;99:6967–6972. doi: 10.1073/pnas.102172399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fargiano AA, Desai KV, Green JE. Interrogating mouse mammary cancer models: Insights from gene expression profiling. J Mammary Gland Biol Neoplasia. 2003;8:321–334. doi: 10.1023/b:jomg.0000010032.05234.6f. [DOI] [PubMed] [Google Scholar]
- 39.Janne OA, Moilanen AM, Poukka H, Rouleau N, Karvonen U, Kotaja N, Hakli M, Palvimo JJ. Androgen-receptor-interacting nuclear proteins. Biochem Soc Trans. 2000;28:401–405. [PubMed] [Google Scholar]
- 40.Kaur M, Agarwal R. Transcription factors: Molecular targets for prostate cancer intervention by phytochemicals. Curr Cancer Drug Targets. 2007;7:355–367. doi: 10.2174/156800907780809732. [DOI] [PubMed] [Google Scholar]
- 41.Shankar DB, Sakamoto KM. The role of cyclic-AMP binding protein (CREB) in leukemia cell proliferation and acute leukemias. Leuk Lymphoma. 2004;45:265–270. doi: 10.1080/1042819031000151095. [DOI] [PubMed] [Google Scholar]
- 42.Kim J, Jia L, Stallcup MR, Coetzee GA. The role of protein kinase A pathway and cAMP responsive element-binding protein in androgen receptor-mediated transcription at the prostate-specific antigen locus. J Mol Endocrinol. 2005;34:107–118. doi: 10.1677/jme.1.01701. [DOI] [PubMed] [Google Scholar]
- 43.Unni E, Sun S, Nan B, McPhaul MJ, Cheskis B, Mancini MA, Marcelli M. Changes in androgen receptor nongenotropic signaling correlate with transition of LNCaP cells to androgen independence. Cancer Res. 2004;64:7156–7168. doi: 10.1158/0008-5472.CAN-04-1121. [DOI] [PubMed] [Google Scholar]
- 44.Ricote M, Garcia-Tunon I, Bethencourt F, Fraile B, Onsurbe P, Paniagua R, Royuela M. The p38 transduction pathway in prostatic neoplasia. J Pathol. 2006;208:401–407. doi: 10.1002/path.1910. [DOI] [PubMed] [Google Scholar]
- 45.Hirano D, Okada Y, Minei S, Takimoto Y, Nemoto N. Neuroendocrine differentiation in hormone refractory prostate cancer following androgen deprivation therapy. Eur Urol. 2004;45:586–592. doi: 10.1016/j.eururo.2003.11.032. discussion 592. [DOI] [PubMed] [Google Scholar]
- 46.Wafa LA, Cheng H, Rao MA, Nelson CC, Cox M, Hirst M, Sadowski I, Rennie PS. Isolation and identification of L-dopa decarboxylase as a protein that binds to and enhances transcriptional activity of the androgen receptor using the repressed transactivator yeast two-hybrid system. Biochem J. 2003;375:373–383. doi: 10.1042/BJ20030689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Masumori N, Thomas TZ, Chaurand P, Case T, Paul M, Kasper S, Caprioli RM, Tsukamoto T, Shappell SB, Matusik RJ. Aprobasin-large T antigen transgenic mouse line develops prostate adenocarcinoma and neuroendocrine carcinoma with metastatic potential. Cancer Res. 2001;61:2239–2249. [PubMed] [Google Scholar]
- 48.Kaplan-Lefko PJ, Chen TM, Ittmann MM, Barrios RJ, Ayala GE, Huss WJ, Maddison LA, Foster BA, Greenberg NM. Pathobiology of autochthonous prostate cancer in a pre-clinical transgenic mouse model. Prostate. 2003;55:219–237. doi: 10.1002/pros.10215. [DOI] [PubMed] [Google Scholar]
- 49.Hu Y, Ippolito JE, Garabedian EM, Humphrey PA, Gordon JI. Molecular characterization of a metastatic neuroendocrine cell cancer arising in the prostates of transgenic mice. J Biol Chem. 2002;277:44462–44474. doi: 10.1074/jbc.M205784200. [DOI] [PubMed] [Google Scholar]
- 50.Mosca A, Berruti A, Russo L, Torta M, Dogliotti L. The neuroendocrine phenotype in prostate cancer: Basic and clinical aspects. J Endocrinol Invest. 2005;28:141–145. [PubMed] [Google Scholar]
- 51.Friedrich B, Gronberg H, Landstrom M, Gullberg M, Bergh A. Differentiation-stage specific expression of oncoprotein 18 in human and rat prostatic adenocarcinoma. Prostate. 1995;27:102–109. doi: 10.1002/pros.2990270207. [DOI] [PubMed] [Google Scholar]
- 52.Curmi PA, Nogues C, Lachkar S, Carelle N, Gonthier MP, Sobel A, Lidereau R, Bieche I. Overexpression of stathmin in breast carcinomas points out to highly proliferative tumours. Br J Cancer. 2000;82:142–150. doi: 10.1054/bjoc.1999.0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saal LH, Johansson P, Holm K, Gruvberger-Saal SK, She QB, Maurer M, Koujak S, Ferrando AA, Malmstrom P, Memeo L, Isola J, Bendahl PO, Rosen N, Hibshoosh H, Ringner M, Borg A, Parsons R. Poor prognosis in carcinomais associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proc Natl Acad Sci USA. 2007;104:7564–7569. doi: 10.1073/pnas.0702507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deeb KK, Michalowska AM, Yoon CY, Krummey SM, Hoenerhoff MJ, Kavanaugh C, Li MC, Demayo FJ, Linnoila I, Deng CX, Lee EY, Medina D, Shih JH, Green JE. Identification of an integrated SV40 T/t-antigen cancer signature in aggressive human breast, prostate, and lung carcinomas with poor prognosis. Cancer Res. 2007;67:8065–8080. doi: 10.1158/0008-5472.CAN-07-1515. [DOI] [PubMed] [Google Scholar]
- 55.Alli E, Bash-Babula J, Yang JM, Hait WN. Effect of stathmin on the sensitivity to antimicrotubule drugs in human breast cancer. Cancer Res. 2002;62:6864–6869. [PubMed] [Google Scholar]
- 56.Nishio K, Nakamura T, Koh Y, Kanzawa F, Tamura T, Saijo N. Oncoprotein 18 overexpression increases the sensitivity to vindesine in the human lung carcinoma cells. Cancer. 2001;91:1494–1499. doi: 10.1002/1097-0142(20010415)91:8<1494::aid-cncr1157>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 57.Mistry SJ, Bank A, Atweh GF. Targeting stathmin in prostate cancer. Mol Cancer Ther. 2005;4:1821–1829. doi: 10.1158/1535-7163.MCT-05-0215. [DOI] [PubMed] [Google Scholar]
- 58.Polzin RG, Benlhabib H, Trepel J, Herrera JE. E2F sites in the Op18 promoter are required for high level of expression in the human prostate carcinoma cell line PC-3-M. Gene. 2004;341:209–218. doi: 10.1016/j.gene.2004.06.052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.